Outcome for children with childhood acute lymphoblastic leukemia (ALL) who relapse is poor. To gain insight into the mechanisms of relapse, we analyzed gene-expression profiles in 35 matched diagnosis/relapse pairs as well as 60 uniformly treated children at relapse using the Affymetrix platform. Matched-pair analyses revealed significant differences in the expression of genes involved in cell-cycle regulation, DNA repair, and apoptosis between diagnostic and early-relapse samples. Many of these pathways have been implicated in tumorigenesis previously and are attractive targets for intervention strategies. In contrast, no common pattern of changes was observed among late-relapse pairs. Early-relapse samples were more likely to be similar to their respective diagnostic sample while we noted greater divergence in gene-expression patterns among late-relapse pairs. Comparison of expression profiles of early- versus late-relapse samples indicated that early-relapse clones were characterized by overexpression of biologic pathways associated with cell-cycle regulation. These results suggest that early-relapse results from the emergence of a related clone, characterized by the up-regulation of genes mediating cell proliferation. In contrast, late relapse appears to be mediated by diverse pathways.
Introduction
In spite of the significant progress in the improvement of cure rates for childhood acute lymphoblastic leukemia (ALL), 20% of children will suffer a recurrence, making relapsed ALL the fifth most common childhood cancer. Unfortunately, retrieval therapy is inadequate in most cases, and most of these children succumb to their disease. The failure of intensive chemotherapy to cure most children, as well as the toxicity of these approaches, mandates a search for new treatment approaches.
Numerous clinical and biologic factors are helpful in predicting outcome at initial diagnosis, but few prognosticators exist at relapse. The duration of first remission is the most important prognostic variable. Namely, patients relapsing early, while on therapy, or shortly after completing treatment (< 36 months from initial diagnosis), have long-term outcomes far worse than those with later relapses (≥ 36 months from diagnosis). Only 10% of patients with early bone marrow relapse are long-term survivors.1-3 In addition, dismal outcomes have been observed at relapse in patients with a T-cell phenotype.4
Recent advances in microarray technology have made it possible to obtain a molecular portrait of cancer.5,6 The goals of this study were to identify pathways that potentially account for drug resistance at relapse and provide an explanation for the observed differences in outcome among patients who relapse early versus late following diagnosis, to provide insight into the origin of the relapsed clone, and to discover pathways that are attractive targets for future therapy. To accomplish these goals we examined gene-expression profiles in 2 cohorts of samples, a matched-pair cohort of diagnosis/relapse samples from the same patient, and a large group of relapse samples from children enrolled in a contemporary Children's Oncology Group (COG) protocol for relapsed ALL, AALL01P2.
Patients, methods, and materials
Patient samples
Ficoll-enriched, cryopreserved bone marrow samples (peripheral blood with > 80% circulating blasts from a small subset) were obtained from 35 patients where matched samples from initial diagnosis and first marrow relapse were available. More than half (23) of the patients had early relapses (< 36 months from initial diagnosis), while 12 had late relapses (≥ 36 months from diagnosis). The majority of patients had a B-precursor phenotype (n = 32); 3 patients had T-cell ALL (T-ALL; Table 1). These patients were treated on contemporary cooperative group protocols from 1999 to 2004. Patient characteristics are detailed in Table S2 (available on the Blood website; click on the Supplemental Materials link at the top of the online article). An independent set of 29 marrow samples acquired at the time of initial diagnosis (balanced for National Cancer Institute [NCI] standard and high risk) and 19 samples acquired at relapse were used for verification of target gene expression by quantitative real-time polymerase chain reaction (PCR).
Cohorts of patients used for the various analyses
. | No. matched pairs . | No. samples at relapse . |
|---|---|---|
| Early relapse | ||
| B-precursor | 21 | 32 |
| T-cell | 2 | 5 |
| Late relapse | ||
| B-precursor | 11 | 22 |
| T-cell | 1 | 1 |
| Total | 35 | 60 |
. | No. matched pairs . | No. samples at relapse . |
|---|---|---|
| Early relapse | ||
| B-precursor | 21 | 32 |
| T-cell | 2 | 5 |
| Late relapse | ||
| B-precursor | 11 | 22 |
| T-cell | 1 | 1 |
| Total | 35 | 60 |
A cohort of 60 patients at relapse (37 early, 23 late) enrolled in the current Children's Oncology Group protocol, AALL01P2, for patients with first bone marrow relapse were obtained. The majority of patients had a B-precursor immunophenotype (n = 54). Six patients had T-ALL. Treatment for all these patients consisted of 3 35-day blocks of chemotherapy, detailed in Table S1. All patient samples used in these analyses were acquired from the Children's Oncology Group cell bank, and patients (or parents) had provided informed consent for use of these samples for research studies.
RNA extraction, amplification, and DNA arrays
RNA was isolated by Qiagen RNEasy Mini kits (Valencia, CA) and quality was verified by an Agilent 2100 Bioanalyzer (Agilent, Palo Alto, CA). Total RNA (1 μg) was used as a template in a double amplification protocol using RiboAmp RNA amplification kits (Arcturus, Mountain View, CA) according to the manufacturer's recommendation. In vitro transcription was completed with biotinylated UTP and CTP for labeling using the ENZO BioArray HighYield RNA Transcript Labeling kit (Enzo Diagnostics, Farmingdale, NJ), with a representative yield of 40 to 50 μg. A portion of the labeled cRNA (20 μg) was fragmented and hybridized to Affymetrix U133A microarrays according to Affymetrix protocols (Santa Clara, CA). These arrays contain 22 283 probe sets, representing approximately 13 000 genes. After hybridization, DNA arrays were stained with streptavidin-phycoerythrin and scanned using a GeneArray scanner (Agilent).
Data analysis
The data discussed in this publication have been deposited in the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo) and are accessible through GEO Series accession number GSE3912. Image and expression data files were generated with Affymetrix MAS 5.0. The average percentage of probesets with “present calls” (qualitative detection of transcripts) was 33.6%, indicating that the array hybridizations were of good quality. Probe-level analysis including intensity-dependent normalization was performed using the method of robust multiarray analysis (RMA) described by Irizarry et al.7 The analytic software package Genetraffic (Iobion Informatics, La Jolla, CA) was used for RMA. Clustering and visualization were done using Cluster and TreeView software (Eisen Laboratory, Stanford University, Stanford, CA). For each probe set, the arithmetic mean of the expression values of all the hybridization was used to calculate the baseline expression. The probe sets that were flagged as absent in more than 70% of the samples were discarded as a means to filter noise in the data. Unsupervised analysis by hierarchic clustering was performed after application of the variance filter mentioned.
VxInsight, a higher dimensional unsupervised method, was used for discovering inherent relationships between the samples8-11 (http://hsc.unm.edu/crtc/WillmanResearch/Pages/UNMHSC_HPC_SNL_Methodology.htm).
Multiple supervised analytic methods were used to select genes that were differentially expressed between selected cohorts. To identify differentially expressed genes between paired samples at diagnosis and relapse, a paired t test was performed followed by adjustment of the P values for multiple simultaneous inferences by 2 methods. We calculated a false discovery rate (FDR) for each gene according to the method proposed by Benjamini and Hochberg.12 Secondly an adjusted P value was calculated using Hochberg's step-up Bonferroni method,13 which is denoted by HOC P in Table S5B. All the significant genes were sorted on the overall ranks, which were determined by the t-test statistic. Significance analysis of microarrays (SAM)14 was used to select genes differentially expressed between early and late relapse cohorts using a symmetric threshold that better selects for both “positive” and “negative” genes. In order to find significant genes that were associated with the time to relapse we used linear regression to fit the time to relapse data (in log scale), with 1 gene being a predictor. The t test for testing the significance of coefficient was estimated and the P value was used to measure the significance of the gene. The 2 methods mentioned were used to adjust the P values. Tables listing the significant genes for all the analyses along with FDR and P values are available in Tables S3 and S5.
Functional grouping of significant genes
Genes whose expressions were significantly different in the early and late relapse cohort with a FDR less than 5% were selected for classification into functional groups using the Gene Ontology tool through the Affymetrix Netaffyx application (www.affymetrix.com/analysis/netaffx).
PCR
The array expression patterns of several target genes (BIRC5, PTTG1, TOP2A, CCNB1, SCGF, and BCL7A) were subsequently verified by real-time quantitative PCR on an independent cohort of patients. Relative transcript levels were determined by a comparative threshold cycles for amplification (CT) method with normalization to β2-microglobulin (ΔCT). The ΔCT value is inversely proportional to the expression of the particular gene. All reactions were run in triplicate, and the integrity of product was checked by a melting curve analysis at the end of the amplification procedure. Primer sequences are as follows: β2-microglobulin: ATGTGTCTGGGTTTCATCCATCC (sense), AGTCACATGGTTCACACGGCA (antisense); BIRC5: CATCTCTACATTCAAGAACTGG (sense), GGTTAATTCTTCAAACTGCTTC (antisense); PTTG1: TTTCTGCCAAAAAGATGACT (sense), GAGACTGCAACAGATTGGAT (antisense); TOP2A: CTGATTCAGAGGGGATATGA (sense), CCACAAATCTGATGGACTCT (antisense); CCNB1: TGACTTTGCTTTTGTGACTG (sense), GTGTCCATTCACCATTATCC (antisense); SCGF: TGAGGACATCGTCACTTACA (sense), GAGAGCAGGAAGCACTTGT (antisense); and BCL7A: GACATGCATGACGATAACAG, CTGCCGATCTACTTTCTCTG (antisense).
Nested reverse transcriptase–PCR reactions were completed on samples from 35 patients (pairs) to detect the TEL/AML1 fusion transcript as previously described.15
Results
Changes in biologic pathways at relapse
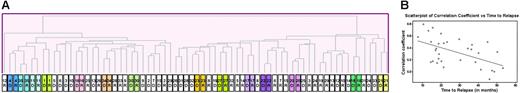
Matched-diagnosis and relapse samples from the same patient offer the best opportunity to study underlying mechanisms leading to emergence of resistant clones. An initial unsupervised hierarchic clustering of 70 samples (35 patients at diagnosis and at relapse) using the Pearson correlation showed that in 14 of 35 cases the diagnosis and relapse sample from the same patient clustered next to one another (Figure 1A). For example, the gene-expression profile of the diagnosis and relapse samples were very similar for the 3 T-cell (no. 4, no. 11, and no. 35), and infant (no. 16) cases. Figure 1B demonstrates the Pearson correlation coefficient (CC) of each of the pairs of patient samples arranged in order of time to relapse; a higher correlation coefficient indicates that the diagnosis and relapse sample are more alike. It is notable that there is a definite trend (see regression line; P = .002) toward a lower correlation coefficient among the later relapse pairs, indicating more dissimilarity.
Unsupervised analysis of paired diagnostic and relapse samples. (A) Samples (70; 35 patients at diagnosis and relapse) were clustered by hierarchic clustering using the Pearson correlation coefficient as the similarity measure. The diagnostic and relapse samples from multiple patients clustered closely together. D indicates sample from initial diagnosis; R, sample from relapse. (B) The Pearson correlation coefficient (CC) of the paired sample for each patient (32 pre-B) is represented as a gray circle; the higher the CC, the more similar the paired samples. Patients have been arranged from left to right according to the length of time to relapse. There is a clear trend toward decreasing CC as the time to relapse increases, as indicated by the regression line (P value of F test = .002).
Unsupervised analysis of paired diagnostic and relapse samples. (A) Samples (70; 35 patients at diagnosis and relapse) were clustered by hierarchic clustering using the Pearson correlation coefficient as the similarity measure. The diagnostic and relapse samples from multiple patients clustered closely together. D indicates sample from initial diagnosis; R, sample from relapse. (B) The Pearson correlation coefficient (CC) of the paired sample for each patient (32 pre-B) is represented as a gray circle; the higher the CC, the more similar the paired samples. Patients have been arranged from left to right according to the length of time to relapse. There is a clear trend toward decreasing CC as the time to relapse increases, as indicated by the regression line (P value of F test = .002).
Probe sets (120 total; 48 up-regulated at diagnosis, 78 upregulated at relapse) were identified using the paired t test to be significantly different at relapse compared to initial diagnosis in the B-precursor cases (FDR < 10%; Figure 2A). Multiple genes involved in cell-cycle regulation, protein biosynthesis, DNA replication and repair, and antiapoptosis were differentially expressed at relapse. The top 20 genes are shown in Table 2. A similar analysis was performed after including the 3 patients with T-cell ALL. Using an identical cutoff of FDR less than 10%, 78% of the genes were common to both analyses. Given differences in the clinical outcomes of early versus late relapse, early- and late-relapse pairs were analyzed separately. A number of probe sets (73 sets; FDR < 10%) were differentially expressed between the initial diagnosis/early relapse matched sample pairs (n = 23; Table S3C). Many (52%) of these genes were identified in the matched pair analysis of all 35 pairs. In contrast, we were unable to identify any genes with significant differences in expression among the initial diagnosis/late-relapse matched pairs (n = 12).
List of the top 20 genes identified in a matched-pair analysis differentially expressed between initial diagnosis and relapse (32 precursor B-ALL)
Rank . | Time of high expression . | FDR, % . | P . | Affymetrix ID . | Symbol . | Gene title . | Reported function . |
|---|---|---|---|---|---|---|---|
| 1 | Diagnosis | 0.66 | .000001 | 204642_at | EDG1 | Endothelial differentiation, sphingolipid G-protein-coupled receptor, 1 | Cell adhesion, signal transduction |
| 2 | Diagnosis | 0.66 | .000002 | 203795_s_at | BCL7A | B-cell CLL/lymphoma 7A | Putative tumor suppressor |
| 3 | Relapse | 0.97 | .000003 | 203554_x_at | PTTG1 | Pituitary tumor-transforming 1 | DNA replication and repair; cell cycle |
| 4 | Relapse | 2.11 | .00001 | 200834_s_at | RPS21 | Ribosomal protein S21 | Protein biosynthesis |
| 5 | Relapse | 2.11 | .000016 | 200763_s_at | RPLP1 | Ribosomal protein, large, P1 | Protein biosynthesis |
| 6 | Relapse | 2.11 | .000017 | 213166_x_at | FLJ14346 | Hypothetical protein FLJ14346 | Unknown |
| 7 | Relapse | 2.11 | .000019 | 201757_at | NDUFS5 | NADH-coenzyme Q reductase | Mitochondrial electron transport |
| 8 | Relapse | 2.11 | .000021 | 200980_s_at | PDHA1 | Pyruvate dehydrogenase (lipoamide) alpha 1 | Glycolysis; protein polyubiquitination |
| 9 | Diagnosis | 2.11 | .000023 | 203796_s_at | BCL7A | B-cell CLL/lymphoma 7A | Putative tumor suppressor |
| 10 | Relapse | 2.11 | .000026 | 203989_x_at | F2R | Coagulation factor II (thrombin) receptor | Cell-cycle regulation, apoptosis |
| 11 | Relapse | 2.11 | .000029 | 200061_s_at | RPS24 | Ribosomal protein S24 | Protein biosynthesis |
| 12 | Relapse | 2.11 | .000029 | 201001_s_at | UBE2V1 | Ubiquitin-conjugating enzyme E2 variant 1 | Cell-cycle progression; DNA repair |
| 13 | Relapse | 2.11 | .000031 | 212834_at | DDX52 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 52 | Nucleic acid binding, helicase activity |
| 14 | Diagnosis | 2.12 | .000034 | 213875_x_at | C6orf62 | Chromosome 6 open reading frame 62 | Unknown |
| 15 | Diagnosis | 2.51 | .000044 | 208892_s_at | DUSP6 | Dual-specificity phosphatase 6 | Cell-cycle progression; DNA repair |
| 16 | Diagnosis | 2.51 | .000048 | 206864_s_at | HRK | Harakiri, BCL2 interacting protein (contains only BH3 domain) | Induction of apoptosis |
| 17 | Relapse | 2.51 | .000049 | 217491_x_at | COX7C | Cytochrome c oxidase subunit VIIc | Generation of precursor metabolites and energy |
| 18 | Relapse | 2.51 | .000052 | 201002_s_at | UBE2V1 | Ubiquitin-conjugating enzyme E2 variant | Cell-cycle progression; DNA repair |
| 19 | Relapse | 2.51 | .000054 | 202279_at | C14orf2 | Chromosome 14 open reading frame 2 | Unknown |
| 20 | Diagnosis | 2.51 | .000057 | 49485_at | PRDM4 | PR domain containing 4 | Transcription regulation |
Rank . | Time of high expression . | FDR, % . | P . | Affymetrix ID . | Symbol . | Gene title . | Reported function . |
|---|---|---|---|---|---|---|---|
| 1 | Diagnosis | 0.66 | .000001 | 204642_at | EDG1 | Endothelial differentiation, sphingolipid G-protein-coupled receptor, 1 | Cell adhesion, signal transduction |
| 2 | Diagnosis | 0.66 | .000002 | 203795_s_at | BCL7A | B-cell CLL/lymphoma 7A | Putative tumor suppressor |
| 3 | Relapse | 0.97 | .000003 | 203554_x_at | PTTG1 | Pituitary tumor-transforming 1 | DNA replication and repair; cell cycle |
| 4 | Relapse | 2.11 | .00001 | 200834_s_at | RPS21 | Ribosomal protein S21 | Protein biosynthesis |
| 5 | Relapse | 2.11 | .000016 | 200763_s_at | RPLP1 | Ribosomal protein, large, P1 | Protein biosynthesis |
| 6 | Relapse | 2.11 | .000017 | 213166_x_at | FLJ14346 | Hypothetical protein FLJ14346 | Unknown |
| 7 | Relapse | 2.11 | .000019 | 201757_at | NDUFS5 | NADH-coenzyme Q reductase | Mitochondrial electron transport |
| 8 | Relapse | 2.11 | .000021 | 200980_s_at | PDHA1 | Pyruvate dehydrogenase (lipoamide) alpha 1 | Glycolysis; protein polyubiquitination |
| 9 | Diagnosis | 2.11 | .000023 | 203796_s_at | BCL7A | B-cell CLL/lymphoma 7A | Putative tumor suppressor |
| 10 | Relapse | 2.11 | .000026 | 203989_x_at | F2R | Coagulation factor II (thrombin) receptor | Cell-cycle regulation, apoptosis |
| 11 | Relapse | 2.11 | .000029 | 200061_s_at | RPS24 | Ribosomal protein S24 | Protein biosynthesis |
| 12 | Relapse | 2.11 | .000029 | 201001_s_at | UBE2V1 | Ubiquitin-conjugating enzyme E2 variant 1 | Cell-cycle progression; DNA repair |
| 13 | Relapse | 2.11 | .000031 | 212834_at | DDX52 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 52 | Nucleic acid binding, helicase activity |
| 14 | Diagnosis | 2.12 | .000034 | 213875_x_at | C6orf62 | Chromosome 6 open reading frame 62 | Unknown |
| 15 | Diagnosis | 2.51 | .000044 | 208892_s_at | DUSP6 | Dual-specificity phosphatase 6 | Cell-cycle progression; DNA repair |
| 16 | Diagnosis | 2.51 | .000048 | 206864_s_at | HRK | Harakiri, BCL2 interacting protein (contains only BH3 domain) | Induction of apoptosis |
| 17 | Relapse | 2.51 | .000049 | 217491_x_at | COX7C | Cytochrome c oxidase subunit VIIc | Generation of precursor metabolites and energy |
| 18 | Relapse | 2.51 | .000052 | 201002_s_at | UBE2V1 | Ubiquitin-conjugating enzyme E2 variant | Cell-cycle progression; DNA repair |
| 19 | Relapse | 2.51 | .000054 | 202279_at | C14orf2 | Chromosome 14 open reading frame 2 | Unknown |
| 20 | Diagnosis | 2.51 | .000057 | 49485_at | PRDM4 | PR domain containing 4 | Transcription regulation |
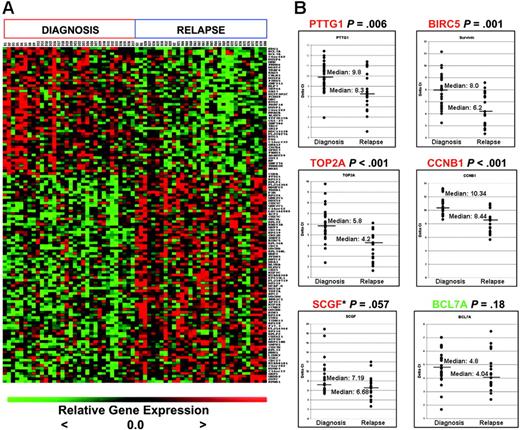
We next validated the expression of a subset of differentially regulated genes that were identified from the matched-pair analyses on an independent sample set which consisted of unmatched marrow samples at initial diagnosis (n = 29) and relapse (n = 19) (Figure 2B) by quantitative real-time PCR. These 19 relapse samples were randomly selected from the cohort of 60 patients treated at relapse in COG AALL01P2 but did not include the relapse samples from the 35 pairs. We selected those genes implicated previously in transformation as well as those that might be suitable targets for future therapeutic modulation, such as BIRC5,16 PTTG1,17,18 TOP2A,19 CCNB1,20 SCGF,21 and BCL7A22 for validation. Although some of these genes were not included in the top 20 gene list, all genes were significantly differentially expressed (P < .005; see gene list in Table S3B). PTTG1, BIRC5, TOP2A, and CCNB1 transcript levels were again shown to be significantly up-regulated at relapse. The P value for SCGF was .057, while the expression of BCL7A (down-regulated at relapse in the paired analysis) did not reach statistical significance. We are unable to discern whether these discrepancies are due to the fact that the independent samples were unmatched.
Genes differentially expressed at diagnosis and relapse in B-precursor ALL. (A) Heatmap of top 126 probe sets (48 high at diagnosis, 78 high at relapse; FDR < 10%). (B) Quantitative real time–PCR (qRT-PCR) validation of selected targets on independent samples (29 initial diagnosis, 19 relapse). The y-axis represents normalized ΔCT values (CT of gene of interest – CT of housekeeping gene). A high ΔCT signifies low expression, and vice versa. CT indicates threshold cycles for amplification. *SCGF was not expressed in 2 of the diagnosis samples; thus, an arbitrary CT value of 40 was used. Median expression is indicated by the horizontal bars.
Genes differentially expressed at diagnosis and relapse in B-precursor ALL. (A) Heatmap of top 126 probe sets (48 high at diagnosis, 78 high at relapse; FDR < 10%). (B) Quantitative real time–PCR (qRT-PCR) validation of selected targets on independent samples (29 initial diagnosis, 19 relapse). The y-axis represents normalized ΔCT values (CT of gene of interest – CT of housekeeping gene). A high ΔCT signifies low expression, and vice versa. CT indicates threshold cycles for amplification. *SCGF was not expressed in 2 of the diagnosis samples; thus, an arbitrary CT value of 40 was used. Median expression is indicated by the horizontal bars.
Intrinsic biologic classes at relapse
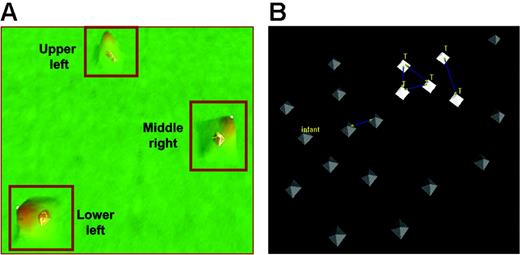
These results indicate that a common pattern of gene expression can be identified in blasts at relapse, especially among samples from patients who relapse early in therapy. To determine further the heterogeneity of blasts at relapse we performed an unsupervised analysis of gene-expression profiles of 60 relapse marrow samples (54 B-precursor ALL and 6 T-ALL) from patients enrolled in COG AALL01P2, a recently completed protocol for children with relapsed ALL. This cohort included 17 relapse samples from the previous cohort of 35 paired samples. Patient samples were clustered into 3 distinct groups: UL (upper left), LL (lower left), and MR (middle right) (Figure 3; Table 3). The MR group was dominated by early-relapse cases compared with the other 2 groups (mean time to relapse, 24.1 months for MR vs 40.9 months [UL] and 43.8 months [LL]). Five of 6 T-ALL cases, all of whom relapsed early, were included in the MR group. In contrast, the UL and LL groups had an equal distribution of early and late relapse sample and average times to relapse. The genes discriminating these 3 groups are included in Table S4.
Characteristics of the patients included in the 3 clusters
Cluster . | No. early/no. late (total) . | Average time to relapse, mo . | No. failures after block 1 therapy . | Average MRD (M1 patients), % . | Phenotype . | Additional information . |
|---|---|---|---|---|---|---|
| Upper left | 9/8 (17) | 40.9 | 1 | 1 | 17 precursor B | — |
| Middle right | 17/3 (20) | 24.1 | 7 | 1.80 | 15 precursor B; 5 T-ALL | 1 infant |
| Lower left | 11/12 (23) | 43.8 | 3 | 0.63 | 22 precursor B; 1 T-ALL | 2 infants; 1 Ph+ infant; 1 Ph+ |
Cluster . | No. early/no. late (total) . | Average time to relapse, mo . | No. failures after block 1 therapy . | Average MRD (M1 patients), % . | Phenotype . | Additional information . |
|---|---|---|---|---|---|---|
| Upper left | 9/8 (17) | 40.9 | 1 | 1 | 17 precursor B | — |
| Middle right | 17/3 (20) | 24.1 | 7 | 1.80 | 15 precursor B; 5 T-ALL | 1 infant |
| Lower left | 11/12 (23) | 43.8 | 3 | 0.63 | 22 precursor B; 1 T-ALL | 2 infants; 1 Ph+ infant; 1 Ph+ |
Most of the patients in the middle right group are early relapses.
— indicates not applicable.
VxInsight analysis for 60 patients at relapse. (A) The VxInsight program located and positioned clusters and used a mountainous terrain metaphor to display the results. Mountains are shown over the clusters such that the height of each mountain represents the number of elements in that cluster. VxInsight defined 3 clusters from the entire cohort of 60 patients. (B) A detailed view of the middle-right cluster shows that all the early-relapse T-cell patients are closely clustered together. Blue lines indicate the strongest similarity links.
VxInsight analysis for 60 patients at relapse. (A) The VxInsight program located and positioned clusters and used a mountainous terrain metaphor to display the results. Mountains are shown over the clusters such that the height of each mountain represents the number of elements in that cluster. VxInsight defined 3 clusters from the entire cohort of 60 patients. (B) A detailed view of the middle-right cluster shows that all the early-relapse T-cell patients are closely clustered together. Blue lines indicate the strongest similarity links.
Gene-expression patterns of early versus late relapse
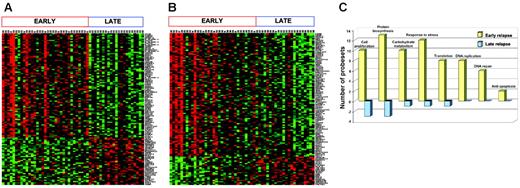
Because the timing of relapse is the most important prognostic factor for the success of retrieval therapy, we identified the genes that best distinguished early (< 36 months from diagnosis) and late (≥ 36 months from diagnosis) B-precursor marrow relapse (n = 54). Using a cutoff of FDR of 2.5% or less, 115 significant genes (79 high in early relapse, 36 high in late relapse) were identified (Figure 4A). The procedure was reapplied to the entire cohort (n = 60; 6 T-cell samples included), and these data are included in Figure S1. The top 100 genes from both lists (eg, B-precursor alone vs B-precursor and T-ALL) were compared, and 74% of the genes were common. Most clinical protocols assign therapy based on the timing of relapse and use 36 months from initial diagnosis as the cut-off for definition. However, since time to relapse is a continuous variable, we also used a linear regression model to fit the time to relapse data, and 118 genes were identified (FDR ≤ 1.5%, 96 positive and 22 negative genes; Figure 4B). Both analyses show that early-relapse samples were characterized by an up-regulation of a number of genes that may confer proliferative and survival advantages to the cell (Figure 4C). Specifically, genes involved in biosynthesis and metabolism, DNA replication/repair, and inhibition of apoptosis were among those groups up-regulated in early versus late relapse. Thus, gene expression analysis on both cohorts of samples (diagnosis/relapse pairs and samples at relapse) shows that early relapse is characterized by the emergence of a highly proliferative clone that is distinct from relapsed clones that are detected at later time points from initial diagnosis.
Discussion
Recurrent ALL remains a major challenge in spite of the dramatically improved survival for newly diagnosed patients.1,4 Further intensification of chemotherapy is unlikely to cure additional patients, and understanding the cellular mechanisms that lead to resistance will result in better therapy and prevention strategies. While many previous studies have examined biologic differences between leukemic blasts at diagnosis and relapse, the current study has important advantages over previous reports. Matched pairs at diagnosis and relapse provide the best opportunity to study emergence of resistance, as each patient acts as his/her own control. Common changes that contribute to drug resistance can thus be uncovered. In previous studies, unlinked cohorts of relapse and new diagnosis samples were examined. An inherent problem with such an approach is the unbalanced assortment of favorable and unfavorable genetic subtypes such that differences noted between the 2 groups may be linked to these differences in the underlying disease subtypes rather than to true resistance pathways. To verify differences in biologic pathways at relapse and to distinguish further differences in relapse mechanisms between patients who relapse early versus those who suffer a recurrence late after initial diagnosis, we also examined a large number of samples from children enrolled in an ongoing study for relapsed ALL. These children had received therapy from current intensive protocols at the time of their initial diagnosis.
Genes differentially expressed between early and late relapse in patients (n = 54) with precursor B-ALL. (A) Patients were classified into 2 groups—early relapse (< 36 months from diagnosis) and late relapse (≥ 36 months)—using binary classification: 114 probe sets were chosen by SAM, with an FDR of 2.5% or less. (B). A linear regression model of survival analysis was used to show the possible association of the gene expression values with time to relapse. A number of probe sets (118) were chosen, with an FDR of 1.5% or less. (C) Functional categorization: 263 probe sets that were differentially expressed between early and late relapse with an FDR less than 5% were grouped according to function using the GeneOntology Biological Process classification. Yellow bars represent the number of genes relatively overexpressed, and blue bars represent genes relatively underexpressed at relapse.
Genes differentially expressed between early and late relapse in patients (n = 54) with precursor B-ALL. (A) Patients were classified into 2 groups—early relapse (< 36 months from diagnosis) and late relapse (≥ 36 months)—using binary classification: 114 probe sets were chosen by SAM, with an FDR of 2.5% or less. (B). A linear regression model of survival analysis was used to show the possible association of the gene expression values with time to relapse. A number of probe sets (118) were chosen, with an FDR of 1.5% or less. (C) Functional categorization: 263 probe sets that were differentially expressed between early and late relapse with an FDR less than 5% were grouped according to function using the GeneOntology Biological Process classification. Yellow bars represent the number of genes relatively overexpressed, and blue bars represent genes relatively underexpressed at relapse.
Many of the genes that we identified by comparing matched diagnosis and relapse samples have been implicated in malignant transformation and/or drug resistance previously. For example, survivin (BIRC5), which is a member of the inhibitor of apoptosis (IAP) family regulating cell division and inhibiting caspase function,16 was up-regulated at relapse. High expression of survivin has been shown to be a marker of poor prognosis, and targeting survivin by antisense oligonucleotides and other methods has been shown to induce apoptosis in cell lines and suppress tumor growth in xenograft models.23 In another study, survivin and cyclinB1 (CCNB1), another one of the genes that is up-regulated at relapse, were included in a panel of a 16-gene recurrence score that could predict recurrence in breast cancer.24
Genes involved in cell proliferation, protein biosynthesis, carbohydrate metabolism, and DNA replication/repair were among those highly expressed in relapsed versus newly diagnosed blasts. Topoisomerase II alpha (TOP2A) encodes an enzyme that controls and alters the topologic state of DNA during transcription. It is the target for the topoisomerase II inhibitor chemotherapeutic agents and TOP2A expression has been shown to be associated with increased proliferation and poor outcome in a variety of tumors.19,25 Pituitary tumor transforming gene 1, or securin, encodes a p53-interacting protein. The interaction prevents the binding of p53 to DNA inhibiting its transcriptional activity. Securin also inhibits the ability of p53 to induce cell death.17 Thus, the oncogenic action of increased expression of securin may make it an attractive target for therapeutic intervention.
A multitude of growth factors orchestrate stem-cell self-renewal and differentiation. Stem-cell growth factor (SCGF) is a member of the C-type lectin superfamily and is a novel human growth factor that supports growth of primitive hematopoietic progenitor cells.21,26 SCGF was another gene which was significantly overexpressed at relapse. Some of the genes down-regulated at relapse compared with initial diagnosis included proapoptotic genes (Harakiri), antiproliferative genes (BTG1 and BTG2) and a putative tumor suppressor (BCL7A). However, using our independent group of samples we could not verify differential expression of BCL7A. Whether this disparity is the result of true false-positive associations using array technology or because our independent sample set was not matched remains to be determined. Like other microarray databases, further validation of the gene sets described in this study will be needed before investigators focus on individual pathways to understand better resistance mechanisms and importantly before therapeutic strategies are developed.
In contrast to previous reports we did not observe increased expression of cyclin D127 and dihydrofolate reductase (DHFR),28 or decreased expression of the reduced folate carrier (RFC),29 BAX,30 and p16INK4A at relapse.31,32 Beesley et al33 recently reported a transcriptional profile of relapsed ALL in 15 matched pairs (11 B-precursor and 4 T-ALL patients). GRP58, the gene that was ranked second in their analysis, was also significant in our pairs (P = .034). We were unable to identify other genes from their top 20 to be statistically significant in our analysis. The reason for the poor concordance between the 2 studies may be due to the small number of patients and the fact that their pairs were generated from samples from children diagnosed as far back as 1984, an era when children were treated with significantly less therapy. In addition, the most significant changes we identified at relapse were noted in patients who relapsed early, and their distribution of cases was not reported. Our pairwise analysis was designed to identify common changes that contribute to drug resistance regardless of biologic subgroup. Although our cohort included a representative mix of cytogenetic subgroups, the number of individual samples in each category is too small to determine if there are additional changes specific to certain subgroups.
The most notable finding of our analysis was that relapse blasts express many genes involved in cell proliferation, a result that is in agreement with previous reports.33,34 Most of the targets that were identified in a pairwise analysis of all 35 patients were contributed by the early-relapse pairs (subgroup analysis), but were not identified in the late-relapse pairs. The distinct differences in early versus late relapse were also underscored by our direct comparison of early- versus late-relapse samples from patients enrolled in COG AALL01P2, where early relapse samples again showed much greater representation of genes involved in proliferation, cell-cycle control, and cellular metabolism. Unsupervised analysis of matched pairs showed that a subset of relapsed samples had a gene-expression profile that was significantly distinct from their respective initial diagnostic sample; this was generally true in the late-relapse group. Although these analyses are limited by the smaller number of samples in the late-relapse cohort, these findings are suggestive of a model whereby late relapse is due to the acquisition of diverse secondary events that might occur in a distinct subpopulation such as a leukemic stem cell. Direct evidence for this model has come from the study of relapsed TEL-AML1+ ALL samples,35 where the analysis of deletions at the nontranslocated TEL allele shows that the relapsed clone is related, but distinct from the clone at initial diagnosis. It has been hypothesized that relapse in essence represents a de novo ALL originating from a preleukemic stem cell. Our data suggest that this general model may be operative in non–TEL-AML1 subtypes of late ALL relapse as well. In contrast, early-relapse mechanisms appear to be more homogeneous and are suggestive of the selection of a resistant, more proliferative clone. While blasts in cell cycle might be more sensitive to chemotherapeutic agents that target steps in DNA metabolism and cell division, the overexpression of DNA repair (eg, PTTG1, RAD51, and POLE2) and antiapoptotic genes (eg, BIRC5, AATF, API5, and AVEN) might overcome the alterations induced by treatment. The common gene expression signature among early-relapse patients provides a list of targets for novel therapeutic and preventive strategies. In contrast, the diverse nature of later relapse may be more challenging to address using a common strategy.
In summary, we have identified potential genes and pathways at relapse that may play a direct role in drug resistance in childhood ALL and offer insight into the clinical differences that are observed among patients based on the timing of disease recurrence. Our findings support a model whereby the mechanisms of relapse differ for early compared with late disease recurrence. Further preclinical validation of the functional role of some of these genes will contribute to rational efforts to treat and prevent tumor recurrence.
Prepublished online as Blood First Edition Paper, March 28, 2006; DOI 10.1182/blood-2006-02-002824.
Supported by National Cancer Institute (NCI) SPEC U01 CA114762, Director's Challenge Grant U01 CA88361, The Penelope London Foundation, The Friedman Fund for Childhood Leukemia, The Walter Family Pediatric Leukemia Fund, The Triple C Foundation, The Pediatric Cancer Foundation, and the Garrett B. Smith Foundation.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We acknowledge the Microarray Shared Research Facility at the Mount Sinai School of Medicine, New York.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal