Abstract
A quantitative trait locus (QTL) controlling HbF levels has previously been mapped to chromosome 6q23 in an Asian-Indian kindred with β thalassemia and heterocellular hereditary persistence of fetal hemoglobin (HPFH). Five protein-coding genes, ALDH8A1, HBS1L, cMYB, AHI1, and PDE7B reside in this 1.5-megabase (Mb) candidate interval of 6q23. To direct sequencing efforts we compared the expression profiles of these 5 genes between 12 individuals with elevated and 14 individuals with normal HbF levels during adult erythropoiesis by real-time quantitative reverse transcription–polymerase chain reaction (RT-PCR). Two genes, cMYB and HBS1L, demonstrated simultaneous transcriptional down-regulation in individuals with elevated HbF levels. Transfection of K562 cells encoding human cDNA of cMYB and HBS1L genes showed that, although overexpression of ectopic cMYB inhibited γ-globin gene expression, overexpression of HBS1L had no effect. Low levels of cMYB were associated with low cell expansions, accelerated erythroid maturation, and higher number of macrophages in erythroid cell culture. These observations suggest that differences in the intrinsic levels of cMYB may account for some of the variation in adult HbF levels. The possible mechanism of cMYB influencing γ- to β-globin switching is discussed.
Introduction
Despite their apparent genetic simplicity, it has long been appreciated that both β thalassemia and sickle cell disease display a remarkable diversity in the severity of their disease. High levels of HbF have been shown to have a beneficial effect in both of these disorders, although the mechanism for the ameliorating effect differs.1 HbF levels vary considerably not only in patients with these β-globin disorders, but also in normal healthy adults. Studies have shown that the distribution of HbF and F cells (FCs; erythrocytes that contain measurable Hb F) in healthy adults is continuous and positively skewed, with the levels varying by greater than 20-fold.2 Although the majority of adults have HbF levels of less than 0.6% of total hemoglobin, 10% to 15% of individuals have modest increases in levels ranging from 0.8% to 5%. The HbF is unevenly distributed among the erythrocytes in such individuals who are classified as having heterocellular hereditary persistence of fetal hemoglobin (HPFH). Unlike the rare forms of pancellular HPFH which are inherited in a Mendelian fashion as alleles of the β-globin gene cluster,3 the genetic etiology of heterocellular HPFH is complex. Twin studies show that 89% of the quantitative variation in HbF and FC levels in healthy adults is genetically controlled4 with 50% to 60% of the F cell variance resulting from genetic factors not linked to the β-globin locus.5 Linkage has been reported between HbF and FC levels and 3 regions of the genome: on chromosomes Xp22.2-3, 6q23, and 8q.6-8 Both the quantitative trait loci (QTLs) on chromosome 6q and 8q were reported by our group in an extended Asian-Indian family with β thalassemia and heterocellular HPFH. Recently, we have conducted a detailed annotation and mutation analysis of all 5 protein-coding genes in this 6q23 candidate interval, but no causative mutations have been identified in the coding sequence of these genes.9
How the implicated QTL affects HbF and F cell levels is not evident, but it is likely to relate to changes in the functional balance of cellular factors involved in the regulation of globin gene transcription.10 Thus, it is possible that differences in the 6q23 transcriptome exist between erythroid precursors of individuals with and without high HbF levels and that the differential expression relates to DNA variant(s) in regulatory regions of the causative gene. For this reason, we decided to compare the mRNAs of the 5 protein-coding genes in cultured erythroid cells between individuals with and without elevated HbF levels. Analysis of a number of erythroid cultures of individuals has revealed a clear correlation of elevated HbF with low levels of cMYB and HBS1L expression, raising the possibility that one or both genes are involved in the regulation of fetal hemoglobin. Further characterization of erythropoiesis in vitro and analysis of clones isolated from K562 cells transfected with human cDNAs of cMYB or HBS1L demonstrated that cMYB is a key factor for regulation of HbF production and is probably involved in globin gene switching through controlling cell cycles. Proliferative features of the differentiating erythroid cells could be correlated with expression levels of the cMYB and Hb F phenotype.
Materials and methods
Subjects and analysis of blood samples
Subjects were healthy adults, and consisted of 2 groups.
Group 1. These 26 adults were unrelated of diverse ethnic backgrounds and do not belong to the Asian-Indian kindred.7 They were healthy adults with no β thalassemia and were selected on the basis of their HbF levels as measured by high-performance liquid chromatography (BioRad Variant; BioRad, Hemel Hempstead, United Kingdom). Twelve individuals (1 male) had HbF levels ranging from 0.8% to 1.8% and were designated as having heterocellular HPFH. Fourteen individuals (8 males) had HbF levels of 0.6% or less and were designated as the control group (Table S1, available on the Blood website; see the Supplemental Table link at the top of the online article). Blood (50 mL) in EDTA was obtained from these subjects and cultured using a modified 2-phase liquid erythroid cell culture.11 Cells were obtained on more than one occasion from 6 donors to compare and validate growth pattern.
Group 2. These 850 adult women were unrelated and all North European whites from the St Thomas' UK Adult Twin Registry.12 Blood samples were collected in EDTA anticoagulant, and blood counts were determined using a Sysmex (Milton Keynes, United Kingdom) automated blood cell analyser. F cells were enumerated by flow cytometry of 20 000 cells using anti–γ-globin chain antibody conjugated with fluorescein isothiocyanate (FITC).13
The study was approved by the local Ethic Committee (LREC no, 01-332) of King's College Hospital, London.
Cell cultures
Erythroid cells were cultured using a 2-phase liquid system (modified from Fibach et al11 ). Mononuclear cells were isolated from peripheral blood by centrifugation on a gradient of Ficoll-Hypaque and cultured for 7 days in phase I medium which consists of serum-free StemSpan (Stem Cell Technologies, Vancouver, BC) supplemented with 1 μg/mL cyclosporin A, 25 ng/mL interleukin-3, 50 ng/mL human stem cell factor (Sigma, Poole, United Kingdom), and 0.01% bovine serum albumin. Cells were incubated at 37°C, 5% CO2. After 7 days, nonadherent cells were collected and reseeded at a concentration of 2.5 × 105 cells/mL in phase II medium (StemSpan supplemented with 10–7 M dexamethasone σ, 50 ng/mL stem cell factor, and 2 U/mL human recombinant erythropoietin [EPO; Sigma]). The cultures were diluted once or twice to maintain the cell concentration lower than 1 × 106 cells/mL in phase II. Cell samples were collected from phase II cultures on days 0, 2, 3, 4, 5, 6, and 7 and evaluated for number and viability.
K562 cells grew in RPMI 1640 medium supplemented with fetal calf serum plus 4 mM glutamine, 10 U/mL penicillin, and 10 μg/mL streptomycin. Cells (2 × 106) were collected for preparation of RNA.
Flow cytometry
Cells (1 × 106) were collected and washed with phosphate-buffered saline (PBS). Cells were labeled in 100 μL PBS containing 1 μg anti–human CD71 monoclonal antibodies conjugated with FITC (Pharmingen, San Diego, CA) or anti–human glycophorinA(GPA) conjugated with phycoerythrin (PE) (DakoCytomation, Ely, United Kingdom) independently or stained simultaneously with 2 antibodies, anti–CD71-FITC and anti–GPA-PE. Following incubation at room temperature for 30 minutes in the dark, the cells were washed twice with cold PBS and resuspended in 1 mL PBS containing 0.5% formaldehyde. Flow cytometric data of FITC and PE were acquired from a minimum of 10 000 cells using a Becton Dickinson flow cytometer and CellQuest (Becton Dickinson, Oxford, United Kingdom) for data acquisition and analysis.
Morphology analysis
Cells (1.5 × 105) were collected on day 5 of phase II erythroid culture and adjusted to 200-μL volume. Cytospins of these cells were made and stained with modified Wright stain (Sigma Diagnostics, St Louis, MO). The slides were examined microscopically and images were obtained by using an Olympus Leica DMR fluorescence-type (Melville, NY) microscope (40×/0.75 oil immersion objective). Images were captured through a Coolsnap digital camera, and were acquired and analyzed via RS 1.5 image software (Improvision, Covington, United Kingdom).
RNA isolation and quantitative real-time RT-PCR
Total RNA was isolated using Tri-reagents (Sigma) and quantified by absorbance at 260 nm. cDNA was synthesized using SuperScript III reverse transcriptase (Invitrogen, Paisley, United Kingdom) from 1μg total RNA. Primers and probes were designed using Primer Express 2.0 program and synthesized by Applied Biosystems (Warrington, United Kingdom). Sequences for primers and probes are available on request. Quantitative reverse transcription–polymerase chain reaction (RT-PCR) was carried out in an ABI 7900 HT Sequence Detection System using TaqMan master mix and the protocol of the manufacturer (Applied Biosystems). All data were normalized using the endogenous HPRT control. Assays for HPRT are available from theApplied Biosystem database. To quantify gene expression, a relative standard method was used. The quantities of targets and of the endogenous HPRT were determined from the appropriate standard curves. The target amount was then divided by the HPRT amount to obtain a normalized value. One of the experimental samples on day 0 (HPRT normalized) was designated as the calibrator and given a relative value of 1.0. All quantities (HPRT normalized) were expressed as n-fold relative to the calibrator.
Western blotting
Protein samples were prepared from 2 × 106 erythroid cells lysed in 40 μL RIPA buffer (1 × PBS, 1% NP40, 0.5% sodium deoxycholate, and 0.1% SDS) with the addition of the protein inhibitors, 1 μg/mL aprotinin and 100 μg/mL PMSF. Cell lysates were stored at –80°C until further use. Before loading onto sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) gel, the protein homogenates were thawed, and equal volume of 2 × loading buffer and 10% β-mercaptoethanol were added. The samples were further denatured by boiling the cell lysates for 5 minutes. Cell extracts were separated on 8% Tris-glycine SDS-PAGE gels and transferred onto nitrocellulose membranes (Amersham Biosciences, Buckinghamshire, United Kingdom). Membranes were blocked in Tris-buffered saline (TBS) supplemented with 5% nonfat dry milk powder for 1 hour and incubated overnight with mouse monoclonal antibody against c-MYB (Upstate, Hampshire, United Kingdom). Subsequently, the blots were washed 3 times with TBS-0.05% Tween-20 buffer and reacted with horseradish peroxidase–conjugated rabbit anti–mouse IgG antibody (Dako, Cambridgeshire, United Kingdom). The specific proteins were developed using an enhanced chemiluminescent (ECL) detection system (Amersham Biosciences) and exposure to X-ray films. The same blots were stripped by Re-blot Solution (Chemicon, Temecula, CA) and relabeled with antibodies against β-actin as loading controls.
Transfection and gene expression studies of HBS1L and cMYB in K562 cells
cDNAs of HBS1L and cMYB were reverse transcribed from total RNA made from K562 cells using reverse transcriptase III (Invitrogen) and cloned into pEF6/V5-His vector (Invitrogen). Vectors encoding cMYB, HBS1L, or vector only were transfected into K562 cells by electroporation (BioRad Gene Pluser II). Stable expressing clones were selected by culturing the cells in 5 μg/mL blasticidin for 2 weeks. Expression levels of γ-globin, HBS1L, and cMYB genes were evaluated by real-time quantitative RT-PCR and normalized by endogenous HPRT.
Statistical analysis
A Student t test was used to derive the significance of the difference between the mean values. Correlation was assessed by an F test. Linear functions describing the relationship between parameters were obtained using linear regression.
Results
Erythroid cell morphology, growth pattern, maturation, and frequency of macrophages in heterocellular HPFH versus controls
Cell morphologies exhibit striking changes in the 2-phase culture system in cell shape, size, nuclear chromatin, and cytoplasmic volume. In phase I, the culture conditions favor the expansion of CD34+ hematopoietic progenitors but inhibit the proliferation of lymphocytes because of the presence of cyclosporin A. At the early stage of phase I, the majority of cells were small with round nuclei and a thin rim of agranular cytoplasm, consistent with the morphology of lymphocytes. At the end of phase I, the vast majority of the cells are still lymphocytes, but macrophages, recognizable as large cells with large volume ratios of cytoplasm to nucleus, could be observed. A homogeneous population of small blasts larger in size than lymphocytes was also observed among the nonadherent cells, representing CD34+ cells. In phase II, rapid erythroid expansion occurs following exposure to EPO in the culture. In the early stage of phase II (days 1-4), a population of erythroblasts increased dramatically. These precursors of proerythroblasts are large with intensely basophilic cytoplasm and prominent vacuoles. Some cells were observed with cytoplasmic projection, whereas others were round and smooth. From days 5 to 7 of phase II, cell numbers continued to increase, but cell volumes decreased. The large cells with intensely basophilic cytoplasm progressed through different stages from cells with reticular nuclei to hemoglobinized cells with pyknotic nuclei (orthochromatic normoblasts), representing the late erythroblastic cells. Enucleation was rarely observed in culture.
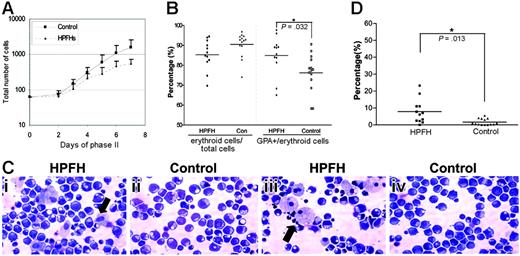
Rapid erythroid expansion was observed in phase II and cell counts were performed on days 0, 2, 3, 4, 5, 6, and 7. An increase in cell number was detected on day 2 with a doubling of the number on day 3, reaching a maximum count on day 6 to day 7, after which the number of cells starts to decline. Throughout phase II, cell count in erythroid cultures of individuals with HPFH was lower than that in the controls (Figure 1A).
Differentiation of the erythroid progenitors in phase II was monitored using 2 surface membrane proteins, transferrin receptor (CD71) and glycophorin A (GPA). CD71 is expressed on all proliferating hematopoietic cells, and, although it is not erythroid lineage specific, it is highly expressed in early erythroid progenitors. GPA, however, is erythroid lineage specific and initially expressed when CD34 is down-regulated. GPA expression represents the later stages of erythroid differentiation compared with CD71. On day 2 of phase II, we observed that 10% to 40% of the cells highly expressed CD71 with 2% to 10% cells expressing GPA. As cells proliferated and differentiated, the percentages of cells simultaneously expressing CD71 and GPA increased rapidly. Initially CD71high expression shifted to CD71med expression. On day 5, a small proportion of erythroid cells was enucleated but was not observed to increase on days 6 and 7, the implication being that enucleated erythrocytes undergo apoptosis, because they are not stable in these culture conditions. On day 7, almost all CD71+ cells also showed GPA positivity, an indication that those cells that were CD71+ only at the earlier stage represented the erythroid population in this culture system. CD71 and GPA expression was analyzed on day 5 samples.
Erythroid cell growth, maturation, frequencies of macrophage during the second phase of erythroid cell culture. (A) Erythroid cell numbers were counted on days 0, 2, 3, 4, 5, 6, and 7 of phase II. Solid line indicates the mean values of cell numbers of the 14 individuals with HbF less than 0.6%. Dashed line represents the mean cell numbers of the 12 individuals with HbF levels of 0.8% to 1.8%. Error bars refer to the SD. (B) Percentages of erythroid cells and GPA-positive cells on day 5 of phase II erythroid cells (CD71pos + CD71posGPApos + GPApos cells). (C) Representative cytospins of erythroid cell cultures on day 5 of phase II from 2 different individuals (i, iii) in HPFH and control (ii,iv) groups. The cytospins were stained with modified Wright-Giemsa. Arrows indicate the macrophages. (D) Percentage of macrophages of the total number of cells (400 cells counted) on day 5 of phase II.
Erythroid cell growth, maturation, frequencies of macrophage during the second phase of erythroid cell culture. (A) Erythroid cell numbers were counted on days 0, 2, 3, 4, 5, 6, and 7 of phase II. Solid line indicates the mean values of cell numbers of the 14 individuals with HbF less than 0.6%. Dashed line represents the mean cell numbers of the 12 individuals with HbF levels of 0.8% to 1.8%. Error bars refer to the SD. (B) Percentages of erythroid cells and GPA-positive cells on day 5 of phase II erythroid cells (CD71pos + CD71posGPApos + GPApos cells). (C) Representative cytospins of erythroid cell cultures on day 5 of phase II from 2 different individuals (i, iii) in HPFH and control (ii,iv) groups. The cytospins were stained with modified Wright-Giemsa. Arrows indicate the macrophages. (D) Percentage of macrophages of the total number of cells (400 cells counted) on day 5 of phase II.
Flow cytometric analysis showed that the HPFH group had fewer erythroid cells (CD71pos + CD71posGPApos + GPApos) on day 5 when compared with the control group, mean of 85.3% versus 90.1% (Figure 1B). However, despite a lower proportion of erythroid population, the ratios of GPA-positive cells to the total erythroid cells were significantly higher in the HPFH group, mean of 84.9% versus 76.5% (P = .032). The relative increase in GPA-expressing cells indicates that the maturation of erythropoiesis in heterocellular HPFH had been accelerated compared with that in the control group.
Although the cell cultures from the different donors exhibited a wide range of cell proliferation in terms of cell number and maturation, the growth and differentiation patterns are reproducible in repeat cultures of different blood donations from the same donor. Thus, the differences in the proliferation patterns and maturation are likely to be related to the intrinsic properties of the cells rather than the culture condition.
We observed different frequencies of macrophages in the erythroid cell cultures between the 2 groups. Cytospin slides were prepared from all samples on day 5 of phase II. Wright-Giemsa–stained cytospins were examined under a microscope, and a total of 400 cells were counted. HPFH erythroid cell cultures showed relatively more macrophages compared with controls (mean of 7.85% of total cells in HPFH versus 1.57% in controls), the difference was statistically significant (P = .013, Figure 1C-D).
Characteristics of globin gene expression by quantitative real-time RT-PCR
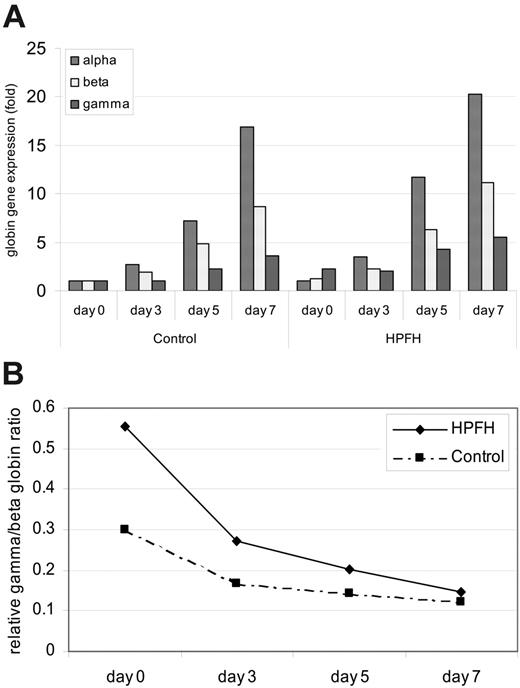
In both groups the relative mRNA transcripts of α-, β-, and γ-globin all increased during the secondary culture after EPO supplement (Figure 2A). α- and β-globin transcripts increased dramatically and their levels were much higher than those of γ-globin transcripts. γ-Globin expression increased by an average of 4- to 5-fold on day 7, compared with an increase of 17- to 20-fold for α-globin, and 9- to 11-fold for β-globin. As erythropoiesis matures, the ratio of γ-to β-globin transcripts decreased (Figure 2B), reflecting the fall in the levels of HbF (α2γ2) as adult erythropoiesis advanced. The average levels for the expressions of the globin genes were generally higher for the HPFH group when compared with control throughout the secondary culture, which is in agreement with the accelerated erythroid maturation observed. The average γ/β-globin transcript ratios were also higher in the heterocellular HPFH group throughout phase II, the highest ratios being observed at the early stage of erythropoiesis in both groups (Figure 2B).
Globin gene expression during phase II of the erythroid cell culture. Gene expression was determined by quantitative real-time RT-PCR, and the levels are related to fold change of the gene expression of one sample after normalization with HPRT gene expression. (A) Levels represent the mean values of the 12 and 14 individuals in the HPFH and control groups, respectively. (B) Relative ratios of gamma to β-globin gene expression. The γ and β values represent the average levels of all the individuals in each group.
Globin gene expression during phase II of the erythroid cell culture. Gene expression was determined by quantitative real-time RT-PCR, and the levels are related to fold change of the gene expression of one sample after normalization with HPRT gene expression. (A) Levels represent the mean values of the 12 and 14 individuals in the HPFH and control groups, respectively. (B) Relative ratios of gamma to β-globin gene expression. The γ and β values represent the average levels of all the individuals in each group.
Differential expression of the genes in the 6q23 candidate interval by quantitative real-time RT-PCR
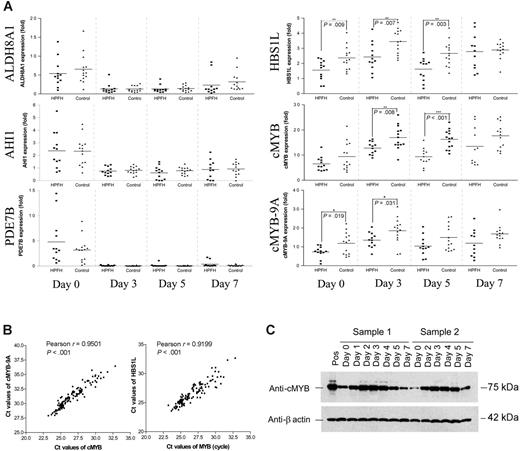
The candidate interval of 1.5 Mb in chromosome 6q23.2 contains 5 protein-coding genes, ALDH8A, HBS1L, cMYB, AHI1, and PDE7B, which we tested. ALDH8A1 expression was low on day 0 (Ct values around 33-36 cycles) and was further down-regulated during the secondary culture. There was no significant difference in ALDH8A1 expression between the 2 groups (Figure 3A). AHI1 has relatively high expression in hematopoietic progenitors on day 0 of phase II in both groups (Ct values around 28-30 cycles) but were down-regulated to similar levels of expression throughout phase II. Again, there was no significant difference in the levels of AHI1 transcripts between the 2 groups (Figure 3A). PDE7B expression was detected at very low levels on day 0 (Ct values around 33-37 cycles) and were hardly detectable from day 3 to day 7 of phase II, an indication that PDE7B is not expressed in the erythroid lineage (Figure 3A).
The mean levels of HBS1L mRNA transcripts detected in the HPFH group were significantly lower compared with controls throughout phase II (P = .009, .007, .003 on days 0, 3, 5, respectively; Figure 3A). Similarly, the mean values of cMYB gene expressions were significantly lower in the HPFH group compared with those in the control group (P = .008 and < .001 on days 3 and 5, respectively; Figure 3A). No difference was detected in the levels of cMYB expression between the male and female individuals within our control group.
The cMYB protein (75 kDa) has an isoform of 89 kDa (cMYB-9A) caused by a splice variant which leads to an insertion of 363 base pairs (bp) between exon 9 and 10 (exon 9A).14,15 Using primer and probes specific for cMYB-9A, we confirmed that the pattern of cMYB-9A expression was similar to that of the total cMYB transcript (ie, lower in the HPFH group when compared with control). The differences in mean values between these 2 groups were statistically significant on days 0 and 3 (P = .019 and .031 on days 0 and 3, respectively; Figure 3A). Furthermore, the expression levels of cMYB and cMYB-9A were highly correlated (Pearson correlation coefficient r = 0.95; Figure 3B).
Both HBS1L and cMYB expression increased on day 3 of phase II when compared with day 0, followed by a decline on day 5, and in both cases, the expression was lower in the HPFH group when compared with control. Analysis of more than 100 erythroid cell samples at different stages of phase II showed that the levels of cMYB and HBS1L mRNA transcripts were highly correlated (Pearson correlation coefficient r = 0.92, P < .001; Figure 3B). Western blot analyses of cellular extracts from the corresponding days of the erythroid cell cultures confirmed the pattern of cMYB gene expression, consistent with the quantitative RT-PCR analysis (Figure 3C).
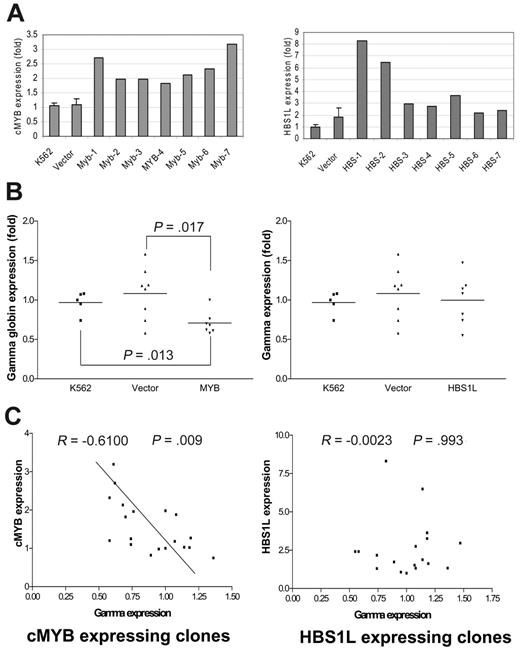
Overexpression of cMYB inhibited the expression of γ-globin in K562 cells, but overexpression of ectopic HBS1L had no effect on γ-globin expression
To determine whether cMYB or HBS1L, or both, are involved in the regulation of γ-globin gene expression, we stably transfected cMYB and HBS1L into K562 cells and analyzed multiple cMYB-expressing, HBS1L-expressing, and vector-alone transfected clones. Both cMYB- and HBS1L-transfected clones expressed far more mRNA transcripts above the endogenous background levels as expected (Figure 4A). Furthermore, cMYB-expressing clones inhibited γ-globin gene expression when compared with the parental cell line (P = .013) or vector controls (P = .017; Figure 4B). However, HBS1L had no effect on γ-globin gene expression, although the clones expressed 2- to 8-fold of HBS1L mRNA transcripts above the background (Figure 4B). Although cMYB and γ-globin mRNA levels were significantly inversely correlated (correlation coefficient =–0.61, P = .009; Figure 4C), there was no correlation between HBS1L and γ-globin mRNA levels (Figure 4C).
Discussion
We have previously mapped a QTL for heterocellular HPFH on chromosome 6q23,7,16 but analysis of the coding sequence of the genes in the region did not reveal any causative mutation in a panel of subjects.9 Using a 2-phase liquid erythroid culture system (modified from Fibach et al11 ), we have observed that 2, cMYB and HBS1L, of the 5 protein-coding genes in the candidate region showed significant alterations in their expression profiles in individuals with heterocellular HPFH when compared with controls. The data suggest that a high HbF phenotype is associated with lower cMYB levels in erythroid cultures.
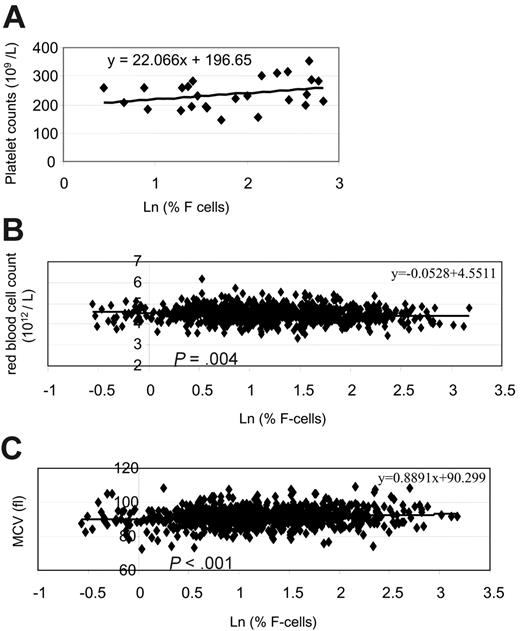
We observed other phenotypes that were consistent with the lower cMYB expression. For example, erythroid cell cultures of individuals with HPFH showed a relatively higher number of macrophages when compared with controls, a feature consistent with that noted in a mouse cMYB knockdown study in which low levels of cMYB favored differentiation of macrophage and megakaryocytes, with perturbed erythroid differentiation.17 cMYB knockout mice die in utero because of a block in erythropoiesis. Apart from primitive erythrocytes, the only mature cells present in the cMYB–/– fetal livers were megakaryocytes and macrophages.18 Further, mice bearing a hypomorphic allele of cMYB induced by N-ethyl-N-nitrosourea mutagenesis had a myeloproliferative syndrome with supraphysiologic expansion of megakaryocyte and platelet production and anemia.19,20 In keeping with these observations, we noted that, although both the HPFH and control groups have platelet levels within the normal range, individuals with HPFH who had lower cMYB expression had relatively higher platelet counts. There was a trend toward a positive correlation between the platelet counts and F cell percentages in this group of 26 subjects, although the results were not significant (Figure 5A).
Relative expression of the genes in the 6q23 candidate interval throughout phase II. Gene transcription was determined by real time RT-PCR. Gene expression levels are related to the gene expression of one sample after HPRT normalization. (A) Relative expression of ALDH8A, AHI1, PDE7B, HBS1L, cMYB, and cMYB-9A. Each data point represents the mean value for each individual in the HPFH and control groups. For day 7, values were obtained for only 11 of the 12 HPFH, and 13 of the 14 controls. There were inadequate cell numbers for analysis for culture of one individual in each group. (B) Correlation of cMYB with cMYB-9A or HBS1L expression. The Ct values are a measure of the amount of template mRNA, the lower the Ct value, the higher the amount of mRNA template. The data points include all values, with more than one value for 6 individuals who had repeat erythroid cultures. (C) Western blot analysis of total cell lysates from phase II erythroid cell cultures. Samples 1 and 2 represent 2 different individuals to illustrate the pattern of the cMYB protein expression in phase II. Pos refers to the cell lysate from K562 cells.
Relative expression of the genes in the 6q23 candidate interval throughout phase II. Gene transcription was determined by real time RT-PCR. Gene expression levels are related to the gene expression of one sample after HPRT normalization. (A) Relative expression of ALDH8A, AHI1, PDE7B, HBS1L, cMYB, and cMYB-9A. Each data point represents the mean value for each individual in the HPFH and control groups. For day 7, values were obtained for only 11 of the 12 HPFH, and 13 of the 14 controls. There were inadequate cell numbers for analysis for culture of one individual in each group. (B) Correlation of cMYB with cMYB-9A or HBS1L expression. The Ct values are a measure of the amount of template mRNA, the lower the Ct value, the higher the amount of mRNA template. The data points include all values, with more than one value for 6 individuals who had repeat erythroid cultures. (C) Western blot analysis of total cell lysates from phase II erythroid cell cultures. Samples 1 and 2 represent 2 different individuals to illustrate the pattern of the cMYB protein expression in phase II. Pos refers to the cell lysate from K562 cells.
Our data and that of others21-24 indicate that hemoglobin switching is likely to be cell-cycle related and linked to differentiation events as summarized in a recent review on the regulation of HbF production in adult erythropoiesis.25 As differentiation advances, a change in globin gene transcription program occurs, from a program of predominant γ-globin expression to one that supports mainly β-globin expression. Although the molecular mechanism is not evident, our observations suggest that reduced cMYB expression increases γ-globin expression via a process of accelerated erythroid maturation. Despite the lower expansion potential, a larger proportion of the erythroblasts in cultures from subjects with HPFH differentiated into GPA-positive population within 5 days of phase II. In addition to the relatively higher proportion of GPA-positive erythroblasts, erythroid cultures from individuals with HPFH also show relatively higher expression of the α-, β-, and γ-globin genes, all implicating an accelerated erythroid differentiation.
Various studies deduced that cMYB blocks differentiation by maintaining proliferation, but cMYB has also been shown to inhibit terminal erythroid differentiation.26 Analysis of knock-down mice/embryos suggested that erythroid development is very sensitive to changes in cMYB levels and that there may be a differential requirement for cMYB in the proliferation of early and late progenitors.26 It would appear that in adults with normal levels of HbF, the levels of cMYB support a balance of proliferation and differentiation such that the majority of erythroid cells mature to erythroblasts that mainly synthesize adult hemoglobin. Few progenitor cells terminate prematurely before the change in program takes place. In individuals with HPFH, with their relatively lower levels of cMYB expression, more of the progenitors and erythroblasts terminate prematurely during the proliferation cycles of adult erythropoiesis, producing more erythroid cells that synthesize predominantly HbF. Thus, in the presence of lower cMYB levels, we would predict a relatively lower erythrocyte count in individuals with HPFH, resulting from the reduced number of proliferation cycles and accelerated erythroid maturation, in combination with higher HbF and F cell levels. Indeed, analysis of 850 healthy adult women showed that F cell percentages are inversely correlated with the erythrocyte counts. Individuals with higher F cell percentages tend to have lower erythrocyte counts (P = .004; Figure 5B). Further, individuals with higher F cell percentages also tend to have higher mean cell volumes (MCVs). indicating a younger erythrocyte population (Figure 5C).
It is evident from several studies that multiple intracellular signaling pathways are involved in the control of globin gene transcription, and cMYB is critical for erythroid development.26-28 The conditions of human erythroid culture could be manipulated to produce low or high fetal hemoglobin production; CD34 cells cultured in EPO alone were associated with low HbF production, whereas CD34 cells cultured in EPO, SCF, and TGF-β were associated with high HbF production.29 In addition, a correlation between cMYB expression patterns and HbF could be demonstrated under these manipulated in vitro conditions.30 It would appear that cMYB expression is affected by these growth factors that are involved in intracellular signal transduction. The cMYB protein could thus be a downstream effector of the various genes controlling HbF levels.
Characterization of cMYB- or HBS1L-expressing clones isolated from K562 cells. (A) K562 refers to the background gene expression; vector refers to the gene expression of clones transfected with vector alone with the error bars referring to the SD. MYB and HBS1L refer to MYB and HBS1L transfectants, respectively. Myb-1 to Myb-7 and HBS-1 to HBS-7 refer to different clones transfected with cMYB or HBS1L cDNA, respectively, with their levels of gene expression indicated. (B) γ-Globin expression of cMYB or HBS1L transfectants. (C) Correlations of γ-globin gene expression with either cMYB or HBS1L expression. The data points include all values from K562, vector-alone, and cMYB or HBS1L transfectants.
Characterization of cMYB- or HBS1L-expressing clones isolated from K562 cells. (A) K562 refers to the background gene expression; vector refers to the gene expression of clones transfected with vector alone with the error bars referring to the SD. MYB and HBS1L refer to MYB and HBS1L transfectants, respectively. Myb-1 to Myb-7 and HBS-1 to HBS-7 refer to different clones transfected with cMYB or HBS1L cDNA, respectively, with their levels of gene expression indicated. (B) γ-Globin expression of cMYB or HBS1L transfectants. (C) Correlations of γ-globin gene expression with either cMYB or HBS1L expression. The data points include all values from K562, vector-alone, and cMYB or HBS1L transfectants.
Equally, cMYB may act more upstream, regulating the activity and expression of other proteins that are involved in various stages of hematopoiesis. The cMYB protein may directly activate the transcription of one of these genes, or the action may be more indirect, through a gene activated within the cMYB transcriptional cascade. Cyclin A1, c-kit, bcl-2, mim-1 have been demonstrated to be direct cMYB target genes,31 and many others have been shown to be regulated by cMYB protein.28,32,33 They are mainly involved in signal transduction and regulatory decisions affecting cell proliferation, differentiation, and apoptosis. Recent studies confirmed that cMYB is strictly required for the expression of c-kit receptor,26 which interacts with SCF, EPO receptor, and EPO in the development of the erythroid program.34,35 The cMYB transcription factor also interacts with a large collection of cofactors in different ways, leading to the regulation of different sets of targets. CBP or CREB-binding protein and p300 are ubiquitous coactivators that also interact with cMYB.36,37 These have histone acetyltransferase activity and presumably play a role in chromatin remodeling.
Thus, it is important to note that our data do not necessarily implicate the cMYB locus as the QTL or the only QTL underlying the high HbF phenotype in the individuals in this study. Heterocellular HPFH is genetically heterogeneous with several loci contributing to the high HbF phenotype.5,38 What our experiments have shown is that cMYB expression in healthy adults is variable and that lower cMYB expression is associated with higher HbF levels. It is likely that cMYB is a quantitative trait gene and that levels of the cMYB product are controlled by cis-regulatory and trans-acting factors.39
But, although we make no assumptions concerning the gene responsible for the high F phenotype in the individuals in this study, among the panel of protein-coding genes within the chromosome 6q23 interval, cMYB must be the prime candidate in the Asian-Indian family.16 The role of cMYB in accelerating erythroid maturation could explain the disproportionately higher HbF levels in subjects with β thalassemia in the Asian-Indian family in which the 6q QTL is a major determinant of the HbF levels.7,40 In this kindred, among individuals possessing the 6q genotype, those without β thalassemia had elevated HbF levels of up to 2.4%, whereas those who are heterozygous for β thalassemia had HbF levels ranging from 3.0% in those with a single copy of the 6q genotype to 24% in those with 2 copies. The proband who was homozygous for β thalassemia and heterozygous for the 6q genotype was transfusion independent with hemoglobin levels of 11 to 12 g/dL, all of which was HbF.40,41 It would appear that the lower cMYB expression implicated from the 6q genotype, together with the rapid erythroid regeneration under the “stressed erythropoiesis,” further increases the proportion of early erythroid progenitors that still possess the primitive fetal globin program, leading to the unusually high fetal hemoglobin levels in those with β thalassemia.
Although cMYB is a prime candidate underlying the high HbF phenotype in the Asian-Indian family, identification of the causative sequence variant(s) could be a daunting challenge. To date, the regulatory sequences controlling cMYB expression are still not completely clear. Our expression profile studies suggest that cMYB may be coregulated with HBS1L. Interestingly, other studies on pancreatic cancer tissues and cell lines using comparative genomic hybridization showed that when cMYB proto-oncogene was overexpressed, HBS1L seemed to be coamplified with cMYB as well.42 Studies have shown that cMYB is a key target for transcriptional activation by long-range upstream and downstream retroviral insertion. Two murine cell lines (p/m16i and G1-500/44i) rearranged at Ahi-1 approximately 35 kilobases (kb) downstream of cMYB, and 2 feline cell lines (FT-1 and FTG) rearranged at fit1 (approximately 100 kb upstream) overexpressed cMYB compared with phenotypically similar cell lines lacking detectable rearrangements.43 In the same study, as the steady-state levels of cMYB RNA declined with the passage of p/m16i cells, expression of the genes flanking cMYB, HBS1L and FLJ20069 (murine ortholog of AHIL), was also down-regulated. Further, it was noted that increased expression of the flanking genes only occurred in the presence of cMYB overexpression. The observations suggest the possibility that regulation of cMYB may affect a wider chromatin domain surrounding the gene. Alternatively, there may be a common transcription factor or a common cis-regulatory element(s) that controls the expression of cMYB and gene(s) in its vicinity.
Correlation of F cell percentages with platelet counts, erythrocyte counts, and erythrocyte MCV. (A) Correlation between platelet counts and Ln (percentage of F cells) in 26 individuals in this study. (B) Correlation between Ln (percentage of F cells) and erythrocyte counts in 850 healthy female adult whites. (C) Correlation between erythrocyte mean cell volume (MCV) values and Ln (percentage of F cells) in 850 healthy female adult whites.
Correlation of F cell percentages with platelet counts, erythrocyte counts, and erythrocyte MCV. (A) Correlation between platelet counts and Ln (percentage of F cells) in 26 individuals in this study. (B) Correlation between Ln (percentage of F cells) and erythrocyte counts in 850 healthy female adult whites. (C) Correlation between erythrocyte mean cell volume (MCV) values and Ln (percentage of F cells) in 850 healthy female adult whites.
Identification of the factors associated with the lower cMYB expression could provide further insights on the molecular basis for globin gene switching. MYB-targeted therapeutics could be a novel strategy for pharmacologic augmentation of fetal hemoglobin for the β hemoglobinopathies.
Prepublished online as Blood First Edition Paper, April 13, 2006; DOI 10.1182/blood-2006-01-008912.
Supported by Medical Research Council (MRC) (grant G0000111, ID51640) (S.L.T.); the TwinsUK project is funded by the Wellcome Trust.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Claire Steward for help in preparation of the manuscript and Helen Rooks for technical help.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal