Abstract
Plasmacytoid dendritic cells (pDCs) are specialized DCs that produce high levels of type I IFN upon viral infection. Despite their key immunoregulatory role, little is known about pDC ontogeny or how developmental events regulate their function. We show that mice expressing low levels of the transcription factor Ikaros (IkL/L) lack peripheral pDCs, but not other DC subsets. Loss of pDCs is associated with an inability to produce type I IFN after challenge with Toll-like receptor-7 and -9 ligands, or murine cytomegalovirus (MCMV) infection. In contrast, conventional DCs are present in normal numbers and exhibit normal responses in vivo after challenge with MCMV or inactivated toxoplasma antigen. Interestingly, IkL/L bone marrow (BM) cells contain a pDC population that appears blocked at the Ly-49Q– stage of differentiation and fails to terminally differentiate in response to Flt-3L, a cytokine required for pDC differentiation. This differentiation block is strictly dependent on a cell-intrinsic requirement for Ikaros in pDC-committed precursors. Global gene expression profiling of IkL/L BM pDCs reveals an up-regulation of genes not normally expressed, or expressed at low levels, in WT pDCs. These studies suggest that Ikaros controls pDC differentiation by silencing a large array of genes.
Introduction
Plasmacytoid dendritic cells (pDCs) are a unique DC subset. pDCs produce large quantities of type I IFN (α, β,or ω) in response to viral infection and bacterial components,1-5 and thus are considered key cells for the immune response to these pathogens. Human and mouse pDCs have been implicated in the activation of natural killer (NK) cells, the differentiation of regulatory T cells and plasma cells, as well as in the polarization of naive T cells into Th1/Th2 responders.6-14 These pleiotropic roles highlight the importance of pDCs in the control of T-cell tolerance, graft-versus-host disease, allergic responses, and lupus erythematosus.15-20 Despite numerous studies, however, pDC function in vivo remains incompletely understood.
Similarly, little is known about the specific signals and factors involved in pDC differentiation.21 Flt-3 signaling has been described as a crucial component of the pDC developmental program.22-24 Recent studies have shown that pDCs develop from Flt-3+ cells within the common lymphoid or common myeloid progenitor populations in mouse bone marrow.25,26 Since many pDCs express some lymphoid-related gene products (such as pTα, V-preB, and EBF) and harbor D-J rearrangements, it has been proposed that pDCs arise from lymphoid progenitors, and are possibly related to B cells. Other pDC subtypes, however, appear to lack lymphoid-associated attributes.27 Thus, pDC development appears complex, as it may be associated with considerable plasticity and as immature pDCs can arise from independent pathways that converge to produce the mature pDC subset.
Several transcription factors have been identified that regulate the development of distinct DC types.28-31 Among these is Ikaros, a zinc finger protein essential for the development of multiple hematopoietic lineages.32-36 Ikaros functions mainly as a repressor.37-39 It binds DNA as homodimers or heterodimers with other members of the Ikaros family such as Aiolos, expressed mainly in B and T cells, and Helios, expressed in early hematopoietic precursors and T cells.40-43 Expression of a dominant-negative form of Ikaros results in a complete loss of all conventional DC (cDC) subsets, while a null mutation in Ikaros leads to the selective loss of CD11c+CD11b+ but not CD11c+CD8α+ cDCs.30 Moreover, human CD34+ BM cells expressing a dominant-negative Ikaros protein can generate DCs from myeloid (CD10–) but not lymphoid (CD10+) progenitors,44 providing further evidence that Ikaros is an important regulator of early DC development. Whether pDCs are affected by Ikaros deficiency is unknown.
Our laboratory has described a mutant mouse line carrying a hypomorphic mutation in the Ikaros locus (IkL/L) in which a β-galactosidase reporter gene was inserted into the 5′ part of the Ikaros locus (exon 3) by homologous recombination in ES cells.35 IkL/L mice express low levels of functional Ikaros protein in their hematopoietic cells. Here we report that IkL/L mice selectively lack pDCs, but not cDCs. Loss of the pDC population results in an inability of these mice to produce type I IFN and control viral replication. Further, we find that pDC development is engaged but incomplete in the BM of IkL/L mice, suggesting that Ikaros plays a key role in the generation of peripheral pDCs from a pDC-committed BM precursor population. Lastly, we show that Ikaros represses the expression of many genes in these pDC precursors. Together, our results show that Ikaros is critical for pDC development and function, and identify the IkL/L mouse line as a potentially valuable in vivo model to study host-pathogen interactions in the absence of pDCs.
Materials and methods
Mice
The IkL/L mouse line was previously described.35 Mice (6-10 weeks old) used in this study were maintained under specific pathogen-free (SPF) conditions, and were backcrossed more than 7 generations onto the C57Bl/6 genetic background. Most experiments used mice from a 10th backcross generation. All gave similar results. B6.Ly5SJL congenics were also maintained under SPF conditions.
Antibodies and flow cytometry
All antibodies were from BD Pharmingen (San Diego, CA) or eBiosciences (San Diego, CA) except for biotin-conjugated Ly-6C (clone ER-MP20; BMA Biomedicals, Augst, Switzerland) and anti–Ly-49Q (MBL, Watertown, MA). Alexa488-conjugated 120G8 was described previously.45 CD11c (N418), CD11b (M1/70), and B220 (RA3-6B2) were purified and conjugated to FITC, PE, allophycocyanin (APC), or biotin according to standard protocols. For fluorescence-activated cell sorting (FACS) analyses, cells were first incubated with anti-CD16/32 to block Fc receptors. Intracellular IL-12 was detected according to Dalod et al.46 Cells were analyzed on a FACSCalibur or LSR II (BD BioSciences, San Jose, CA). Results were analyzed with the FlowJo software (TreeStar, Ashland, OR). Sorting was performed on a FACSVantage SE option DiVa (BD BioSciences).
Cell preparations
Organs were digested by collagenase (liberase CI; Boehringer Mannheim, Mannheim, Germany) as described.47 Cell suspensions were treated with 0.165 MNH4Cl to lyse red blood cells (RBCs). For experiments requiring DC enrichment from the spleen, cells were incubated with anti-CD3 (17A2) and anti-CD19, followed by goat anti–rat IgG–coated Dynabeads (Dynal, Lake Success, NY), and depleted with a Dynal magnet. Depleted cells were then positively purified for CD11c+ cells using CD11c+ Microbeads and MiniMacs (Miltenyi Biotec, Auburn, CA). Purity was more than 95%. For enrichment from the BM, WT cells were depleted of CD19+ cells by the Dynal method, then purified with the indicated antibodies by FACS. IkL/L BM cells were not depleted but were similarly purified. Sort purity was more than 95%.
In vitro stimulation
Cells were cultured in RPMI 1640 (Life Technologies, Bethesda, MD), 10% FCS (Life Technologies), 2 mM l-glutamine, 10 mM HEPES, 50 μM 2-ME, and 80 μg/mL gentamycin. Cells were stimulated at 105 cells/well in 200 μL in 96-well plates. Poly I:C (Invitrogen, Frederick, MD) was used at 20 μg/mL final concentration. The formaldehyde-inactivated human influenza virus, strain NK/TM/138/00 (a kind gift from N. Kuehn, Aventis Pasteur, France), was used at 100 hemagglutinin U/mL. CpG D19 (GGT GCA TCG ATG CAG GGG GG) was phosphorothioate-modified (MWG Biotec, High Point, NC) and used at 10 μg/mL. Murine recombinant Flt-3L (25 ng/mL; R&D Systems, Minneapolis, MN) was used as previously described.23 Supernatants were collected after 24 hours of culture and tested for IFNα by specific enzyme-linked immunosorbent assay (ELISA; PBL Biomedical Labs, Piscataway, NJ), and tested for IL-12p40 or IL-12p70 using IL-12 p70/p40 Duoset ELISA or IL-12 p70-specific Duoset ELISA (both from R&D Systems), respectively.
In vivo treatments
Mice were anesthetized and injected intravenously in the retro-orbital vein with 200 μL CpG 1668 (TCC ATG ACG TTC CTG ATG CT), prepared as described.2 R848 (Invivogen, San Diego, CA) was used at 5 μg/mouse in PBS. Poly I:C (Invitrogen) was used at 50 μg/mouse in PBS. Control mice were injected with PBS. Sera were analyzed using the ELISAs described for in vitro stimulation. Infections were initiated with 2 × 104 to 5 × 104 plaque-forming units of a salivary gland–extracted GFP-recombinant murine cytomegalovirus (MCMV) Smith strain (RVG-102) injected intraperitoneally on day 0. Control mice were injected with the medium in which the virus stocks were diluted. At 1.5 days after challenge, frequencies of splenic pDCs, MCMV-infected DCs, and IL-12+ DCs were analyzed as described. IFNγ was measured using the IFNγ-specific Duoset ELISA (R&D Systems).
BM chimeras
B6.Ly5SJL congenics were irradiated (9 gray) 24 hours before reconstitution. WT (2 × 106) or IkL/L (5 × 106) BM cells, both expressing the Ly5B6 allele, were harvested from donor mice and injected intravenously into the recipients. For the double chimera experiments, WT or IkL/L BM cells were mixed with freshly-isolated, nonirradiated B6.Ly5SJL cells before injection, at a 1:1 ratio for WT cells (1 × 106:1 × 106) and a 5:1 ratio for IkL/L cells (5 × 106:1 × 106). Ly5B6 cells were analyzed 6 to 10 weeks after transfer using an anti-Ly5B6 (Ly5.2) allele-specific antibody (clone 104). Reconstitution efficiency ranged between 30% and 50% for IkL/L and between 50% and 70% for WT donor cells.
RT-PCR
Total RNA was extracted from sorted populations (2 × 105 cells) using Trizol (Invitrogen) and reverse transcribed using AMV reverse transcriptase. Reversetranscription–polymerase chain reaction (RT-PCR) was performed with cDNA from the equivalent of 2 × 104 cells with the following primers: β-actin, 5′-GTGACGAGGCCCAGAGCAAGAG, 5′-AGGGGCCGGACTCATCGTACTC; Ikaros, 5′-GGAGGCACAAGTCTGTTGAT, 5′-GTTGGCACTGTCATAGGGCA; Aiolos, 5′-ATCGAAGCAGTGCCGCTTCTCACC, 5′-GTGTGCGGGTTATCCTGCATTAGC; and Helios, 5′-TGGAAACAGACGCAATTGAT, 5′-CATGGCAACCCATGTGAAT. Thirty-five cycles were used for Ikaros, Aiolos, and Helios, and 28 for β-actin.
Microarray experiments and analysis
Three WT and 3 IkL/L samples of 2 × 105 CD11c+120G8+ BM cells were sorted and RNA extracted with the Qiagen micro RNAeasy kit (Valencia, CA), yielding approximately 30 ng total RNA for each sample. Quality and absence of genomic DNA contamination was assessed with a Bioanalyser (Agilent, Palo Alto, CA). Probes were synthesized using 2 successive rounds of cRNA amplification, according to standard Affymetrix protocols, and hybridized to mouse 430 2.0 chips (Affymetrix, Santa Clara, CA). Raw data were transformed with the Mas5 algorithm, which yields a normalized expression value, and “absent” and “present” calls. Target intensity was set to 100 for all chips. Using Mas5, we performed all possible pairwise analyses, which yield “increased” (I), “decreased” (D), “marginally increased,” “marginally decreased,” or “not changed” calls, as well as fold change values. To select probe sets specifically increased or decreased in IkL/L samples compared with WT, we imposed that an I or D call be met by all possible IkL/L/WT pairwise comparisons. To evaluate the number of changes that might be due to experimental or biologic noise, we performed similar analyses, comparing “nonspecific” groups of 3 samples in which given WT and mutant samples had been permuted between groups (eg, comparing a first group containing 2 WT and 1 IkL/L samples with a second group containing 2 IkL/L and 1 WT samples). All 9 possible nonspecific combinations were analyzed. In 6 combinations, no probe sets were increased or decreased in all pairwise comparisons. In 3 combinations, fewer than 15 genes exhibited variations.
IkL/L mice show selective loss of pDCs. (A) Splenocytes from the indicated mice were stained with Alexa488 anti-120G8; PE anti-CD3, CD19, and NK1.1; APC anti-CD11c; PE-Cy7 anti-CD11b; APC-Cy5.5 anti-B220; and biotin anti-CD8α. Anti-CD8α antibodies were revealed with SA-PE-TexasRed. The resulting cells were analyzed on an LSR II flow cytometer by collecting 200 000 events for each file. Nonviable cells were eliminated from subsequent analyses by staining cells with DAPI. Numbers indicate percentage of events within the indicated gate as a function of the corresponding parent gate for each plot. The absolute number of events within each CD11c+ (CD3– CD19– NK1.1–) gate are as follows: WT, 889; Ik+/L, 795; and IkL/L, 1105. (B) Lymph node cells from the identical mice were stained and analyzed as in panel A. The absolute number of events within each CD11c+ (CD3– CD19– NK1.1–) gate are as follows: WT, 237; Ik+/L, 241; and IkL/L, 164. Absolute numbers of DCs within each subset in 3 mice per group are given in Table 1. These data are representative of 4 experiments.
IkL/L mice show selective loss of pDCs. (A) Splenocytes from the indicated mice were stained with Alexa488 anti-120G8; PE anti-CD3, CD19, and NK1.1; APC anti-CD11c; PE-Cy7 anti-CD11b; APC-Cy5.5 anti-B220; and biotin anti-CD8α. Anti-CD8α antibodies were revealed with SA-PE-TexasRed. The resulting cells were analyzed on an LSR II flow cytometer by collecting 200 000 events for each file. Nonviable cells were eliminated from subsequent analyses by staining cells with DAPI. Numbers indicate percentage of events within the indicated gate as a function of the corresponding parent gate for each plot. The absolute number of events within each CD11c+ (CD3– CD19– NK1.1–) gate are as follows: WT, 889; Ik+/L, 795; and IkL/L, 1105. (B) Lymph node cells from the identical mice were stained and analyzed as in panel A. The absolute number of events within each CD11c+ (CD3– CD19– NK1.1–) gate are as follows: WT, 237; Ik+/L, 241; and IkL/L, 164. Absolute numbers of DCs within each subset in 3 mice per group are given in Table 1. These data are representative of 4 experiments.
Results
Selective loss of peripheral pDCs in mice with reduced Ikaros activity
As DC development has been shown to be affected upon loss of Ikaros activity,30,44 we examined distinct DC subpopulations in the spleens of WT, heterozygote Ik+/L, and homozygote IkL/L mice, using a combination of antibodies including the pDC-specific antibody, 120G8.45 WT, Ik+/L, and IkL/L spleens contained similar proportions of “myeloid” (CD11c+CD11b+) and “lymphoid” (CD11c+CD8α+) cDC populations (Figure 1A). In striking contrast, the pDC population (CD11c+120G8+B220+) was significantly reduced in the spleens of Ik+/L animals and absent in those of IkL/L mice. Similar results were obtained in the lymph node and peripheral blood (Figure 1B, and not shown). A detailed account of pDC and cDC (myeloid and lymphoid) frequencies in collagenase-treated WT and IkL/L organs is provided in Table 1. These data indicate that peripheral pDCs are severely diminished in mice deficient for Ikaros, and that this phenotype is dose dependent on Ikaros.
Impact of lkL/L mutation on peripheral dendritic cell subsets
. | Spleen . | . | Lymph node† . | . | ||
|---|---|---|---|---|---|---|
| Subset*genotype . | %‡ . | No. of cells, × 10-4 . | %‡ . | No. of cells, × 10-3 . | ||
| pDC | ||||||
| WT | 0.29 (0.04) | 25.7 (0.48) | 0.08 (0.009) | 3.41 (0.40) | ||
| IkL/+ | 0.08 (0.01) | 6.7 (1.0) | 0.04 (0.006) | 2.05 (0.27) | ||
| IkL/L | 0.002 (0.003) | 0.11 (0.19) | 0.001 (0.002) | 0.041 (0.081) | ||
| CD8α+ | ||||||
| WT | 0.43 (0.21) | 37.9 (1.8) | 0.07 (0.01) | 2.78 (0.45) | ||
| IkL/+ | 0.34 (0.02) | 28.2 (0.15) | 0.07 (0.005) | 3.08 (0.37) | ||
| IkL/L | 0.60 (0.17) | 41.7 (9.9) | 0.08 (0.01) | 3.09 (0.43) | ||
| CD11b+ | ||||||
| WT | 0.54 (0.20) | 52.0 (13.1) | 0.09 (0.01) | 3.68 (0.36) | ||
| IkL/+ | 0.46 (0.04) | 37.8 (3.4) | 0.09 (0.01) | 3.98 (0.16) | ||
| IkL/L | 0.66 (0.10) | 45.8 (3.4) | 0.10 (0.01) | 3.36 (0.31) | ||
. | Spleen . | . | Lymph node† . | . | ||
|---|---|---|---|---|---|---|
| Subset*genotype . | %‡ . | No. of cells, × 10-4 . | %‡ . | No. of cells, × 10-3 . | ||
| pDC | ||||||
| WT | 0.29 (0.04) | 25.7 (0.48) | 0.08 (0.009) | 3.41 (0.40) | ||
| IkL/+ | 0.08 (0.01) | 6.7 (1.0) | 0.04 (0.006) | 2.05 (0.27) | ||
| IkL/L | 0.002 (0.003) | 0.11 (0.19) | 0.001 (0.002) | 0.041 (0.081) | ||
| CD8α+ | ||||||
| WT | 0.43 (0.21) | 37.9 (1.8) | 0.07 (0.01) | 2.78 (0.45) | ||
| IkL/+ | 0.34 (0.02) | 28.2 (0.15) | 0.07 (0.005) | 3.08 (0.37) | ||
| IkL/L | 0.60 (0.17) | 41.7 (9.9) | 0.08 (0.01) | 3.09 (0.43) | ||
| CD11b+ | ||||||
| WT | 0.54 (0.20) | 52.0 (13.1) | 0.09 (0.01) | 3.68 (0.36) | ||
| IkL/+ | 0.46 (0.04) | 37.8 (3.4) | 0.09 (0.01) | 3.98 (0.16) | ||
| IkL/L | 0.66 (0.10) | 45.8 (3.4) | 0.10 (0.01) | 3.36 (0.31) | ||
Mean absolute cell numbers derived from 3 mice were calculated by multiplying the total cell numbers by the percent cells within each population. Standard deviations are indicated in parentheses.
DC subsets were defined as illustrated in Figure 1.
Data are derived from a single axillary lymph node from each mouse following liberase treatment.
Mean percent of gated population of three 7-week-old mice per group. Standard deviations are indicated with parentheses.
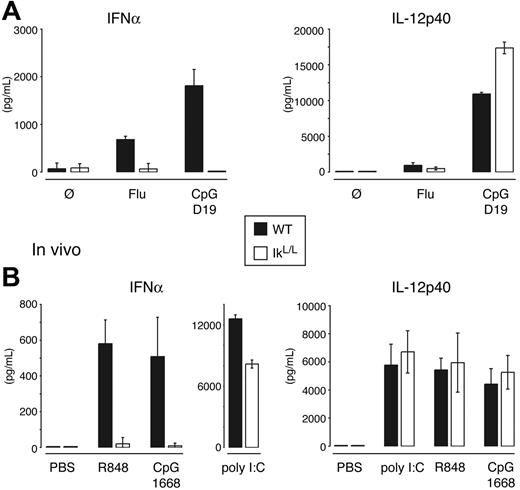
To determine if pDCs are functionally lacking in these animals, we used both in vitro and in vivo assays to test the capacity of IkL/L mice to produce IFNα in response to stimuli known to activate pDCs through the triggering of Toll-like receptors (TLRs). The TLR ligands included influenza virus, TLR9-binding CpG oligonucleotides, and the TLR7 ligand R848, all of which can also induce both pDCs and cDCs to produce IL-12.1,48 As shown in Figure 2A, when CD11c+ WT and IkL/L spleen cells were cultured with influenza virus or CpG D19, WT cells produced IFNα but IkL/L cells did not. Likewise, injection of CpG 1668 or R848 induced IFNα production in WT but not mutant animals (Figure 2B). In contrast, normal IL-12 production was observed in all conditions, suggesting that TLR and/or cDC responses were not grossly altered in IkL/L mice. The in vivo IFNα response to polyinosinic: polycytidylic acid (poly I:C), which activates a variety of cell types through TLR3,49 was also unaffected in IkL/L mice. Thus, IkL/L mice specifically lack pDCs, but not cDCs, both at the phenotypic and functional levels.
IkL/L mice do not produce IFNα upon stimulation with influenza virus or TLR ligands. (A) Purified CD11c+ spleen cells were stimulated with inactivated influenza virus (Flu) or CpG D19 for 24 hours, and supernatants were tested for the indicated cytokines. Results represent 1 of 2 experiments, on cells pooled from 5 mice/genotype per experiment. (B) Mice were injected with poly I:C, R848, or CpG 1668. Sera were collected at 2 hours (R848) and 4 hours (PBS, poly I:C, CpG) after injection and tested for the indicated cytokines. Results are representative of 2 experiments, with 3 mice/genotype per condition for each experiment. Means and SDs are shown for 1 representative experiment.
IkL/L mice do not produce IFNα upon stimulation with influenza virus or TLR ligands. (A) Purified CD11c+ spleen cells were stimulated with inactivated influenza virus (Flu) or CpG D19 for 24 hours, and supernatants were tested for the indicated cytokines. Results represent 1 of 2 experiments, on cells pooled from 5 mice/genotype per experiment. (B) Mice were injected with poly I:C, R848, or CpG 1668. Sera were collected at 2 hours (R848) and 4 hours (PBS, poly I:C, CpG) after injection and tested for the indicated cytokines. Results are representative of 2 experiments, with 3 mice/genotype per condition for each experiment. Means and SDs are shown for 1 representative experiment.
Impaired pDC response to MCMV infection
The lack of a peripheral pDC compartment implies that IkL/L mice may be susceptible to systemic infections. As pDCs have been reported to be the major IFNαβ producer during murine cytomegalovirus (MCMV) infection,46 we tested the physiological relevance of pDC deficiency by evaluating the capacity of IkL/L mice to mount early innate immune responses to MCMV. Animals were challenged with GFP-recombinant MCMV, and titers of IFNα, IL-12, and IFNγ were measured at 1.5 days, the peak of the in vivo response.50 The intensity of viral replication was also evaluated by measuring GFP expression in splenic leukocytes.
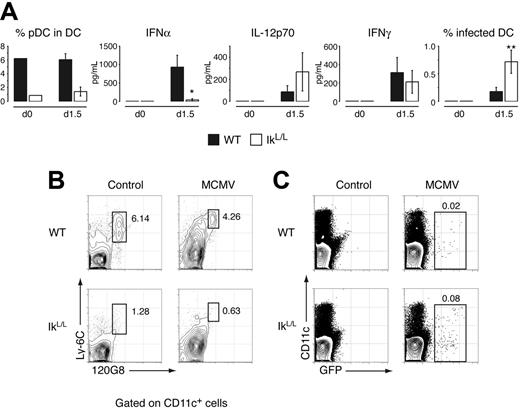
While WT mice produced robust levels of IFNα upon infection, IFNα levels were strikingly lower (> 10-fold less) in the IkL/L serum (Figure 3A), spleen homogenate, and ex vivo–conditioned media of CD11c-enriched DCs (not shown). Furthermore, infected IkL/L mice continued to lack peripheral pDCs (Figure 3A), indicating that MCMV infection and its associated acute inflammatory context could not restore a peripheral pDC compartment in these animals. IkL/L spleen cells contained a larger proportion of GFP-positive cells following MCMV challenge (Figure 3C), particularly within the CD11c+ population (Figure 3A), suggesting that IkL/L animals were also less efficient at controlling viral replication. This overall response may be due to the lack of pDCs and/or compromised function in other leukocytes, as Ikaros is expressed in all hematopoietic cells. However, serum IL-12 and IFNγ titers were not significantly different between IkL/L and WT animals (Figure 3A), suggesting normal antiviral responses from cDCs and NK cells, innate cell types known to be involved in the early defense against MCMV. Together, these data confirm that pDCs are major IFNα/β producers and contribute to an efficient early response to MCMV infection.
Antiviral response to MCMV infection. (A) Animals were injected intraperitoneally with GFP-recombinant MCMV. At 1.5 days after challenge, the frequencies of pDCs and MCMV-infected DCs were measured in the spleen, and cytokines were titrated in the serum. *P = .005; **P = .001. (B) Distribution of pDCs (Ly-6C+ 120G8+) in WT and IkL/L spleens following infection. Contour plots are gated on CD11c+ cells. Numbers indicate percentage of gated cells in each plot. (C) Proportion of GFP+ WT and IkL/L spleen cells after infection. Results shown are representative of 5 experiments, each with 3 to 12 mice per group. Means and SDs are shown for 1 representative experiment.
Antiviral response to MCMV infection. (A) Animals were injected intraperitoneally with GFP-recombinant MCMV. At 1.5 days after challenge, the frequencies of pDCs and MCMV-infected DCs were measured in the spleen, and cytokines were titrated in the serum. *P = .005; **P = .001. (B) Distribution of pDCs (Ly-6C+ 120G8+) in WT and IkL/L spleens following infection. Contour plots are gated on CD11c+ cells. Numbers indicate percentage of gated cells in each plot. (C) Proportion of GFP+ WT and IkL/L spleen cells after infection. Results shown are representative of 5 experiments, each with 3 to 12 mice per group. Means and SDs are shown for 1 representative experiment.
In vivo IkL/L cDC responses are normal
To further confirm that IkL/L cDCs function normally in vivo, we tested their capacity to produce IL-12 and up-regulate maturation markers in 2 systems—MCMV infection and stimulation with Toxoplasma gondii soluble tachyzoite antigen (STAg).51 MCMV infection induces both CD8α+ and CD11b+ cDCs to produce IL-12 through TLR9 triggering.52 STAg challenge specifically induces CD8α+ cDCs to make IL-12 via TLR11 activation.53 As shown in Figure S1 (available at the Blood website; see the Supplemental Materials link at the top of the online article), the frequencies of IL-12–producing cells within the cDC subsets were similar between IkL/L and WT mice, either 1.5 days after MCMV infection (Figure S1A) or 6 hours after stimulation with STAg (Figure S1B). Furthermore, all cDCs up-regulated CD86 and MHC class II expression in both conditions. Thus, IkL/L cDCs behaved normally in 2 experimental situations, involving distinct TLRs on different cDC subsets. These data further demonstrate that TLR signaling is unaffected and that cDCs are phenotypically and functionally normal in IkL/L mice.
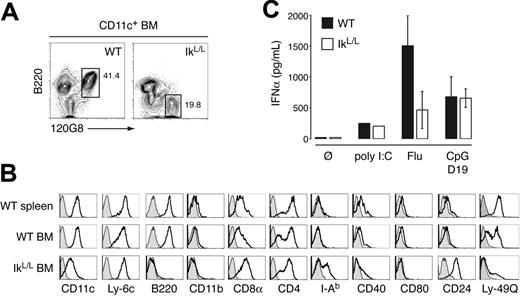
A pDC population in the BM of IkL/Lmice. (A) CD11c+ BM cells were gated and analyzed for B220 and 120G8 expression. Numbers indicate percentage of cells in the gated contour plots. (B) 120G8+ spleen and BM cells were analyzed for their expression of the indicated surface antigens. The pDC population is indicated by the white histograms. Gray histograms indicate the isotype controls. Results are representative of 3 experiments. (C) Sorted CD11c+ 120G8+ BM cells were cultured with poly I:C, inactivated influenza virus, or CpG D19 for 24 hours, and supernatants were tested for the indicated cytokines. Results are representative of 2 experiments, on pools of cells from 5 mice/genotype per experiment. Means and SDs are shown for 1 representative experiment.
A pDC population in the BM of IkL/Lmice. (A) CD11c+ BM cells were gated and analyzed for B220 and 120G8 expression. Numbers indicate percentage of cells in the gated contour plots. (B) 120G8+ spleen and BM cells were analyzed for their expression of the indicated surface antigens. The pDC population is indicated by the white histograms. Gray histograms indicate the isotype controls. Results are representative of 3 experiments. (C) Sorted CD11c+ 120G8+ BM cells were cultured with poly I:C, inactivated influenza virus, or CpG D19 for 24 hours, and supernatants were tested for the indicated cytokines. Results are representative of 2 experiments, on pools of cells from 5 mice/genotype per experiment. Means and SDs are shown for 1 representative experiment.
A pDC population in the IkL/L bone marrow
As pDC development normally occurs in the BM, we examined the IkL/L BM for pDC differentiation. WT BM contained a population of CD11c+B220+Ly-6C+120G8+ cells that was similar in phenotype and frequency to splenic pDCs, although B220 expression was more heterogeneous in the BM population (Figure 4A; Table 1). Surprisingly, 120G8+ cells were also detected in similar numbers in the IkL/L BM; these cells expressed low levels of CD11c and were positive for Ly-6C, but negative for B220. Loss of the B220 marker was not a general phenomenon in IkL/L mice, as it is expressed at normal levels by IkL/L B cells.35 We compared the expression of other pDC-related markers on IkL/L BM 120G8+ cells and WT BM and splenic pDCs (Figure 4B). Notably, a larger proportion of IkL/L 120G8+ cells expressed high levels of CD8α, CD4, CD24, and MHC class II, but were mostly negative for CD40, CD80, and CD86 (Figure 4B and not shown). Interestingly, Ly-49Q expression was also altered in IkL/L 120G8+ cells. Ly-49Q, an ITIM-bearing inhibitory receptor, is highly expressed by WT splenic pDCs and can be used to discriminate 2 pDC populations in the WT BM, where Ly-49Q– cells appear more immature and may represent the immediate precursors of the Ly-49Q+ population (Figure 4B).54-56 IkL/L BM 120G8+ cells were mostly negative for Ly-49Q, suggesting a block in pDC differentiation at an immature stage. Finally, both WT and IkL/L BM 120G8+ cells exhibited plasmacytoid morphology, as determined by May-Grünwald/Giemsa staining and transmission electron microscopy (not shown).
A hallmark of pDC identity is the capacity to produce IFNα. We therefore tested the ability of IkL/L BM 120G8+ cells to produce IFNα upon activation with influenza virus and CpG D19. IkL/L 120G8+ cells produced IFNα in both cases (Figure 4C), while CD11c+CD11b+ BM cells from either genotype did not (not shown). Interestingly, CpG D19 induced WT levels of IFNα from IkL/L cells, while the response to influenza virus was suboptimal, indicating that the mutant cells respond more fully to some stimuli than others. These data provide strong functional evidence that the 120G8+ cells in the IkL/L BM are pDC-lineage cells.
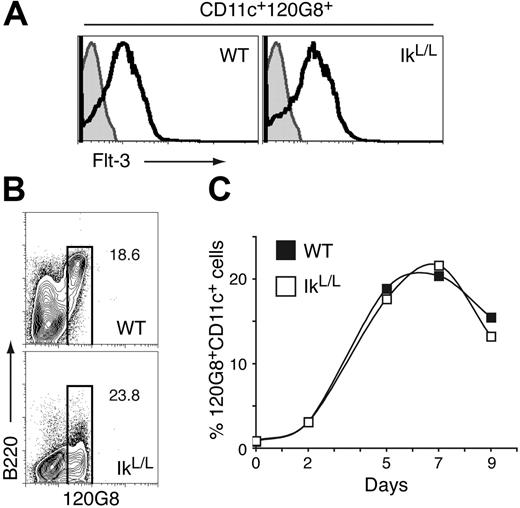
IkL/L pDC precursors respond to Flt-3 signaling. (A) 120G8+ BM cells were analyzed for Flt-3 expression. The pDC population is indicated by the white histograms. Gray histograms indicate the isotype controls. Results are representative of 5 experiments. (B-C) Unfractionated BM cells were cultured with Flt-3L over a 9-day period. (B) Contour plots show B220 and 120G8 expression of CD11c+ cells after 7 days of culture. (C) Graph shows the percentage of 120G8+ cells among total cells for the indicated number of days. One representative experiment of 5 is shown.
IkL/L pDC precursors respond to Flt-3 signaling. (A) 120G8+ BM cells were analyzed for Flt-3 expression. The pDC population is indicated by the white histograms. Gray histograms indicate the isotype controls. Results are representative of 5 experiments. (B-C) Unfractionated BM cells were cultured with Flt-3L over a 9-day period. (B) Contour plots show B220 and 120G8 expression of CD11c+ cells after 7 days of culture. (C) Graph shows the percentage of 120G8+ cells among total cells for the indicated number of days. One representative experiment of 5 is shown.
BM pDC development can be enhanced by Flt-3/Flt-3 ligand (Flt-3L) interactions,22-24 and Ikaros appears to be required for Flt-3 expression at the mRNA level in hematopoietic progenitors.34 We therefore asked if IkL/L BM 120G8+ cells were blocked in their differentiation due to a loss of Flt-3 signaling. To address this issue, we analyzed Flt-3 expression, and responsiveness to Flt-3L in IkL/L BM 120G8+ cells. IkL/L 120G8+ BM cells expressed strong levels of surface Flt-3 (Figure 5A), which at times were higher than WT levels, depending on the mouse (not shown). Importantly, culture of unfractionated BM cells with Flt-3L led to the ready emergence of CD11c+120G8+ cells in both WT and IkL/L cultures, with similar frequency and kinetics (Figure 5B-C). These results indicate that Flt-3 signaling is functional in IkL/L pDC precursors, even though the cells derived from IkL/L BM still failed to up-regulate B220.
Together, our results show that a pDC population resides in the bone marrow of IkL/L mice. These cells produce IFNα and express markers indicative of the pDC lineage, but do not terminally differentiate even upon Flt-3 signaling.
The pDC defect is intrinsic to IkL/L pDC progenitor cells
As Ikaros is expressed by all hematopoietic cells, it was important to distinguish if the block in pDC development in IkL/L mice was due to a cell-intrinsic defect within the pDC lineage or to an indirect effect due to diminished Ikaros levels in another hematopoietic population. To allow the development of IkL/L BM cells in a WT environment, we established single and double (mixed) bone marrow chimeras. In the single chimeras, we used unfractionated WT or IkL/L BM cells (both expressing the Ly5B6 allele) to reconstitute lethally irradiated B6.Ly5SJL recipients. These experiments tested the capacity of IkL/L hematopoietic progenitors to differentiate in an environment containing WT stromal cells. In the double chimeras, we used WT or IkL/L BM cells mixed with B6.Ly5SJL congenic BM cells to reconstitute irradiated B6.Ly5SJL recipients. This latter combination tested the capacity of IkL/L hematopoietic progenitors to differentiate in an environment containing both WT stromal and hematopoietic elements. Double chimeras also allowed us to determine if IkL/L hematopoietic cells can interfere with WT pDC development, or reciprocally, if the presence of WT (B6.Ly5SJL) BM cells can rescue IkL/L pDC development. Ly5B6 spleen and BM cells were analyzed 6 to 10 weeks after reconstitution.
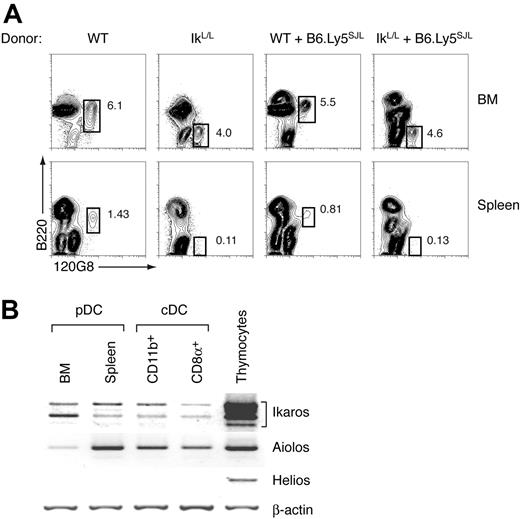
As shown in Figure 6A, chimeras reconstituted with WT BM cells exhibited normal pDC compartments (B220+120G8+ cells) in both the BM and spleen. In striking contrast, both single chimeras reconstituted with IkL/L BM cells and double chimeras exhibited the same block in IkL/L pDC differentiation at the B220–120G8+ stage in the BM, with few if any detectable IkL/L pDCs in the spleen. WT B6.Ly5SJL pDCs differentiated normally in the double chimeras reconstituted with WT and IkL/L BM cells, eliminating the possibility that IkL/L BM cells might exert a dominant-negative intercellular effect on pDC differentiation (not shown). These results demonstrate that the pDC block is due to a cell-intrinsic defect in IkL/L hematopoietic cells.
Ikaros expression in pDCs
To determine the relative expression of Ikaros and other family members during pDC maturation, we analyzed its expression, versus that of Aiolos and Helios, in pDCs and cDCs by RT-PCR. Ikaros was expressed in WT BM and splenic pDCs, and in splenic cDCs (Figure 6B), although at much lower levels than in thymocytes. Interestingly, Aiolos was expressed by all splenic DCs, but at barely detectable levels in the BM pDC population. Helios expression was not detected in any of the DC populations tested. These results suggest that Ikaros is the predominant family member expressed in BM pDCs.
The defect in pDC differentiation is cell intrinsic. (A) BM from WT (Ly5B6; 2 × 106), IkL/L (Ly5B6; 5 × 106), or mixtures of WT:B6.Ly5SJL (1 × 106: 1 × 106) or IkL/L:B6.Ly5SJL (5 × 106:1 × 106) were used to reconstitute lethally-irradiated B6.Ly5SJL recipients. Recipients were analyzed 6 to 10 weeks after BM transfer. All plots are gated on small Ly5B6+CD11c-enriched cells. Numbers indicate the percentage of gated cells in each plot. One representative experiment of 4 is shown. (B) Ikaros expression in DC subsets. WT BM and splenic pDCs (CD11c+ B220+ 120G8+), and splenic CD11b+ cDCs (CD11c+ CD11b+120G8–) and CD8α+ cDCs (CD11c+CD8α+CD11b–120G8–) were analyzed by RT-PCR for their expression of Ikaros, Aiolos, and Helios. Whole WT thymocytes were used as a control. One representative experiment of 3 is shown.
The defect in pDC differentiation is cell intrinsic. (A) BM from WT (Ly5B6; 2 × 106), IkL/L (Ly5B6; 5 × 106), or mixtures of WT:B6.Ly5SJL (1 × 106: 1 × 106) or IkL/L:B6.Ly5SJL (5 × 106:1 × 106) were used to reconstitute lethally-irradiated B6.Ly5SJL recipients. Recipients were analyzed 6 to 10 weeks after BM transfer. All plots are gated on small Ly5B6+CD11c-enriched cells. Numbers indicate the percentage of gated cells in each plot. One representative experiment of 4 is shown. (B) Ikaros expression in DC subsets. WT BM and splenic pDCs (CD11c+ B220+ 120G8+), and splenic CD11b+ cDCs (CD11c+ CD11b+120G8–) and CD8α+ cDCs (CD11c+CD8α+CD11b–120G8–) were analyzed by RT-PCR for their expression of Ikaros, Aiolos, and Helios. Whole WT thymocytes were used as a control. One representative experiment of 3 is shown.
Ikaros represses non-pDC gene expression
To gain insight into the mechanism of Ikaros function during pDC differentiation, we compared the gene expression profiles of WT and IkL/L BM CD11c+120G8+ pDCs. Three independent RNA samples were prepared for each genotype, and analyzed on pangenomic Affymetrix arrays. Most genes were similarly expressed between WT and IkL/L samples (note the global shape of the scatterplots in Figure 7A), including those previously found to be highly expressed in pDCs (ie, SpiB, Bcl-11a, and Siglec_H). These data provide further evidence that IkL/L BM pDCs belong to the pDC lineage. However, there were striking differences in the IkL/L transcriptome. Three-hundred and seventy genes were strongly overexpressed in IkL/L BM pDCs with 137 of these up-regulated more than 4-fold (Figure 7B; Table 2; Table S1). Furthermore, 52 of these genes were normally not expressed in WT BM pDCs (Figure 7C; Table 2 asterisks). In contrast, most of the 247 down-regulated genes showed only a modest decrease in expression (Figure 7B; Table S2), and only 4 of them were turned off in IkL/L BM pDCs. These experiments suggest that Ikaros deficiency results in a widespread and strong derepression of specific genes in immature pDCs. Interestingly, some of these deregulated transcripts are T- or B-cell specific. IkL/L BM pDCs strongly express Lck, pKCθ,pTα, TCR-Vβ13, and Thy1, all T-cell–specific, as well as λ5 and VpreB1, which encode the surrogate light chains of the pre-BCR. Also intriguing is the strong induction of CD56 (NCAM1, Table 2), a diagnostic marker for human plasmacytoid leukemia not normally expressed in premalignant pDCs.57 CD56 might therefore be a marker for immature pDCs. Collectively, our data indicate that Ikaros functions to repress a specific set of genes during pDC differentiation, by direct or indirect means.
Top up- or down-regulated genes in lkL/L BM pDCs
Gene name . | Gene symbol . | Fold change, log2 . |
|---|---|---|
| Up-regulated genes | ||
| Neural cell adhesion molecule 1 (CD56)* | Ncam1 | 8.04 |
| Sulfatase 2* | Sulf2 | 7.58 |
| Troponin T2, cardiac* | Tnnt2 | 7.12 |
| Myosin 1B* | Myo1b | 6.77 |
| Low-density lipoprotein receptor-related protein 1 | Lrp1 | 6.24 |
| Immunoglobulin lambda-like polypeptide 1 (lambda 5)* | Igll1 | 6.17 |
| Von Willebrand factor homolog* | Vwf | 5.66 |
| RIKEN cDNA D030029J20 gene* | D030029J20Rik | 5.54 |
| Cytochrome P450, family 2, subfamily j, polypeptide 6* | Cyp2j6 | 5.48 |
| Matrix metallopeptidase 14 (membrane inserted)* | Mmp14 | 5.20 |
| Lipoma HMGIC fusion partner* | Lhfp | 5.03 |
| Lipoma HMGIC fusion partner-like 2* | Lhfp12 | 5.00 |
| Serologically defined colon cancer antigen 33-like* | MG1:2153084 | 5.00 |
| Pre-B lymphocyte gene 1 | Vpreb1 | 4.96 |
| Dedicator of cytokinesis 1 | Dock1 | 4.90 |
| C1q and tumor necrosis factor-related protein 1* | Clqtnf1 | 4.83 |
| Catenin (cadherin associated protein), delta 1* | Ctnnd1 | 4.67 |
| Uveal autoantigen with coiled-coil domains and ankyrin repeats* | Uaca | 4.58 |
| Protein phosphatase 1, regulatory (inhibitor) subunit 9A* | Ppplr9a | 4.48 |
| Interleukin-18 receptor 1* | Il18r1 | 4.33 |
| SAM and SH3 domain-containing 1* | Sash1 | 4.18 |
| Parathyroid hormone receptor 1* | Pthr1 | 4.10 |
| AXL receptor tyrosine kinase* | Axl | 4.07 |
| FK506 binding protein 9* | Fkbp9 | 4.06 |
| RIKEN cDNA 4732416N19 gene* | 4732416N19Rik | 4.04 |
| Lymphocyte protein tyrosine kinase* | Lck | 4.01 |
| Fibroblast growth factor 13* | Fgfl3 | 4.00 |
| DNA segment, Chr 15, ERATO Doi 366, expressed* | D15Erid366e | 3.92 |
| Sulfotransferase family 1A, phenol-preferring, member 1* | Sultlal | 3.92 |
| cDNA sequence BC031593* | BC031593 | 3.88 |
| Solute carrier family 27 (fatty acid transporter), member 3* | Slc27a3 | 3.87 |
| Retinoic acid-induced 14* | Rail4 | 3.86 |
| Nerve growth factor receptor (TNFR superfamily, member 16) | Ngfr | 3.81 |
| Selenoprotein M* | MG1:2149786 | 3.76 |
| T-cell receptor beta, variable 13* | Tcrb-V13 | 3.73 |
| Heterogeneous nuclear ribonucleoprotein L-like | Hnrpll | 3.72 |
| Protein kinase C, theta* | Prkcq | 3.67 |
| RIKEN cDNAC730015A04 gene* | C730015A04Rik | 3.60 |
| Fucosyltransferase 10, mRNA* | Fut10 | 3.59 |
| Nuclear receptor subfamily 1, group H, member 4 (FXR)* | Nrlh4 | 3.57 |
| Cytochrome P450, family 7, subfamily b, polypeptide 1 | Cyp7b1 | 3.57 |
| Cadherin 22 | Cdh22 | 3.54 |
| Ajuba* | Jub | 3.53 |
| Kit ligand* | Kitl | 3.47 |
| Galectin-related interfiber protein* | Grifin | 3.46 |
| Hairy and enhancer of split 1 | Hes1 | 3.44 |
| Epithelial membrane protein 1* | Emp1 | 3.42 |
| Cyclin D2 | Ccnd2 | 3.31 |
| RIKEN cDNA B230206N24 gene* | B230206N24Rik | 3.23 |
| Dedicator of cytokinesis 9* | Dock9 | 3.20 |
| Serine (or cysteine) peptidase inhibitor, clade H, member 1 | Serpinh1 | 3.17 |
| Dynamin 1* | Dnml | 3.17 |
| RIKEN cDNA 2010300C02 gene* | 2010300C02Rik | 3.16 |
| cDNA sequence BC025600 | BC025600 | 3.14 |
| ATP-binding cassette transporter subfamily A member 9 | Abca9 | 3.08 |
| Nuclear factor I/X | Nfix | 3.03 |
| SH3-domain binding protein 5 (BTK-associated) | Sh3bp5 | 3.01 |
| Prion protein | Prnp | 3.01 |
| Down-regulated genes | ||
| Obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF* | Obscn | -4.06 |
| CD22 antigen | Cd22 | -3.26 |
| Paired-Ig-like receptor A1 | Pira1 | -2.93 |
| Odd Oz/ten-m homolog 4 (Drosophila)* | Odz4 | -2.92 |
| Regulator of G-protein signaling 1 | Rgs1 | -2.82 |
| GTP binding protein (gene overexpressed in skeletal muscle) | Gem | -2.68 |
| G-protein-coupled receptor 65 | Gpr65 | -2.67 |
| Sterile alpha motif domain containing 9-like | Samd91 | -2.67 |
| Killer cell lectinlike receptor, subfamily A, member 17 (Ly49Q) | Klra17 | -2.52 |
| Hypothetical protein 9530028C05 | 9530028C05 | -2.47 |
| Hydroxysteroid 11-beta dehydrogenase 1 | Hsd11b1 | -2.47 |
| Paired-Ig-like receptor A2 | Pira2 | -2.42 |
| CD8 antigen, beta chain 1 | Cd8b1 | -2.39 |
| Expressed sequence A1447904 | A1447904 | -2.36 |
| ATPase, Na+/K+ trasporting, alpha 3 polypeptide | Atpla3 | -2.32 |
| cDNA sequence BC022623 | BC022623 | -2.29 |
| Properdin factor, complement | Pfc | -2.19 |
| Serine (or cysteine) peptidase inhibitor, clade 1, member 1 | Serpinil | -2.12 |
| CEA-related cell adhesion molecule 1 | Ceacam1 | -2.10 |
| Chemokine (C-C motif) receptor 9 | Ccr9 | -2.07 |
Gene name . | Gene symbol . | Fold change, log2 . |
|---|---|---|
| Up-regulated genes | ||
| Neural cell adhesion molecule 1 (CD56)* | Ncam1 | 8.04 |
| Sulfatase 2* | Sulf2 | 7.58 |
| Troponin T2, cardiac* | Tnnt2 | 7.12 |
| Myosin 1B* | Myo1b | 6.77 |
| Low-density lipoprotein receptor-related protein 1 | Lrp1 | 6.24 |
| Immunoglobulin lambda-like polypeptide 1 (lambda 5)* | Igll1 | 6.17 |
| Von Willebrand factor homolog* | Vwf | 5.66 |
| RIKEN cDNA D030029J20 gene* | D030029J20Rik | 5.54 |
| Cytochrome P450, family 2, subfamily j, polypeptide 6* | Cyp2j6 | 5.48 |
| Matrix metallopeptidase 14 (membrane inserted)* | Mmp14 | 5.20 |
| Lipoma HMGIC fusion partner* | Lhfp | 5.03 |
| Lipoma HMGIC fusion partner-like 2* | Lhfp12 | 5.00 |
| Serologically defined colon cancer antigen 33-like* | MG1:2153084 | 5.00 |
| Pre-B lymphocyte gene 1 | Vpreb1 | 4.96 |
| Dedicator of cytokinesis 1 | Dock1 | 4.90 |
| C1q and tumor necrosis factor-related protein 1* | Clqtnf1 | 4.83 |
| Catenin (cadherin associated protein), delta 1* | Ctnnd1 | 4.67 |
| Uveal autoantigen with coiled-coil domains and ankyrin repeats* | Uaca | 4.58 |
| Protein phosphatase 1, regulatory (inhibitor) subunit 9A* | Ppplr9a | 4.48 |
| Interleukin-18 receptor 1* | Il18r1 | 4.33 |
| SAM and SH3 domain-containing 1* | Sash1 | 4.18 |
| Parathyroid hormone receptor 1* | Pthr1 | 4.10 |
| AXL receptor tyrosine kinase* | Axl | 4.07 |
| FK506 binding protein 9* | Fkbp9 | 4.06 |
| RIKEN cDNA 4732416N19 gene* | 4732416N19Rik | 4.04 |
| Lymphocyte protein tyrosine kinase* | Lck | 4.01 |
| Fibroblast growth factor 13* | Fgfl3 | 4.00 |
| DNA segment, Chr 15, ERATO Doi 366, expressed* | D15Erid366e | 3.92 |
| Sulfotransferase family 1A, phenol-preferring, member 1* | Sultlal | 3.92 |
| cDNA sequence BC031593* | BC031593 | 3.88 |
| Solute carrier family 27 (fatty acid transporter), member 3* | Slc27a3 | 3.87 |
| Retinoic acid-induced 14* | Rail4 | 3.86 |
| Nerve growth factor receptor (TNFR superfamily, member 16) | Ngfr | 3.81 |
| Selenoprotein M* | MG1:2149786 | 3.76 |
| T-cell receptor beta, variable 13* | Tcrb-V13 | 3.73 |
| Heterogeneous nuclear ribonucleoprotein L-like | Hnrpll | 3.72 |
| Protein kinase C, theta* | Prkcq | 3.67 |
| RIKEN cDNAC730015A04 gene* | C730015A04Rik | 3.60 |
| Fucosyltransferase 10, mRNA* | Fut10 | 3.59 |
| Nuclear receptor subfamily 1, group H, member 4 (FXR)* | Nrlh4 | 3.57 |
| Cytochrome P450, family 7, subfamily b, polypeptide 1 | Cyp7b1 | 3.57 |
| Cadherin 22 | Cdh22 | 3.54 |
| Ajuba* | Jub | 3.53 |
| Kit ligand* | Kitl | 3.47 |
| Galectin-related interfiber protein* | Grifin | 3.46 |
| Hairy and enhancer of split 1 | Hes1 | 3.44 |
| Epithelial membrane protein 1* | Emp1 | 3.42 |
| Cyclin D2 | Ccnd2 | 3.31 |
| RIKEN cDNA B230206N24 gene* | B230206N24Rik | 3.23 |
| Dedicator of cytokinesis 9* | Dock9 | 3.20 |
| Serine (or cysteine) peptidase inhibitor, clade H, member 1 | Serpinh1 | 3.17 |
| Dynamin 1* | Dnml | 3.17 |
| RIKEN cDNA 2010300C02 gene* | 2010300C02Rik | 3.16 |
| cDNA sequence BC025600 | BC025600 | 3.14 |
| ATP-binding cassette transporter subfamily A member 9 | Abca9 | 3.08 |
| Nuclear factor I/X | Nfix | 3.03 |
| SH3-domain binding protein 5 (BTK-associated) | Sh3bp5 | 3.01 |
| Prion protein | Prnp | 3.01 |
| Down-regulated genes | ||
| Obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF* | Obscn | -4.06 |
| CD22 antigen | Cd22 | -3.26 |
| Paired-Ig-like receptor A1 | Pira1 | -2.93 |
| Odd Oz/ten-m homolog 4 (Drosophila)* | Odz4 | -2.92 |
| Regulator of G-protein signaling 1 | Rgs1 | -2.82 |
| GTP binding protein (gene overexpressed in skeletal muscle) | Gem | -2.68 |
| G-protein-coupled receptor 65 | Gpr65 | -2.67 |
| Sterile alpha motif domain containing 9-like | Samd91 | -2.67 |
| Killer cell lectinlike receptor, subfamily A, member 17 (Ly49Q) | Klra17 | -2.52 |
| Hypothetical protein 9530028C05 | 9530028C05 | -2.47 |
| Hydroxysteroid 11-beta dehydrogenase 1 | Hsd11b1 | -2.47 |
| Paired-Ig-like receptor A2 | Pira2 | -2.42 |
| CD8 antigen, beta chain 1 | Cd8b1 | -2.39 |
| Expressed sequence A1447904 | A1447904 | -2.36 |
| ATPase, Na+/K+ trasporting, alpha 3 polypeptide | Atpla3 | -2.32 |
| cDNA sequence BC022623 | BC022623 | -2.29 |
| Properdin factor, complement | Pfc | -2.19 |
| Serine (or cysteine) peptidase inhibitor, clade 1, member 1 | Serpinil | -2.12 |
| CEA-related cell adhesion molecule 1 | Ceacam1 | -2.10 |
| Chemokine (C-C motif) receptor 9 | Ccr9 | -2.07 |
Genes that up-regulated more than 3 logs or down-regulated more than 2 logs are shown. Fold change values correspond to the average of the values for all the IkL/L/WT comparisons.
Genes that are absent in all WT (ie, up-regulated genes in IkL/L samples) or IkL/L (ie, down-regulated genes in IkL/L samples) samples.
Discussion
Our data show that pDCs are selectively and severely reduced in mice with diminished Ikaros activity and that this phenotype is dose dependent on Ikaros. Loss of the pDC population, but not other DC subsets or hematopoietic lineages in IkL/L mice,35,36 allowed us to confirm the highly specific IFNα-producing role of pDCs to influenza virus, and to TLR7 and TLR9 ligands. Moreover, we confirm that pDCs are essential in the early response to viral infection, as MCMV-infected IkL/L mice failed to produce high levels of IFNα and control viral replication.
Although pDCs are absent in IkL/L mice, cDCs appear normal. Their numbers are slightly reduced in IkL/L collagenase-treated organs, but these cells respond normally when stimulated in vivo with MCMV or STAg, producing IL-12 and up-regulating maturation markers. Thus, the IkL/L mouse line provides a powerful tool to study pDC requirement in vivo, without previous external manipulation. Indeed, IkL/L mice have been valuable in revealing a second and novel wave of IFNα production following MCMV infection that is pDC independent.52 Some caution, however, should be exercised when using this mouse line as a “pDC-less” model, as other hematopoietic lineages including T and B cells and neutrophils are also affected by the IkL/L mutation, and IkL/L mice develop thymic lymphomas by 10 to 12 weeks of age.35,36,58
Significant gene derepression in IkL/L BM pDCs. (A) Representative scatterplots comparing the gene expression profile between 2 independent WT samples (top), versus a WT and an IkL/L sample (bottom). The right panels are enlargements of boxed areas in the left panels. Red indicates increased genes; blue, decreased genes. (B) Genes that either “increased” or “decreased” in all of the 9 IkL/L/WT comparisons were selected and grouped according to the magnitude of change (average of the “fold change” [in log2] from all 9 comparisons). Note that the decreased genes exhibited mostly modest variations, while many increased genes exhibited large variations. ▦ correspond to the estimated number of nonspecific changes (“Materials and methods”). (C) Number of up-regulated genes in IkL/L samples whose expression was not detected in WT samples (▪) or were expressed in WT but not detected in IkL/L samples (□).
Significant gene derepression in IkL/L BM pDCs. (A) Representative scatterplots comparing the gene expression profile between 2 independent WT samples (top), versus a WT and an IkL/L sample (bottom). The right panels are enlargements of boxed areas in the left panels. Red indicates increased genes; blue, decreased genes. (B) Genes that either “increased” or “decreased” in all of the 9 IkL/L/WT comparisons were selected and grouped according to the magnitude of change (average of the “fold change” [in log2] from all 9 comparisons). Note that the decreased genes exhibited mostly modest variations, while many increased genes exhibited large variations. ▦ correspond to the estimated number of nonspecific changes (“Materials and methods”). (C) Number of up-regulated genes in IkL/L samples whose expression was not detected in WT samples (▪) or were expressed in WT but not detected in IkL/L samples (□).
The lack of peripheral pDCs in IkL/L mice is likely due to a developmental block in the BM, where we have identified a putative pDC precursor population. These cells resemble WT BM pDCs in several aspects: (1) they express 120G8, Ly-6C, Flt-3, CD8α, and CD4, and exhibit a gene expression program similar to that of WT pDCs; (2) they present a typical plasmacytoid morphology; and (3) they produce IFNα after stimulation with CpG oligonucleotides and, to a lesser extent, influenza virus. Since IkL/L BM pDCs express less TLR7 at the mRNA level (Table S2), and TLR7 mediates influenza-induced IFNα production,59,60 our transcriptome data may explain why IkL/L pDCs respond less well to influenza stimulation. Moreover, IkL/L BM cells give rise to 120G8+ cells with similar kinetics as WT BM cells following Flt-3L stimulation in vitro, indicating that IkL/L progenitors can proceed through the initial steps of pDC differentiation. Interestingly, IkL/L BM 120G8+ cells are mostly Ly-49Q–, like the immature pDC subset recently identified in WT BM, which may represent the immediate precursors of the more mature Ly-49Q+ pDCs found in the BM and the periphery.54-56 It is therefore tempting to speculate that IkL/L pDCs are blocked at this immature Ly-49Q– stage of differentiation. However, IkL/L BM 120G8+ cells are also different in other respects—they lack B220, and express some markers of activation (CD8α, MHC class II, CD40) but not others (CD80, CD86). Collectively, our data suggest that commitment to the pDC lineage occurs in the IkL/L BM, but that differentiation is blocked.
Our results show that pDC-committed precursors in the BM show a cell-intrinsic requirement for Ikaros, as IkL/L BM pDCs do not terminally differentiate in vivo even if given a WT BM and stromal microenvironment, as shown in single and double chimera experiments. Although Flt-3/Flt-3L signaling has been shown to be essential for optimal pDC development22-24 and Flt-3 expression appears dependent on Ikaros,34 our studies suggest that this pathway is not defective in IkL/L BM pDCs, and notably, that Flt-3/Flt-3L signaling alone is not sufficient for pDC development. Indeed, IkL/L BM cells respond to Flt-3L stimulation in vitro but never give rise to mature cells. Moreover, cDC development and function appear normal in IkL/L mice, and their differentiation also depends on Flt-3/Flt-3L.61
How Ikaros regulates pDC differentiation may be linked to its proposed function as a transcriptional repressor. Our gene expression analyses show that a significant number of genes, not normally expressed in WT pDCs, are strongly up-regulated in IkL/L BM pDCs. These data suggest that Ikaros may act as a key repressor of non-pDC–specific genes during pDC differentiation, similar to Pax5 function in early B-cell development.62 Of importance, several T- and B-lineage–specific genes are highly expressed in IkL/L pDCs, suggesting that Ikaros is required to suppress lymphoid-related genes in pDCs. It is also noteworthy that the majority of deregulated genes code for adhesion molecules, cell surface receptors, extracellular matrix proteins, and secreted factors (Table 2; Figure S2), suggesting that an altered response to the BM environment may play an important role in the differentiation block seen here. Further functional investigation will be required to dissect the roles of these genes in pDC differentiation.
Our observation that most of the deregulated genes show increased mRNA expression supports the hypothesis that Ikaros acts mainly as a repressor, through its association with the NURD histone deacetylase complex or the CtBP and Sin3 corepressors. The fact that few transcriptional activators are strongly induced in IkL/L pDCs (Table 2) argues in favor of a direct influence by Ikaros on the affected genes. Indeed, 2 genes have previously been shown to be targets of Ikaros-mediated repression in other systems: λ5, which contains several Ikaros binding sites in its promoter responsible for silencing a reporter gene during the proB/preB transition in transgenic mice,63 and Hes-1, which was shown to be up-regulated in Ikaros-deficient leukemic T cells and whose promoter contains a regulatory element recognized by Ikaros and RBP-Jκ.58
IkL/L mice carry a β-galactosidase reporter gene inserted into exon 3 of the Ikaros locus (exon 3).35 We have shown that low levels of Ikaros transcripts are expressed in the hematopoietic cells of these mice that lack the altered exon and exhibit in-frame splicing between exons 2 and 4. As a result, residual quantities of smaller-sized Ikaros proteins (Ik*) are synthesized. An important issue is whether these Ik* proteins function similarly to WT proteins. Our previous data indicate that both normal Ik and Ik* proteins bind Ikaros target sequences and repress transcription similarly.58 Furthermore, the IkL/L mutation leads to incomplete loss of function, as (1) the phenotype of IkL/L mice is similar but milder than that of Ikaros-null mice, and (2) overexpression of WT or mutant Ikaros proteins can rescue T-cell phenotypes.58 Thus, Ik* proteins probably function normally but are present at low levels in IkL/L cells. Moreover, Wu and colleagues have alluded, in a recent review, that pDCs are also missing in Ikaros-null mice, in addition to their published defects in the cDC compartment (Naik et al21 ). Together, these data suggest that the lack of pDCs in IkL/L mice results from loss of Ikaros function, rather than the manifestation of a novel dominant effect of the Ik* protein.
Our results, together with previous work highlighting the role of Ikaros in hematopoiesis and DC differentiation,30,44 should provide insights into the molecular events required for the development of each DC lineage. Previous data illustrate that mice expressing a dominant-negative Ikaros mutation exhibit a complete block in the development of all DCs, and animals bearing a null mutation produce some CD8α+ but no CD11b+ cDCs.30 Our work extends these findings by showing that pDC development is exquisitely dependent on Ikaros, as IkL/L mice, which express low Ikaros levels, lack pDCs but exhibit only slightly reduced numbers of CD8α+ and CD11b+ cDCs. Furthermore, heterozygote Ik+/L mice that express intermediate levels of Ikaros show a dramatic reduction in pDC numbers but present normal numbers of cDCs. As BM pDCs uniquely express low levels of Aiolos and Helios, Ikaros may play an essential, nonredundant role in pDC differentiation. Collectively, these results demonstrate that Ikaros and/or related family members are essential for the development of all DC populations. Our results suggest that different DC subsets require graded levels of Ikaros activity for their development: very low levels suffice for CD8α+ cDCs, moderate levels are needed for CD11b+ cDCs, and high levels are required for pDCs.
In conclusion, we have shown that our mouse model bearing a hypomorphic mutation for Ikaros specifically lacks plasmacytoid dendritic cells, but not conventional dendritic cells, in the periphery. We have demonstrated that high levels of Ikaros are not necessary for engagement of BM precursors into the pDC lineage but are required for terminal differentiation of BM pDCs. This block is linked to a failure of IkL/L pDCs to silence the expression of many genes, some of which encode proteins important in intercellular crosstalk. Further studies are now required to determine the specific contribution of these genes to pDC differentiation.
Authorship
Contribution: D.A., M.D., C.A.-P., T.D., S.H.R., P.K., and S.C. designed and performed experiments; G.T. and C.A.B provided valuable reagents and intellectual input; and M.D., P.K., and S.C. wrote and edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
D.A., M.D., P.K., and S.C. contributed equally to this work.
Prepublished online as Blood First Edition Paper, August 15, 2006; DOI 10.1182/blood-2006-03-007757.
The online version of this article contains a data supplement.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
We thank G. Yap (Brown University) for the kind gift of STAg; M. Sellars for critical reading of the paper; S. Duhautbois-Boine, G. Kimmich, and G. Brizard for help; C. Thibault, C. Grussenmeyer, and D. Dembélé for microarray experiments and analysis; J. Barths, C. Ebel, and I. Durand for flow cytometry; and F. Memedov and M. Gendron for animal husbandry.
This work was supported by institute funds from INSERM, CNRS, and the Hôpital Universitaire de Strasbourg; a grant from the Association pour la Recherche sur le Cancer (ARC); and the Centre National de Ressources en Génomique (Programme Affymetrix) (P.K. and S.C.). M.D. was supported by an ATIPE (Action Thématique et Incitative sur Programme) grant from the CNRS and a grant from the ARC. S.H.R. was supported by the CNRS, the Fondation pour la Recherche Médicale, and the Philippe Foundation. D.A. was supported by National Institutes of Health (NIH) grants AI52861 and AI58066, and is the recipient of a Career Development Program Award from the Leukemia and Lymphoma Society. C.A.B. was supported by NIH grant AI55677.


![Figure 7. Significant gene derepression in IkL/L BM pDCs. (A) Representative scatterplots comparing the gene expression profile between 2 independent WT samples (top), versus a WT and an IkL/L sample (bottom). The right panels are enlargements of boxed areas in the left panels. Red indicates increased genes; blue, decreased genes. (B) Genes that either “increased” or “decreased” in all of the 9 IkL/L/WT comparisons were selected and grouped according to the magnitude of change (average of the “fold change” [in log2] from all 9 comparisons). Note that the decreased genes exhibited mostly modest variations, while many increased genes exhibited large variations. ▦ correspond to the estimated number of nonspecific changes (“Materials and methods”). (C) Number of up-regulated genes in IkL/L samples whose expression was not detected in WT samples (▪) or were expressed in WT but not detected in IkL/L samples (□).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/108/13/10.1182_blood-2006-03-007757/7/m_zh80240604910007.jpeg?Expires=1769855548&Signature=AzQOtJN7kAVlT1o0PTPgjO3XozesefZka4M1NGrDUwoJL2yWMzC~PwyTGDZOIeRajMyElgFVN1mE~USJDMsDZGjGLeU78XD8ECVhtoM54zUbnh02oxAipo2YFLlQT8FBhBCafTZxMUn2evGzvwrvV~MvC9Z6tjWvv~LULjdzUCy5us00T3qZEX1NwV4oYUAT70D5oPgqNlnu4HgYECpWJlmJvrYEO7O33kNMQuOwKqxK5TJtAeZ-bojPRYJ2J-iv4AipCwDMITU2Qqz~3TGMQ-Zz6HyUt~pdCPJfjU~OBciC0QjKBjBXbQFtRvIO0khGCAAfDrW60bRJ4gkqM2hvJQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal