Abstract
The histone deacetylase inhibitors (HDA-CIs) butyrate and trichostatin A activate γ-globin expression via a p38 mitogen-activating protein kinase (MAPK)-dependent mechanism. We hypothesized that down-stream effectors of p38 MAPK, namely activating transcription factor-2 (ATF-2) and cyclic AMP response element (CRE) binding protein (CREB), are intimately involved in fetal hemoglobin induction by these agents. In this study, we observed increased ATF-2 and CREB1 phosphorylation mediated by the HDACIs in K562 cells, in conjunction with histone H4 hyperacetylation. Moreover, enhanced DNA-protein interactions occurred in the CRE in the Gγ-globin promoter (G-CRE) in vitro after drug treatments; subsequent chromatin immunoprecipitation assay confirmed ATF-2 and CREB1 binding to the G-CRE in vivo. Enforced expression of ATF-2 and CREB produced Gγ-promoter trans-activation which was abolished by a 2-base pair mutation in the putative G-CRE. The data presented herein demonstrate that γ-gene induction by butyrate and trichostatin A involves ATF-2 and CREB1 activation via p38 MAPK signaling.
Introduction
The growth factor erythropoietin (Epo) exerts its effects on commitment, proliferation, and differentiation of erythroid progenitors and globin chain synthesis through Janus kinase 2/Stat5 signaling and crosstalk with mitogen-activated protein kinase (MAPK) pathways.1-3 p38 MAPK signaling is required for Epo mRNA stability and hemoglobin synthesis.4,5 The reversible inhibition of p38 MAPK using SB203580 blocked Epo-dependent accumulation of mouse globin chains,6 and studies in p38α-/- knockout mice showed a failure of definitive βmaj-globin gene expression. These studies confirm an Epo-p38 MAPK-dependent mechanism for hemoglobin synthesis.7
The HDACI sodium butyrate (NaB) induces differentiation in erythroleukemia cells via Stat58,9 and p38 MAPK signaling.10,11 Butyrate is a clinically useful fetal hemoglobin (HbF) inducer which has been used to treat individuals with sickle cell disease12 and thalassemia13 ; however, the molecular mechanism for NaB-mediated HbF induction is poorly understood. Recent data from Weinberg et al14 showed that HbF induction by arginine butyrate is due in part to posttranslational mechanisms and increased γ-globin mRNA translation.
Several HDACIs, including trichostatin A (TSA),10,11 MS-275, and scriptaid,15,16 induce γ-globin expression via p38 MAPK signaling. These studies suggest that different pharmacologic agents converge on the p38 MAPK pathway to activate γ-globin expression. Four major MAPK pathways have been characterized: ERK1/2, ERK5/BMK1, cJun amino-terminal signal kinases (JNK), and p38.17-20 Studies using erythroid progenitors,21,22 knockout mice,7 and K562 stable lines10 suggest that p38α is the primary mediator of globin gene regulation.
The downstream effector molecules of p38 MAPK signaling include MAPK-activated protein kinases 1 and 2,23,24 PRAK,25 ATF-1-4, CREB1, CREB2, and CREM.26,27 Commonly, p38 phosphorylates ATF-2 and CREB to augment gene transcription. We recently demonstrated a p38 MAPK-dependent mechanism for NaB and TSA-induced γ-globin expression.10 Mechanistically, both agents bind a central zinc atom in HDAC to produce hyperacetylation of histone H3 (H3) and H428,29 to activate gene transcription.30,31 We previously proposed a dual mechanism for γ-globin gene activation by HDACIs involving (1) hyperacetylation of histone tails to produce open chromatin configurations and (2) transcription factor binding.16
We expanded our investigations to identify the effector molecules involved in γ-globin trans-activation by NaB and TSA via p38 MAPK signaling. CREB1 and ATF-2 phosphorylation was increased by the HDACIs in K562 cells. The CRE in the Gγ-globin promoter was bound by ATF-2 and CREB1 in vitro and in vivo and was required for promoter trans-activation in a luciferase reporter system. Furthermore, both trans-factors augmented endogenous γ-gene expression in primary erythroid cells. These data demonstrate that γ-globin induction by NaB and TSA involves trans-activation by ATF-2 and CREB1 via p38 MAPK cell signaling.
Materials and methods
Cell culture
K562 cells were cultured in Iscove Modified Dulbecco medium containing 10% fetal bovine serum (Atlanta Biologicals, Atlanta GA), penicillin (100 U/mL), and streptomycin (0.1 mg/mL) at 37°C and 5% CO2. Cells were treated for 12 to 48 hours with NaB (2 mM), TSA (0.5 μM), or anisomycin (100 ng/mL) alone or following pretreatment for 1 hour with 10 μM SB203580 (Calbiochem, San Diego, CA) which was dissolved in 100% dimethyl sulfoxide (DMSO). Control studies were completed with 0.4% DMSO (final concentration added with SB203580). Drug inducers and antibiotics were purchased from Sigma (St Louis, MO).
Acid extraction of histone proteins
K562 cells were disrupted using a dounce homogenizer, and then nuclei were washed with lysis buffer. After centrifugation, acid extraction of nuclear proteins was carried out by adding 0.4 N H2SO4 at 4°C. The supernatant was mixed with acetone, stored at -20°C overnight, and then proteins were collected for Western blot analysis.
Western blot analysis
K562 cells from the different experimental conditions were mixed with lysis buffer (Promega, Madison, WI) to isolate total proteins. Protein (100 μg) was separated by 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis, transferred to nitrocellulose membranes, and then analyzed with mouse monoclonal phosphorylated ATF-2 (p-ATF-2; Thr 71) and total ATF-2 (t-ATF-2) antibodies (Santa Cruz Biotechnology, Santa Cruz, CA; SC-8398 and SC-242). Similar studies were performed with rabbit polyclonal p-CREB1 (Ser 133) and t-CREB1 antibodies (SC-7978-R and SC-58). The membranes were stripped and reprobed with the respective total antibody. Acetylated histone H4 (ac-H4) levels were measure in 30 μg histone protein using rabbit polyclonal ac-H4 (06-598) and t-H4 (05-858) antibodies (Upstate, Lake Placid, NY). Horseradish peroxidase secondary antibodies were used for chemiluminiscent detection of protein (enhanced chemiluminescence [ECL] kit; Amersham Biosciences, Piscataway NJ), and band intensities were measured using the ChemiDoc System (Bio-Rad, Hercules, CA).
Electrophoretic mobility shift assay (EMSA)
Nuclear proteins were extracted from untreated K562 cells or those induced for 48 hours with 2 mM NaB or 0.5 μM TSA.32 Oligonucleotide probes were end-labeled with [γ-32P]-ATP using T4 kinase. Binding reactions were performed with 4 μg nuclear protein and radiolabeled probes for 20 minutes. Competition studies were conducted with 50-fold excess of unlabeled oligonucleotide. Supershift experiments were carried out with p-CREB1 (c1) and p-ATF-2 (a2) TransCruz antibodies (Santa Cruz Biotechnology) or IgG (Diapharma, Westchester, OH). Binding reactions were also completed with 2 to 3 μg histidine-tagged CREB1 (HisCREB1) and HisATF-2 protein (Panomics, Redwood City, CA). All binding reactions were performed with poly (dI-dC) at a concentration of 0.50 μg/mL. DNA-protein complexes were resolved in 4% native polyacrylamide gels then exposed to x-ray film.
Chromatin immunoprecipitation (ChIP) assay
After NaB and TSA inductions, aliquots of K562 cells were used for γ-globin quantifications by real-time polymerase chain reaction (PCR) (see next section) and ChIP assay. Data were used from samples with more than 3-fold γ-globin induction to ensure that changes in chromatin configurations had been achieved by the inducing agents. ChIP assay was completed using a standard kit from Upstate. Briefly, K562 cells were crosslinked with 1% formaldehyde, and then nuclei were isolated using lysis buffer (10 mM Tris-HCL, 10 mM NaCl, 0.2% Nonidet P-40) containing protease inhibitors (1 μg/mL leupeptin, 1 mM phenymethlsulfonyl fluoride, 1 μg/mL aprotinin). DNA was isolated using lysis buffer 2 (50 mM Tris-HCl, 10 mM EDTA, 1% sodium dodecyl sulfate) and sonicated on ice for 15 cycles (10 seconds each) (SONICATOR 3000; Misonix, Farmingdale, NY). The supernatant containing input DNA was used to generate standard curves for real-time PCR. Chromatin immunoprecipitation was performed with the p-CREB1, p-ATF-2, ac-H4, and TFIID antibodies; IgG was analyzed along with a no antibody negative control. The immunoprecipitated complexes were eluted in buffer (1% sodium dodecyl sulfate, 0.1 M NaHCO3) and then heated at 65°C to reverse crosslinking. The DNA was precipitated by standard methods.
Quantitative PCR (qPCR) analysis
For chromatin quantification, standard curves were generated with 1:10 serial dilutions of input DNA. Primers were designed to amplify the -1350 to -1100 region of the Gγ-promoter, 5′-AAGCCTTACACAGGATTATGAAGTCTG-3′ (forward), 5′-ACATGGCAGGAAGTATTCATGCTG-3′ (reverse); the Aγ-globin region between -1343 to -1095, 5′-AAGCTTTACACAGGATCATGAAGG-3′ (forward) and 5′ CAGGGTAGGAAGTATTTATGGTGG 3′ (reverse); and TATA box between -53 to +69. qPCR was performed at an annealing temperature of 60°C for 22 cycles. Chromatin was quantified using Sybergreen iQ Supermix (Bio-Rad) and 100 nM of each primer set.
For globin gene expression studies, total RNA was isolated using RNA Stat-60 (TEL-TEST “B” Inc, Friendswood, TX) according to the manufacturer's instructions. cDNA was prepared from total RNA (100 ng) using the Improm-II reverse transcriptase system (Promega). The γ-globin, β-globin, and glyceraldehyde-3-phosphate dehydrogenase (GAPD) mRNA levels were quantified by qPCR (iCycler iQ; Bio-Rad). Standard curves were generated using a Topo7-based plasmid carrying the γ-globin (Topo7-γ-globin), Topo7-β-globin, or Topo7-GAPD cDNA which produced 219-bp (base pair), 198-bp, and 230-bp PCR products, respectively. Ten-fold dilutions of globin (2000-2.0 pg) and GAPD (200-0.2 pg) plasmids were used to generate standard curves. The qPCR studies were performed with previously published γ-globin and GAPD primers33 at 100-nM concentrations.
To quantify Gγ-globin and Aγ-globin mRNA levels separately, a common forward primer 5′-GAATTCACCCCTGAGGTG-3′ and gene-specific reverse primers (Gγ-globin, 5′-CCTATCCTTGAAAGCTCTAC-3′, or Aγ-globin, 5′-CTGAATCATGGGCAACAG-3′) were used to generate 107-bp and 91-bp PCR products, respectively. The other conditions were as described for chromatin quantification.
Reporter plasmid constructs
Truncation mutants of the Gγ-globin promoter were generated by PCR amplification using a common reverse primer at position +36 and 5′ primers at -1500, -1350, and -1180 which were subcloned into pGL3-basic (Promega) to produce -1500GγLuc, -1350GγLuc, and -1180GγLuc. A control -1500AγLuc reporter carrying the -1500 to +36 Aγ-promoter sequence was also produced. Two mutant reporters with the -1225 G→A and an AC→TG mutation at -1226 were produced by site-directed mutagenesis (Bio S&T, Montreal, Quebec, Canada); all reporter constructs were verified by direct sequencing. Affinity-purified plasmid DNA (Qiagen Maxiprep, Valencia, CA) was used for transient transfection studies.
Transient transfections
All luciferase reporters (10 μg) were cotransfected into 107 K562 cells with 2.5 μg pSV-β-galactosidase vector (Promega) at 260 V and 975 μF (GenePulser; Bio-Rad); enzyme activity was measured using the β-galactosidase Enzyme Assay System (Promega) per the manufacturer's protocol. For enforced expression studies, K562 cells were transfected with 30 μg of the constitutively active expression vectors pCMV-CREB (Clontech, Mountainview, CA) or pDNA3.1-ATF-2 (Invitrogen, Carlsbad, CA). Total protein was isolated at 12 and 24 hours, and Western blot analysis was performed to measure p-CREB1 (Ser 133) and p-ATF-2 (Thr 71) levels. Dose-response studies were also completed with 10 to 50 μg of expression vector added for 24 hours or 50 μg of the empty base plasmid pCMV (Strategene, La Jolla, CA) and pDNA3.1. The latter plasmid was produced using standard cloning methods to remove the ATF-2 cDNA, and then the base vector was circularized with T4 ligase. Luciferase activity was measured using a TD-20/20 Luminometer (Turner Biosystems, San Diego, CA) and the Luciferase Assay System kit (Promega); all readings were normalized by β-galactosidase activity and total protein.
Two-phase liquid culture system
For primary erythroid studies, peripheral blood was drawn from healthy individuals in accordance with guidelines of the Institutional Review Board at the University of Texas at Dallas. Mononuclear cells were isolated using Histapaque-1077 (Sigma), and liquid cultures were established using the Fibach method.34 During phase 1, cells were treated with interleukin-3 and -6, stem cell factor, and granulocyte-monocyte colony-stimulating factor at 50 ng/mL each, followed by a change of medium on day 7 and the addition of Epo (4 IU) and stem cell factor (10 ng/mL) to initiate phase 2. Primary cells were transfected with the pCMV-CREB or pDNA3.1-ATF-2 vectors using a CD34-Nucleofector kit designed for use with the Nucleofector device (Amaxa, Gaithersburg, MD) per the manufacturer's protocol. Briefly, 5 million cells were isolated on day 4 of phase 2 cultures, and then were mixed with 100 μL Nucleofector solution with 5 μg of each expression vector or 2 μg of the control pmaxGFP (green fluorescence protein; Amaxa) reporter to monitor transfection efficiency. Cells were electroporated on the recommended setting U-08 for erythroid progenitors and then were grown for 24, 48, and 72 hours. The quantification of γ-globin, β-globin, and GAPD mRNA levels was completed by qPCR. The percentage of GFP-positive cells was used to determine transfection efficiency and normalization of gene expression levels.
Statistical analysis
The data are reported as the mean plus or minus standard error of the mean (SEM) for at least 4 experiments. Statistical analysis of the raw data was performed by the 2-tailed t test. The Student t test was used to measure differences in samples. A probability of less than .05 (P < .05) was considered significant.
Results
HDACIs induce phosphorylation of p38 MAPK downstream effectors
The end result of cell signaling is activation of gene expression. Studies aimed at identifying the p38 MAPK effectors that trans-activate γ-globin expression in response to NaB and TSA induction were completed. Candidate molecules activated by p38 include MAPK-activated protein kinases 1 and 2,23,24 ATF-1-4, CREB, CREM,26,27 cJun,35 Stat3,36 and ELK-1.37 Commonly, p38 MAPK mediates ATF-2 and CREB activation to augment gene transcription. Thus, we determined the phosphorylation levels for ATF-2 and CREB under different experimental conditions.
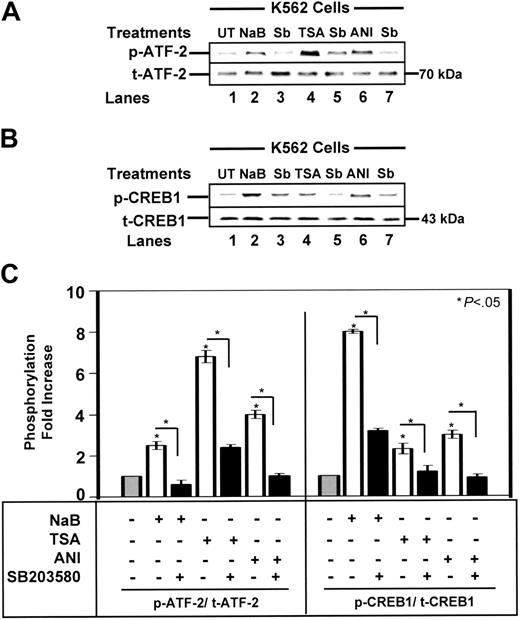
Western blot analysis was performed with protein extracts from K562 cells (Figure 1A-B). We observed a 6.8-fold increase in p-ATF-2 in TSA-treated cells, compared with a 2.5-fold increase by NaB and a 4-fold increase after treatment with the p38 MAPK activator, anisomycin (Figure 1C, left). SB203580 pretreatment inhibited ATF-2 phosphorylation from 64% to 75%. NaB and TSA also triggered an 8.0-fold and 2.3-fold increase in p-CREB1 levels (Figure 1C, right) which was blocked by SB203580, demonstrating p38 MAPK-dependent activation of ATF-2 and CREB by the HDACIs.
Nuclear protein binding to the G-CRE requires p38 MAPK signaling
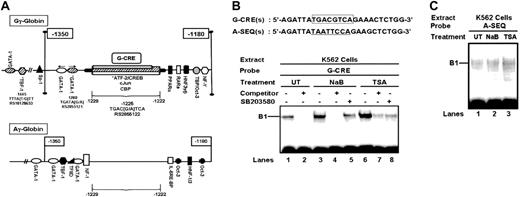
Because CREB1 and ATF-2 were activated by the HDACIs, a homology search using the Transcription Element Search System (TESS)38 database was performed to identify potential binding sites in the γ-globin promoters. A sequence located between nucleotides -1222 and -1229 (G-CRE) containing overlapping ATF-2 (-TGACGT-), CREB (-TGACGTCA-) and cJun (T(G/A)ACGTCA) binding sites was identified in the Gγ-promoter (Figure 2A); a similar motif was not present in the Aγ-globin promoter. We also identified other binding sites between -1180 and -1500 in both γ-globin promoters (Figure 2A; Table 1).
Aγ-globin versus Gγ-globin transcription binding sites
Transcription factor . | Position* . | Binding site . |
|---|---|---|
| Aγ-globin promoter | ||
| DTF-1 | – 1480 | gatgttgc |
| NF-ATc | – 1464 | ttttcc |
| Ets-2 | – 1462 | ttcctt |
| RSRFC4 | – 1453 | taatttta |
| EVE | – 1437 | tcattttaat |
| Oct-3 | – 1437 | cattttaat |
| USF | – 1421 | catgtg |
| Elk-1 | – 1412 | gcaggagtg |
| PEA3 | – 1413 | caggatgt |
| INSAF | – 1395 | acatgg |
| E12 | – 1360 | catctg |
| Gγ-globin promoter | ||
| GATA-1 | – 1488 | tgataa |
| NF1 | – 1465 | ctttcc |
| NF-ATp | – 1464 | ctttcct |
| Ets-2 | – 1463 | ttcctt |
| IRF | – 1462 | ctttccttt |
| TBP† | – 1445 | tttaT/Gtt |
| GR | – 1429 | agaaca |
| E2BP | – 1419 | tgcaatat |
| PPAR | – 1396 | aggtca |
| GATA-1 | – 1386 | gtatct |
| Sp1 | – 1365 | ccgcct |
| UME6 | – 1364 | tagccgccta |
Transcription factor . | Position* . | Binding site . |
|---|---|---|
| Aγ-globin promoter | ||
| DTF-1 | – 1480 | gatgttgc |
| NF-ATc | – 1464 | ttttcc |
| Ets-2 | – 1462 | ttcctt |
| RSRFC4 | – 1453 | taatttta |
| EVE | – 1437 | tcattttaat |
| Oct-3 | – 1437 | cattttaat |
| USF | – 1421 | catgtg |
| Elk-1 | – 1412 | gcaggagtg |
| PEA3 | – 1413 | caggatgt |
| INSAF | – 1395 | acatgg |
| E12 | – 1360 | catctg |
| Gγ-globin promoter | ||
| GATA-1 | – 1488 | tgataa |
| NF1 | – 1465 | ctttcc |
| NF-ATp | – 1464 | ctttcct |
| Ets-2 | – 1463 | ttcctt |
| IRF | – 1462 | ctttccttt |
| TBP† | – 1445 | tttaT/Gtt |
| GR | – 1429 | agaaca |
| E2BP | – 1419 | tgcaatat |
| PPAR | – 1396 | aggtca |
| GATA-1 | – 1386 | gtatct |
| Sp1 | – 1365 | ccgcct |
| UME6 | – 1364 | tagccgccta |
Transcription Element Search System (TESS) analysis of the Aγ-globin and Gγ-globin promoters (www.cbil.upenn.edu/cgi-bin/tess/tess). Binding site positions are identified relative to the cap site as recorded in the 81 706-kilobase β-locus sequence (GenBank accession no., gi:28380636, NG_000007).
DTF-1 indicates Drosophila transcription factor-1; NF-ATc, nuclear factor of activated T cells; Ets-related protein; RSRFC4, serum response factor–related protein; EVE, even-skipped; Oct-3, octomer 3; USF, upstream stimulatory factor; PEA3, polyoma virus enhancer A3; INSAF, insulin activator factor; E12, immunoglobulin enhancer binding factor; IRF, interferon-related factor-1; TBP, TATA binding protein; GR, glucocorticoid receptor; E2BP, estrogen binding protein; PPAR, peroxisome proliferators-activated receptor α; UME6, upstream repression sequence 1 binding protein 6
The binding sites shown are from base – 1500 to – 1350 relative to the Aγ-globin and Gγ-globin cap sites
T/G indicates an SNP in the TBP binding site
HDACIs mediate phosphorylation of ATF-2 and CREB1. Western blot analysis was performed with mouse monoclonal phosphorylated ATF-2 (p-ATF-2) and rabbit polyclonal p-CREB1 antibodies and protein isolated from K562 cells. Total nonphosphorylated CREB (t-CREB) and t-ATF-2 levels were analyzed as loading controls (see “Material and methods”). (A) A representative blot for ATF-2 phosphorylation by drug treatments is shown. (B) A representative blot for drug-mediated CREB1 phosphorylation is shown. (C) The quantitative data for p-ATF-2 (Thr 71) and p-CREB1 (Ser 133) levels after NaB (2 mM), TSA (0.5 μM), or 100 ng/mL anisomycin (ANI) treatment alone (□) or after pretreatment with 10 μM SB203580 (▪) are shown. Data are presented as the fold increase in phosphorylated protein ± SEM. The star above the graph represents a comparison between untreated ( ) and treated cells; the bracket and star represent a comparison between the different treatment conditions as indicated. Significance was achieved at the P < .05 level.
) and treated cells; the bracket and star represent a comparison between the different treatment conditions as indicated. Significance was achieved at the P < .05 level.
HDACIs mediate phosphorylation of ATF-2 and CREB1. Western blot analysis was performed with mouse monoclonal phosphorylated ATF-2 (p-ATF-2) and rabbit polyclonal p-CREB1 antibodies and protein isolated from K562 cells. Total nonphosphorylated CREB (t-CREB) and t-ATF-2 levels were analyzed as loading controls (see “Material and methods”). (A) A representative blot for ATF-2 phosphorylation by drug treatments is shown. (B) A representative blot for drug-mediated CREB1 phosphorylation is shown. (C) The quantitative data for p-ATF-2 (Thr 71) and p-CREB1 (Ser 133) levels after NaB (2 mM), TSA (0.5 μM), or 100 ng/mL anisomycin (ANI) treatment alone (□) or after pretreatment with 10 μM SB203580 (▪) are shown. Data are presented as the fold increase in phosphorylated protein ± SEM. The star above the graph represents a comparison between untreated ( ) and treated cells; the bracket and star represent a comparison between the different treatment conditions as indicated. Significance was achieved at the P < .05 level.
) and treated cells; the bracket and star represent a comparison between the different treatment conditions as indicated. Significance was achieved at the P < .05 level.
We previously proposed a dual mechanism for γ-gene activation by HDACIs involving histone hyperacetylation and transcription factor-mediated gene activation.16 To test this model we measured ac-H4 levels under the different experimental conditions. We observed a 3.1-fold and an 1.8-fold increase in ac-H4 levels induced by NaB and TSA, respectively (data not shown), and no change was produced by anisomycin. The Western blot data suggested that once hyperacetylation has been achieved by the HDACIs, ATF-2 and/or CREB1 might trans-activate γ-globin.
We next performed DNA-protein binding studies to determine whether CREB1 or ATF-2 could bind in the G-CRE and Aγ (A-SEQ) globin promoters. A single DNA-protein complex was established with the G-CRE probe and untreated K562 cell nuclear extract which was competed with unlabeled oligonucleotide (Figure 2B, lanes 1-4, 6, and 7). Similar results were obtained with a consensus CREB probe (data not shown). Protein binding in the B1 complex increased with nuclear extracts from K562 cells treated with NaB and TSA which decreased after SB203580 pretreatment (Figure 2B, lanes 1, 3, 5, 6, and 8) confirming the requirement for p38 signaling to augment protein binding in the G-CRE. EMSA studies with A-SEQ did not produce a significant DNA-protein complex (Figure 2C) nor did cold A-SEQ oligonucleotide compete for protein binding to the G-CRE (data not shown). These data support our dual mechanism model for Gγ-globin induction by NaB and TSA involving H4 hyperacetylation and altered transcription factor binding to the G-CRE.
Transcription factor binding to the G-CRE is altered by HDACIs through p38 signaling. (A) Schematic diagram of the upstream γ-globin promoters. The transcription factor binding sites between nucleotides -1350 and -1180 in the Gγ-globin and Aγ-globin promoters are shown in relative order based on a Transcription Element Search System (TESS)38 database analysis. Note the significant difference in the number of binding sites between nucleotides -1229 and -1222. Single nucleotide polymorphisms (SNPs) (*) and the RS identifiers from the SNP database39 are shown. CBP indicates CREB binding protein; HNF, hepatic nuclear factor; IL-6RE-BP, interleukin-6 response element binding protein; NF1, nuclear factor 1; NF-Y, nuclear factor-Y; Oct3, Octomer 3; PPAR, peroxisome proliferation-activated receptor; RARα, retinoic acid receptor α; TFIID, TATA-associated factor IID; TBF, TATA binding protein. (B) The nucleotide sequences of the sense strand for the 2 probes used in electrophoretic mobility shift assay (EMSA) are shown. The G-CRE is outlined in the box. The analogous region from the Aγ-globin promoter (A-SEQ) is underlined. The EMSA gel obtained with the G-CRE probe and untreated (UT), NaB-, or TSA-treated nuclear extract is shown. Reactions were also completed in the presence (+) or absence (-) of SB203580 pretreatment or G-CRE cold competitor at 50-fold excess. (C) Shown is an EMSA gel for the A-SEQ probe tested with UT, NaB-, or TSA-treated nuclear extract.
Transcription factor binding to the G-CRE is altered by HDACIs through p38 signaling. (A) Schematic diagram of the upstream γ-globin promoters. The transcription factor binding sites between nucleotides -1350 and -1180 in the Gγ-globin and Aγ-globin promoters are shown in relative order based on a Transcription Element Search System (TESS)38 database analysis. Note the significant difference in the number of binding sites between nucleotides -1229 and -1222. Single nucleotide polymorphisms (SNPs) (*) and the RS identifiers from the SNP database39 are shown. CBP indicates CREB binding protein; HNF, hepatic nuclear factor; IL-6RE-BP, interleukin-6 response element binding protein; NF1, nuclear factor 1; NF-Y, nuclear factor-Y; Oct3, Octomer 3; PPAR, peroxisome proliferation-activated receptor; RARα, retinoic acid receptor α; TFIID, TATA-associated factor IID; TBF, TATA binding protein. (B) The nucleotide sequences of the sense strand for the 2 probes used in electrophoretic mobility shift assay (EMSA) are shown. The G-CRE is outlined in the box. The analogous region from the Aγ-globin promoter (A-SEQ) is underlined. The EMSA gel obtained with the G-CRE probe and untreated (UT), NaB-, or TSA-treated nuclear extract is shown. Reactions were also completed in the presence (+) or absence (-) of SB203580 pretreatment or G-CRE cold competitor at 50-fold excess. (C) Shown is an EMSA gel for the A-SEQ probe tested with UT, NaB-, or TSA-treated nuclear extract.
p-CREB1 (Ser 133) and p-ATF-2 (Thr 71) bind the G-CRE in vitro and in vivo
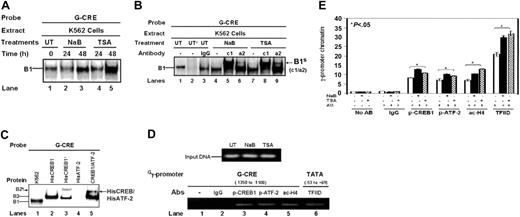
Additional EMSA studies were completed to confirm p-ATF-2 and p-CREB1 binding in the G-CRE. Extract from K562 cells treated for 24 and 48 hours with NaB and TSA produced a time-dependent increase in protein binding to the G-CRE (Figure 3A). Subsequent studies with a p-CREB1 (Ser 133) (c1) antibody produced a supershifted complex compared with the no antibody or IgG controls (Figure 3B, B1s (c1/a2), lanes 4, 5, 7, and 8). Moreover, a p-ATF-2 (Thr 71)-specific antibody (a2) produced a supershifted band with TSA-treated nuclear extract (Figure 3B, lanes 7 and 9), consistent with the Western blot data for TSA (Figure 1). These studies confirm the presence of p-CREB1 and p-ATF-2 in the B1 complex established on the G-CRE.
To determine whether CREB and ATF-2 bind the G-CRE as a heterodimeric complex, HisCREB1 and HisATF-2 protein was tested. As shown in Figure 3C, a single B2 DNA-protein complex formed with HisCREB1 protein which traveled slower than the B1 complex because of the histidine tag (Figure 3C, lanes 1 and 2). When CREB1 antibody was added, a supershifted complex B2S was observed (Figure 3C, lane 3). Binding studies with 3 μg HisATF-2 protein failed to produce a complex (Figure 3C, lane 4). This is consistent with published reports of the inability of ATF-2 to bind DNA in the nonphosphorylated state40 compared with CREB1 where DNA binding is observed independent of Ser 133 activation.41 To determine whether ATF-2 could heterodimerize with bound CREB, we incubated HisCREB (2 μg) with the G-CRE probe followed by the addition of HisATF-2 (2 μg). Under these conditions we observed a slower-moving complex consistent with the formation of a CREB/ATF-2 heterodimer on the G-CRE (Figure 3C, lane 5).
ChIP assays were also completed to test protein binding to the G-CRE region in vivo. Shown in Figure 3D are the PCR products obtained with input DNAs and each experimental condition. ChIP assay with p-CREB1 and p-ATF-2 antibody produced an 8.5-fold and 7.4-fold enrichment of Gγ-promoter chromatin, respectively, in untreated K562 cells (Figure 3E) compared with no antibody and IgG controls. In K562 cells in which γ-globin expression was induced more than 3-fold after NaB and TSA treatments, p-CREB1 binding was increased further by 4.5-fold and 2.5-fold, respectively (Figure 3E, p-CREB1). Similarly, TSA further increased p-ATF-2 binding 3.2-fold versus a 2.0-fold increase by NaB (Figure 3E, p-ATF-2); ac-H4 levels were increased 3.5-fold by NaB and 6.0-fold by TSA (Figure 3E). Both agents stimulated Gγ-promoter transcription exemplified by the 9- to 11-fold increase in TFIID binding to the TATA box region. Similar studies in the analogous Aγ-globin region showed a lack of chromatin enrichment (data not shown). We concluded from these data that p-CREB1 and p-ATF-2 bind the G-CRE and that the HDACIs altered binding to this region in vivo during γ-globin gene activation.
HDACIs induce Gγ-globin promoter activity in a preferential manner
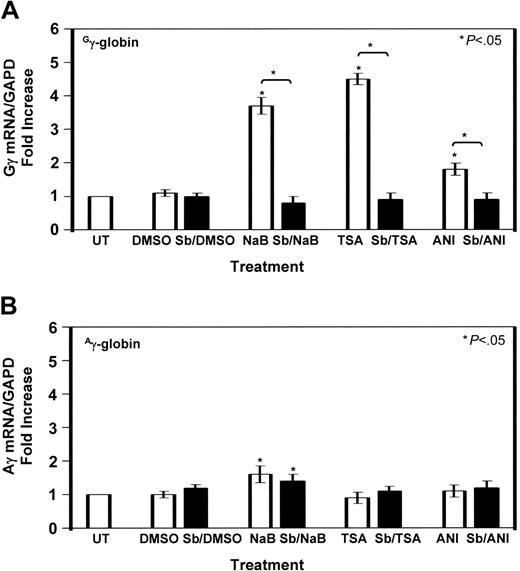
The 2 types of HbF produced during development contain nonallelic γ-globin chains that differ by a single amino acid residue at position 136 (glycine; Gγ versus alanine; Aγ). Previous studies showed that hydroxurea preferentially activates Gγ-globin expression.42 The unique G-CRE motif that was bound by p-ATF-2 and p-CREB1 suggested that p38 MAPK might converge at this element. To test this interpretation, we developed a qPCR assay to measure Gγ-globin and Aγ-globin mRNA levels separately.
K562 cells induced with HDACIs for 48 hours with or without 10 μM SB203580 pretreatment were analyzed. Compared with untreated cells, 2 mM NaB increased Gγ-globin 3.7-fold (Figure 4A, □) which was inhibited 78% by SB203580 (▪). Likewise, TSA (0.5 μM) induced Gγ-globin 4.5-fold which was blocked 80% by SB203580 (Figure 4A, TSA, Sb/TSA); anisomycin (200 ng/mL) increased Gγ-globin mRNA 1.8-fold which was inhibited 50% by SB203580. Although NaB increased Aγ-globin 1.6-fold, pretreatment with SB203580 had no effect. Furthermore, there was not a significant increase in Aγ-globin expression produced by TSA or anisomycin (Figure 4B), suggesting that p38 MAPK signaling stimulated by HDACIs converges at the Gγ-globin promoter. The level of γ-globin transcription did not change significantly in control studies in which K562 cells were treated with 0.4% DMSO.
G-CRE contributes to Gγ-globin promoter activation by HDACIs
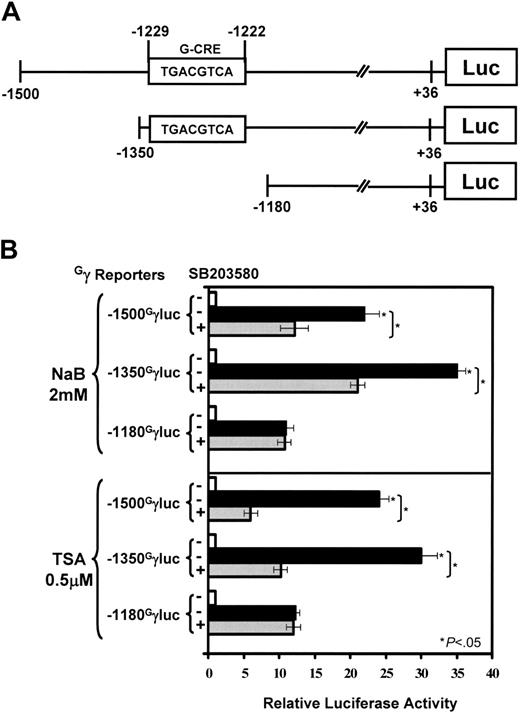
To determine whether the G-CRE is required for Gγ-promoter activation by NaB and TSA, a luciferase reporter system was established (Figure 5A). Data shown in Figure 5B showed the ability of SB203580 to inhibit by 45% the 22-fold increase in Gγ-globin promoter activity mediated by NaB in the -1500GγLuc reporter in K562 cells. Luciferase activity was increased 35-fold by NaB in the -1350GγLuc reporter which was inhibited 40% by SB203580, suggesting that a negative element exists between nucleotides -1500 and -1350. Deletion of the G-CRE in the -1180 reporter attenuated NaB-mediated promoter activation and luciferase activity was no longer subject to SB203580 inhibition.
The same pattern of response was observed for TSA. Luciferase activity increased 24-fold and 30-fold in the -1500 and -1350 reporters, respectively, which was inhibited by SB203580 (Figure 5B, TSA). The -1180GγLuc reporter was induced 12-fold by TSA, representing a 50% loss of inducibility without the G-CRE and a loss of SB203580 inhibition. The truncation mutants support a functional role for the G-CRE and p38 MAPK signaling in HDACI-mediated Gγ-promoter activation.
Enforced CREB and ATF-2 expression selectively trans-activates the Gγ-globin promoter
Although the transfection data suggested an important role for the G-CRE, we lacked direct evidence for trans-activation by target effector molecules; therefore, enforced expression studies were completed with constitutively active pCMV-CREB and pDNA3.1ATF-2 expression vectors. On the basis of published studies showing the ability of exogenous CREB to be activated in vivo independent of external stimuli and cell signaling,43,44 we initiated studies to test whether CREB trans-activates the Gγ-promoter. First, we measured p-CREB1 (Ser 133) and p-ATF-2 (Thr 71) levels at 12 and 24 hours in transfected K562 cells (Figure 6A). The levels of both transcription factors increased significantly after expression vector (30 μg) transfection. Subsequently, we observed a concentration-dependent increase in luciferase activity when 10 to 50 μg pCMV-CREB was transfected; a 39.2-fold increase in promoter activity was produced at the 50-μg concentration with the -1500GγLuc reporter (Figure 6B). Control studies with pCMV (50 μg) did not significantly increase luciferase activity. Similar studies with pDNA3.1-ATF-2 showed increased p-ATF-2 and luciferase activity up to 70-fold at the 50-μg concentration (Figure 6A,C). These studies showed that CREB and ATF-2 trans-activate the Gγ-promoter.
We next tested whether truncation of the G-CRE would abrogate the ability of CREB to trans-activate the Gγ-promoter. The data shown in Figure 6D confirm the ability of pCMV-CREB (30 μg) to increase luciferase activity 8.7-fold in the -1500GγLuc reporter compared with a 1.4-fold increase with pCMV. By contrast, the -1500AγLuc reporter was not trans-activated by CREB. Luciferase activity increased further to 19.4-fold for the -1350GγLuc reporter followed by a lack of trans-activation by CREB when the G-CRE was deleted in -1180GγLuc.
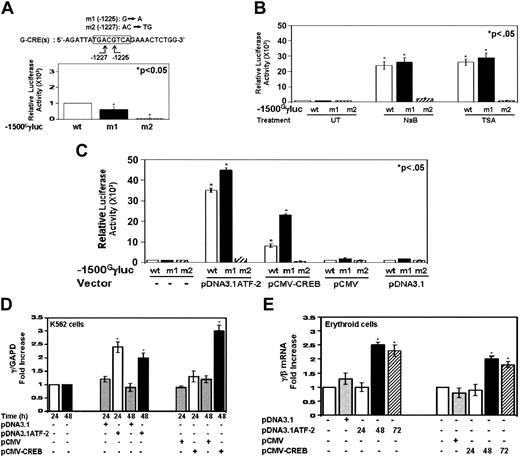
One might argue that important elements may have been deleted by truncation mutants; therefore, to address this question, site-directed mutagenesis was used to create an SNP at nucleotide -1225 (Figure 7A, m1). This alteration in DNA corresponds to a naturally occurring SNP in individuals with sickle cell disease and the Benin β-locus haplotype.45 In addition, a second mutant reporter with the AC→TG mutations at -1226 (m2) was created. The m1 SNP produced a 40% decrease in steady-state Gγ-promoter transcription, whereas the 2-base pair mutation (m2) abolished promoter transcription (Figure 7A). The 40% loss of promoter activity is consistent with the tendency for individuals with the Benin haplotype to have lower HbF levels.46
We next determined the ability of drug inducers to augment γ-promoter transcription in the mutant reporters. Both NaB and TSA induced luciferase activity in the m1 reporter comparable with wild type, suggesting the G→A mutation does not inhibit Gγ-promoter trans-activation by downstream effectors of the HDACIs (Figure 7B, ▪). This is consistent with our TESS analysis which predicted that CREB, ATF-2, or cJun could bind the m1 sequence -TGACATCA-. For the m2 reporter there was a lack of induction by either HDACI. Subsequent cotransfections with the m1 reporter and pDNA3.1ATF-2 and pCMV-CREB produced a 10-fold and 15-fold trans-activation, respectively, above the wild-type reporter (Figure 7C), in contrast to a loss of trans-activation for the m2 reporter.
ATF-2 and CREB bind the G-CRE in vitro and in vivo. (A) K562 cells were treated with NaB (2 mM) or TSA (0.5 μM) for 24 and 48 hours. Nuclear extracts were prepared and analyzed by EMSA using the G-CRE probe. A single DNA-protein complex (B1) was established with the G-CRE probe and untreated extract (0) which increased in intensity as the duration of HDACI treatment increased up to 48 hours. (B) The EMSA gel is shown for antibody studies with untreated K562 cell extract in the absence (UT) or presence (UT+) of G-CRE cold competitor or nuclear extracts isolated from K562 cells treated with NaB and TSA for 48 hours. Studies were completed with a p-CREB1 (Ser 133) (c1), p-ATF-2 (Thr 71) (a2), and IgG antibodies. B1S (c1/a2)-supershift band obtained with c1 or a2 antibody. (C) EMSA studies with histidine-tagged CREB1 (HisCREB1) and HisATF-2-purified protein. Binding studies were performed with the G-CRE probe and t-CREB1 antibody. In lane 5, HisCREB1 was added first followed by HisATF-2 protein to show binding of a heterodimeric complex. (D) ChIP assay to determine in vivo protein binding. Shown in the minigel are the PCR fragments obtained for the input DNA and different antibodies used to analyze the G-CRE and TATA box regions (see “Materials and methods”) in untreated and K562 cells induced with NaB (2 mM) and TSA (0.5 μM). (E) Chromatin quantifications obtained for qPCR analyses are shown in the graph. Immunoprecipitations were performed with IgG, p-CREB1 (Ser 133), p-ATF-2 (Thr 71), ac-H4, and TFIID antibodies along with a no antibody control. Data are presented as the mean fold increase in chromatin enrichment ± SEM. The bracket and star above the graph represent significant differences (P < .05) for the values obtained with the no antibody (No AB) control samples versus the level of chromatin enrichment obtained for immunoprecipitation experiments with the antibody indicated at the bottom of the graph.
ATF-2 and CREB bind the G-CRE in vitro and in vivo. (A) K562 cells were treated with NaB (2 mM) or TSA (0.5 μM) for 24 and 48 hours. Nuclear extracts were prepared and analyzed by EMSA using the G-CRE probe. A single DNA-protein complex (B1) was established with the G-CRE probe and untreated extract (0) which increased in intensity as the duration of HDACI treatment increased up to 48 hours. (B) The EMSA gel is shown for antibody studies with untreated K562 cell extract in the absence (UT) or presence (UT+) of G-CRE cold competitor or nuclear extracts isolated from K562 cells treated with NaB and TSA for 48 hours. Studies were completed with a p-CREB1 (Ser 133) (c1), p-ATF-2 (Thr 71) (a2), and IgG antibodies. B1S (c1/a2)-supershift band obtained with c1 or a2 antibody. (C) EMSA studies with histidine-tagged CREB1 (HisCREB1) and HisATF-2-purified protein. Binding studies were performed with the G-CRE probe and t-CREB1 antibody. In lane 5, HisCREB1 was added first followed by HisATF-2 protein to show binding of a heterodimeric complex. (D) ChIP assay to determine in vivo protein binding. Shown in the minigel are the PCR fragments obtained for the input DNA and different antibodies used to analyze the G-CRE and TATA box regions (see “Materials and methods”) in untreated and K562 cells induced with NaB (2 mM) and TSA (0.5 μM). (E) Chromatin quantifications obtained for qPCR analyses are shown in the graph. Immunoprecipitations were performed with IgG, p-CREB1 (Ser 133), p-ATF-2 (Thr 71), ac-H4, and TFIID antibodies along with a no antibody control. Data are presented as the mean fold increase in chromatin enrichment ± SEM. The bracket and star above the graph represent significant differences (P < .05) for the values obtained with the no antibody (No AB) control samples versus the level of chromatin enrichment obtained for immunoprecipitation experiments with the antibody indicated at the bottom of the graph.
Gγ-globin is preferentially activated by TSA and anisomycin. (A) Quantitative data obtained by qPCR analysis with Gγ-globin-specific primers are shown in the graph (see “Materials and methods”). The fold increase in Gγ-globin/GAPD mRNA ratio (□) is shown for NaB, TSA, and anisomycin treatments in the absence or presence of SB203580 (Sb) pretreatment (▪). Control studies with 0.4% dimethyl sulfoxide (DMSO) were completed. The levels in untreated (UT) K562 cells were normalized to 1. Data are shown as the mean ± SEM. The star above the graph (comparison between UT and treated cells) or the bracket and star (comparison between the different treatment conditions indicated) represent significance at the P < .05 level. (B) Analogous studies were performed to determine Aγ-globin/GAPD mRNA ratios (□) under the same experimental conditions.
Gγ-globin is preferentially activated by TSA and anisomycin. (A) Quantitative data obtained by qPCR analysis with Gγ-globin-specific primers are shown in the graph (see “Materials and methods”). The fold increase in Gγ-globin/GAPD mRNA ratio (□) is shown for NaB, TSA, and anisomycin treatments in the absence or presence of SB203580 (Sb) pretreatment (▪). Control studies with 0.4% dimethyl sulfoxide (DMSO) were completed. The levels in untreated (UT) K562 cells were normalized to 1. Data are shown as the mean ± SEM. The star above the graph (comparison between UT and treated cells) or the bracket and star (comparison between the different treatment conditions indicated) represent significance at the P < .05 level. (B) Analogous studies were performed to determine Aγ-globin/GAPD mRNA ratios (□) under the same experimental conditions.
Enforced CREB and ATF-2 expression activates endogenous γ-gene expression in K562 and primary erythroid cells
To complement our genetic reporter studies we also performed transfections to determine the ability of ATF-2 and CREB to activate the endogenous γ-globin promoter in K562 cells and primary erythroid progenitors. Enforced ATF-2 expression in K562 cells increased the γ/GAPD ratio 2.4-fold by 24 hours. However, 48 hours were required for CREB to induce γ-gene expression 2.8-fold above baseline (Figure 7D); no induction was observed with the control empty base vectors. Similar studies were completed in erythroid progenitors grown in liquid culture,34 using peripheral blood mononuclear cells (Figure 7E). Cells were transfected with 5 μg of each expression vector on day 4 during phase 2, followed by RNA harvest at the times indicated. The pmaxGFP vector was used to monitor transfection efficiency. At the time of transfection we observed 30% to 40% cell death with 70% viability at harvest. ATF-2 produced trans-activation of γ-globin 2.5-fold and 2.3-fold as a ratio to β-globin at 48 and 72 hours, respectively. CREB increased the γ/β ratio 1.8- to 2-fold; the level of β/GAPD increased 2% to 8% under the different conditions (data not shown). These studies confirm the ability of ATF-2 and CREB to activate the endogenous γ-globin gene.
Discussion
Eukaryotic gene transcription is regulated by sequence-specific transcription factors which bind modular promoter elements. Although the Gγ-globin and Aγ-globin genes share extensive homology, there exist differences in the promoters that may provide a basis for novel strategies to induce HbF. This notion is supported by unique mutations in each γ-globin promoter which produces different forms of nondeletion hereditary persistence of HbF.47-49 Milder symptoms in patients with sickle cell disease and high HbF levels underscore the therapeutic potential for γ-gene reactivation.50
The G-CRE is required for Gγ-promoter activation by HDACIs. (A) Shown is a schematic diagram of the reporter plasmids established with Gγ-promoters truncated at nucleotides -1500, -1350, and -1180. (B) K562 cells were transfected with reporters along with β-galactosidase to control for transfection efficiency. Transfected K562 cells were treated with HDACIs in the absence (-) or presence (+) of 10 μM SB203580. The relative luciferase activity was calculated after correcting for total protein and β-galactosidase activity. The untreated samples (-, □) were normalized to 1. Data for NaB or TSA in the absence (▪) or presence ( ) of SB203580 are shown as the mean ± SEM. The star and brackets are the same comparisons as defined in Figure 4A.
) of SB203580 are shown as the mean ± SEM. The star and brackets are the same comparisons as defined in Figure 4A.
The G-CRE is required for Gγ-promoter activation by HDACIs. (A) Shown is a schematic diagram of the reporter plasmids established with Gγ-promoters truncated at nucleotides -1500, -1350, and -1180. (B) K562 cells were transfected with reporters along with β-galactosidase to control for transfection efficiency. Transfected K562 cells were treated with HDACIs in the absence (-) or presence (+) of 10 μM SB203580. The relative luciferase activity was calculated after correcting for total protein and β-galactosidase activity. The untreated samples (-, □) were normalized to 1. Data for NaB or TSA in the absence (▪) or presence ( ) of SB203580 are shown as the mean ± SEM. The star and brackets are the same comparisons as defined in Figure 4A.
) of SB203580 are shown as the mean ± SEM. The star and brackets are the same comparisons as defined in Figure 4A.
The role of cell-signaling mechanisms in drug-mediated HbF induction are becoming better defined. Stat5 signaling is involved in NaB-mediated erythroid differentiation51 and γ-globin induction by short chain fatty acids.9 Moreover, hydroxyurea,52 valproic acid,53 NaB, and TSA10,11 stimulate γ-globin expression via p38 MAPK signaling. The goal of this study was to identify the target trans-factors involved in HDACI-mediated γ-gene reactivation. We explored the hypothesis that downstream effectors of p38 MAPK bind the G-CRE to augment gene transcriptional rates.
NaB induces hyperacetylation of core histones through HDAC inhibition,54 DNA methylation,55 and dephosphorylation of target genes.56 Both NaB and TSA regulate cell-cycle genes such as p21 and cyclin-D1 among others.57-59 Combining HDACIs with histone acetyl transferases can lead to the decompression of local chromatin54,60 and gene activation. We observed a similar mechanism in the γ-globin promoter where increased ac-H4 levels and p-CREB and p-ATF-2 binding were observed in the G-CRE after HDACI treatment. Members of the ATF/CREB family bind the palindromic CRE, -TGAC/GTCA-. The structure of each transcription factor contains a highly divergent N-terminal domain (1-283) and a C-terminal leucine zipper for dimerization and DNA binding (284-341).61 Although CREB binds to DNA in an unphosphorylated state, it cannot activate transcription unless phosphorylation occurs on Ser 133 which facilitates the ability of CREB to interact with CREB binding protein and p300.61,62 ATF-2, a second family member forms homodimers and also heterodimerizes with CREB and cJun to augment CRE-dependent transcription.63 ATF-2 phosphorylation at Thr 69 and Thr 71 is required for DNA binding and transcriptional activation.
Enforced CREB and ATF-2 expression trans-activates Gγ-promoter activity. (A) Western blot analysis showed increased phosphorylated and total CREB1 and ATF-2 protein levels after enforced expression for 12 and 24 hours after transfection. (B) The effect of increasing concentrations of pCMV-CREB on Gγ-promoter transcription was analyzed. Maximal trans-activation was observed at the 50-μg concentration at 24 hours. The base vector was transfected to control for nonspecific trans-activation. The bracket and star above the graph (comparison between the untransfected K562 cells [0] versus those treated with pCMV-CREB) represent significance at the P < .05 level. Data are shown as the mean ± SEM. (C) The effects of enforced ATF-2 (pDNA3.1ATF-2) expression on Gγ-promoter activity were tested. A robust concentration-dependent increase in luciferase activity was observed for both expression vectors. (D) The -1500GγLuc, -1350GγLuc, -1180GγLuc, and -1500AγLuc reporter plasmids (10 μg) were cotransfected into K562 cells with the constitutively active pCMV-CREB expression vector; luciferase activity was analyzed after 24 hours. A pattern of luciferase activity similar to that obtained with the HDACIs was observed. The star above the graph (comparison between untreated and pCMV-CREB-transfected cells) represents significance at the P < .05 level.
Enforced CREB and ATF-2 expression trans-activates Gγ-promoter activity. (A) Western blot analysis showed increased phosphorylated and total CREB1 and ATF-2 protein levels after enforced expression for 12 and 24 hours after transfection. (B) The effect of increasing concentrations of pCMV-CREB on Gγ-promoter transcription was analyzed. Maximal trans-activation was observed at the 50-μg concentration at 24 hours. The base vector was transfected to control for nonspecific trans-activation. The bracket and star above the graph (comparison between the untransfected K562 cells [0] versus those treated with pCMV-CREB) represent significance at the P < .05 level. Data are shown as the mean ± SEM. (C) The effects of enforced ATF-2 (pDNA3.1ATF-2) expression on Gγ-promoter activity were tested. A robust concentration-dependent increase in luciferase activity was observed for both expression vectors. (D) The -1500GγLuc, -1350GγLuc, -1180GγLuc, and -1500AγLuc reporter plasmids (10 μg) were cotransfected into K562 cells with the constitutively active pCMV-CREB expression vector; luciferase activity was analyzed after 24 hours. A pattern of luciferase activity similar to that obtained with the HDACIs was observed. The star above the graph (comparison between untreated and pCMV-CREB-transfected cells) represents significance at the P < .05 level.
ATF-2 and CREB augment endogenous γ-globin transcription in K562 and primary erythroid cells. (A) Site-directed mutagenesis was completed in the G-CRE at base -1225 (G→A) (m1) and bases -1226 and -1227 (AC→TG) to produce the m2 mutant reporter. Steady-state Gγ-promoter activity was decreased for m1 and abolished in the m2 reporter compared with wild type (wt). The star above the graph (comparison between the wt and mutant reporters) represents significance at the P < .05 level. (B) The ability of NaB (2 mM) and TSA (0.5 μM) to activate γ-promoter transcription in the mutant reporters was tested. The reporters were transfected into K562 cells, induced with NaB or TSA for 24 hours, and then luciferase activity was measured. (C) The ability of CREB and ATF-2 to trans-activate the mutant Gγ-promoters was analyzed. Both proteins were able to trans-activate the m1 reporter, whereas the m2 mutation abolished this effect. (D) CREB and ATF-2 activate endogenous γ-globin gene transcription. Studies were completed to determine endogenous γ-globin transcription after enforced protein expression (see “Materials and methods”). The cells were harvested 24 and 48 hours later, and qPCR analysis was completed to quantify γ-globin mRNAlevels as a ratio to the internal control GAPD. Data are shown as the fold increase in γ-globin/GAPD expression relative to untreated sample (mean ± SEM). (E) Enforced CREB and ATF-2 expression activates γ-globin in primary erythroid progenitors. Peripheral blood mononuclear cells were grown in a two-phase liquid culture system. Progenitor cells were transfected on day 4 during phase 2 using a Nucleofector device transfection system (see “Materials and methods”). The level of γ-globin, β-globin, and GAPD gene expression was quantified by qPCR. The ratio of γ/β globin mRNA was calculated after the level of mRNA for each gene was divided by GAPD mRNA; the ratios were normalized to values obtained for untransfected erythroid progenitors.
ATF-2 and CREB augment endogenous γ-globin transcription in K562 and primary erythroid cells. (A) Site-directed mutagenesis was completed in the G-CRE at base -1225 (G→A) (m1) and bases -1226 and -1227 (AC→TG) to produce the m2 mutant reporter. Steady-state Gγ-promoter activity was decreased for m1 and abolished in the m2 reporter compared with wild type (wt). The star above the graph (comparison between the wt and mutant reporters) represents significance at the P < .05 level. (B) The ability of NaB (2 mM) and TSA (0.5 μM) to activate γ-promoter transcription in the mutant reporters was tested. The reporters were transfected into K562 cells, induced with NaB or TSA for 24 hours, and then luciferase activity was measured. (C) The ability of CREB and ATF-2 to trans-activate the mutant Gγ-promoters was analyzed. Both proteins were able to trans-activate the m1 reporter, whereas the m2 mutation abolished this effect. (D) CREB and ATF-2 activate endogenous γ-globin gene transcription. Studies were completed to determine endogenous γ-globin transcription after enforced protein expression (see “Materials and methods”). The cells were harvested 24 and 48 hours later, and qPCR analysis was completed to quantify γ-globin mRNAlevels as a ratio to the internal control GAPD. Data are shown as the fold increase in γ-globin/GAPD expression relative to untreated sample (mean ± SEM). (E) Enforced CREB and ATF-2 expression activates γ-globin in primary erythroid progenitors. Peripheral blood mononuclear cells were grown in a two-phase liquid culture system. Progenitor cells were transfected on day 4 during phase 2 using a Nucleofector device transfection system (see “Materials and methods”). The level of γ-globin, β-globin, and GAPD gene expression was quantified by qPCR. The ratio of γ/β globin mRNA was calculated after the level of mRNA for each gene was divided by GAPD mRNA; the ratios were normalized to values obtained for untransfected erythroid progenitors.
As many as 10 000 CREs have been predicted in the mammalian genome.64 p38 MAPK, ERK, and MAPK-activated protein kinases 224,65 directly phosphorylate CREB to facilitate DNA binding. Alternatively, CREB can be activated via protein kinase A phosphorylation by cAMP66 independent of p38 MAPK signaling. Inoue et al67 demonstrated CREB phosphorylation in K562 cells by the known HbF inducer hemin; however, inhibition of cAMP-mediated protein kinase A activation produced γ-gene transcription, suggesting inhibition of a negative-acting factor. The researchers eliminated the MAPK pathways as negative regulators; thus, studies are required to identify the downstream repressor of γ-gene expression activated through cAMP signaling. We observed a propensity of NaB to activated CREB1 more robustly than ATF-2 and the opposite effect was produced by TSA. This may reflect activation of other signaling pathways such as JNK which is also known to activate ATF-2.68 Whether TSA activates JNK signaling is not known; however, additional experimental data are required to test this speculation.
The HDACIs altered the binding of p-ATF-2 and p-CREB1 to the G-CRE motif located at -1222 in the Gγ-promoter. We observed γ-globin promoter trans-activation by CREB and ATF-2 in transient transfection studies and to a lesser degree the endogenous γ-globin gene in K562 and normal erythroid cells. Because chromatin structure is a major determinant of gene transcriptional rates,69 the robust response observed in the reporter plasmids is likely due to the ability of plasmid DNA to remain extrachromosomal and not be subject to the effects of chromatin remodeling. The opposite would be true for the endogenous γ-genes where chromatin remodeling would be required to activate gene transcription, a known function of HDACIs.70
Limited studies have been published to increase our understanding of molecular mechanisms used by HDACIs to induce HbF. Butyrate response elements (BREs) have been identified in the distal CCAAT box71 and proximal and upstream γ-globin promoter.32,72 However, target trans-activators that interact in these elements have not been identified. By contrast, transcription factors involved in BRE-mediated gene transcription such as human adenosine nucleotide translocase-273 and the WAF/Cip174,75 and HDAC1/Sp-1 complexes76 have been identified in other systems. Moreover, IGF binding protein 3 is activated by NaB through alterations in an Sp1/Sp3 multiprotein repressor complex.77
Our mutational analysis confirmed the functional relevance of the G-CRE in normal Gγ-promoter transcription. Mechanisms for loss of promoter activity in the m2 mutant might include binding of repressor proteins since the -tgtggt- sequence is a consensus motif for acute myeloid leukemia, a transcriptional repressor.78 Moreover, the negative element in the -1500 to -1350 region may contribute to the silencing of promoter activity. Studies by our colleagues Ofori-Acquah et al79 supported an upstream silencer region in the Gγ-promoter. Herein, we fine-mapped a negative element between -1350 and -1500 where 19 cognate DNA binding motifs were identified (Table 1). Of particular interest is the overlapping Ume6/Sp1 binding sites at -1364. The Ume6 protein acts as a repressor through the recruitment of HDAC1 and the Sin3p-Rd3p chromatin remodeling complex.80,81 Preliminary ChIP assay in the -1319 to -1500 region showed 3-fold and 6-fold chromatin enrichment by HDAC1 and Sp-1, supporting in vivo binding of these proteins (B.S.P., unpublished data, October 2005). We speculate that a functional negative element in this region would be unopposed because p-CREB and p-ATF-2 cannot bind the m2 reporter, thus providing a plausible explanation for the total abolishment of Gγ-promoter activity in this reporter. The data presented herein are the first confirmation of trans-activators involved in γ-globin induction by HDACIs.
Prepublished online as Blood First Edition Paper, August 8, 2006; DOI 10.1182/blood-2006-01-023713.
Supported by the National Heart, Lung, and Blood Institute (grant HL69234) (B.S.P.).
The authors declare no competing financial interests.
J.S. and M.S.L. contributed equally to this study.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
We thank Dr Santosh D'Mello and members of his laboratory for suggesting additional studies and critical evaluation of our experimental design and data interpretation.

![Figure 6. Enforced CREB and ATF-2 expression trans-activates Gγ-promoter activity. (A) Western blot analysis showed increased phosphorylated and total CREB1 and ATF-2 protein levels after enforced expression for 12 and 24 hours after transfection. (B) The effect of increasing concentrations of pCMV-CREB on Gγ-promoter transcription was analyzed. Maximal trans-activation was observed at the 50-μg concentration at 24 hours. The base vector was transfected to control for nonspecific trans-activation. The bracket and star above the graph (comparison between the untransfected K562 cells [0] versus those treated with pCMV-CREB) represent significance at the P < .05 level. Data are shown as the mean ± SEM. (C) The effects of enforced ATF-2 (pDNA3.1ATF-2) expression on Gγ-promoter activity were tested. A robust concentration-dependent increase in luciferase activity was observed for both expression vectors. (D) The -1500GγLuc, -1350GγLuc, -1180GγLuc, and -1500AγLuc reporter plasmids (10 μg) were cotransfected into K562 cells with the constitutively active pCMV-CREB expression vector; luciferase activity was analyzed after 24 hours. A pattern of luciferase activity similar to that obtained with the HDACIs was observed. The star above the graph (comparison between untreated and pCMV-CREB-transfected cells) represents significance at the P < .05 level.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/108/10/10.1182_blood-2006-01-023713/4/m_zh80220603790006.jpeg?Expires=1769090606&Signature=CdhGxRHhVuDgLqRD1JhWXzeAE-xrIrwXQSb3tvCcoIuUfKoFK9SwdTdlNptorJOp7lsa9p0QOKIjIsoz9eDm0ywCRwR9~cGT3PyieObVGa5dBB4SdwW~BUL62e4K9AdZ~flrjaZ3GGZBSuSXTitsT7OKWC9Vme99ZBKBizZm0ljeRiYZhrm0tscBbHEJ2GiL0YUd78XGoeiLxsPclAzO8utElWOyYx8-AOAI0YR0NnxjgvqMt1bm1YqiO7Ql-uJQH4xKSKAF2B9Cbz1LiMSHBBmM3Z8HQJWNLIY9tXWXbuZMW-F8~QX4TDbyAnOLAsldfvtijkJTyl9Y4ip6w0jVxw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal