We report on the molecular etiology of an unusual clinical phenotype associating congenital neutropenia, thrombocytopenia, developmental delay, and hypopigmentation. Using genetic linkage analysis and targeted gene sequencing, we defined a homozygous genomic deletion in AP3B1, the gene encoding the β chain of the adaptor protein-3 (AP-3) complex. The mutation leads to in-frame skipping of exon 15 and thus perturbs proper assembly of the heterotetrameric AP-3 complex. Consequently, trafficking of transmembrane lysosomal proteins is aberrant, as shown for CD63. In basal keratinocytes, the incorporated immature melanosomes were rapidly degraded in large phagolysosomes. Despite distinct ultramorphologic changes suggestive of aberrant vesicular maturation, no functional aberrations were detected in neutrophil granulocytes. However, a comprehensive immunologic assessment revealed that natural killer (NK) and NKT-cell numbers were reduced in AP-3-deficient patients. Our findings extend the clinical and molecular phenotype of human AP-3 deficiency (also known as Hermansky-Pudlak syndrome, type 2) and provide further insights into the role of the AP-3 complex for the innate immune system. (Blood. 2006;108:362-369)
Introduction
Congenital neutropenia represents a heterogeneous group of disorders whose molecular etiology and pathophysiology remains enigmatic in most cases. Over the past few years, heterozygous mutations in the gene encoding neutrophil elastase have been associated with cyclic1 and congenital2 neutropenia, yet insights into the pathophysiology remain scarce. Aberrant vesicular trafficking of endosomes has been proposed as one potential mechanism accounting for decreased neutrophil numbers.3
Intracellular protein sorting and trafficking are controlled by means of coat proteins.4 The adaptor protein-3 (AP-3) complex facilitates trafficking of vesicles from the trans Golgi network and/or endosomal compartments.5,6 The AP-3 complex is a heterotetramer consisting of 2 big subunits, β3A (140 kDa) and δ3 (160 kDa), a medium subunit μ3A (47 kDa), and a small σ3A subunit. Three of the 4 subunits are also expressed as neuronal isoforms (β3B, μ3B, σ3B). AP-3 belongs to the family of cytosolic adaptor complexes and is involved in intracellular protein sorting and vesiculation. Details of the function of AP-3 in mammalian cells are poorly understood. A study indicates that AP-3 is associated with budding profiles evolving from a tubular endosomal compartment.7
Various animal models confirm the significance of AP-3 in endosomal trafficking. Mutations in the fruit fly genes coding for the subunits of AP-3 result in defective pigmentation of the eyes.8 The autosomal recessive mouse mutation pearl shows abnormal platelet dense granules, hypopigmentation, and abnormal lysosomal secretion and is associated with mutations in the β3A subunit gene (Ap3b1) of the AP-3 adaptor complex.9 Similar findings are seen in an engineered Ap3b1 knockout mouse strain.10 Interestingly, in contrast to mice whose neutrophil counts appear to be normal, dogs with mutations in AP3B1 have cyclic neutropenia.3 These animal models indicate that AP-3 deficiency results in increased surface expression of lysosomal proteins.
In humans, mutations in AP3B1 cause a complex phenotype known as Hermansky-Pudlak syndrome11 (HPS; Online Mendelian Inheritance in Man [OMIM; http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=OMIM] OMIM catalog no. 203300) type 2 (HPS2), which was first demonstrated by Dell'Angelica et al.12 To date, 4 human patients with HPS2 have been described in the literature.12-15
In general, patients with Hermansky-Pudlak syndrome have hemorrhagic diathesis as a result of prolonged bleeding time and tyrosinase-positive oculocutaneous albinism. Congenital neutropenia appears to be a distinguishing feature of HPS2. Some patients develop lung fibrosis and inflammatory colitis over time, others show defects in CD8+ T-cell-mediated cytotoxicity.15 In all cases reported so far, HPS appears to be inherited as a monogenic, autosomal recessive disease. However, it is a genetically heterogeneous disorder caused by mutations in 8 known genes.12,16-21 For HPS type 1, which is by far the most common form of HPS in humans, there is also allelic heterogeneity leading to a variety of clinical manifestations.16 Like AP3B1, most of the HPS causative genes were identified with the aid of mouse models. Because at least 16 mouse mutants have been shown to mimic the phenotype of HPS,22 it is likely that some of their human orthologs will be found mutated in human patients with HPS in the future.
In this paper, we report on genetic investigations of a consanguineous pedigree first reported by Kotzot et al23 in 1994. In this family, 2 cousins show a phenotype associating coarse facial features, hypopigmentation, mild mental retardation, splenomegaly, neutropenia, and thrombocytopenia.
In view of this unusual clinical presentation, we took a positionally unbiased approach and performed a genetic linkage study across the genome. We identified a homozygous genomic deletion in AP3B1 and functionally characterized AP-3-deficient fibroblasts and neutrophils.
Patients, materials, and methods
Patients
Blood samples and skin biopsies were taken after informed consent. The study was approved by the Internal Review Boards at Hannover Medical School and the University of Freiburg.
Marker selection and genotyping
For the initial genome scan, 261 polymorphic markers (Invitrogen, Karlsruhe, Germany) were genotyped on genomic DNA extracted from whole blood of 11 individuals according to the published conditions. Two additional polymorphic markers on chromosome 5 were later genotyped to better define the upper boundary of the linkage interval and to prove that the AP3B1 gene is inside the linkage interval.
Polymerase chain reaction (PCR) products were analyzed on an ABI 377 sequencer (PE Applied Biosystems, Foster City, CA) with the COLLECTION and ANALYSIS software packages (PE Applied Biosystems). Allele sizes were determined by using the program GENOTYPER (PE Applied Biosystems).
Genetic linkage analysis
Genetic linkage analysis computations were done with FASTLINK24,25 for 1 and 2 markers, and Superlink26,27 for more markers. A fully penetrant autosomal recessive inheritance model was used. The disease allele frequency was set to 0.001, and marker allele frequencies were set all equal because of the small number of individuals genotyped.
Mutation detection
The candidate gene AP3B1 on chromosome 5q was analyzed by direct sequencing of genomic DNA. Results were confirmed by cDNA sequencing. The cDNA of the affected individual no. 30 was amplified with 8 primer sets. For cDNA amplification of exons 11 to 16, the forward primer (5′-AAAGAAAGGGGATGTTTGAACCT) and the reverse primer (5′-TTCGGAACAATAAGCTGCCTAATA) were used at an annealing temperature of 53°C. Sequencing was performed with the forward primer at an annealing temperature of 54°C.
Long-Range PCR was performed with the Takara LA PCR Kit (Takara Bio, Otsu, Japan) by using the forward primer (5′-AAAGCCGCCGAAATGGACATC) and the reverse primer (5′-TTCACGGCAAACCAGCTACTCATC) at an annealing temperature of 68°C. Sequencing of the Long-Range PCR product of the patient was performed with the reverse primer (5′-GCAGGAAAGGCAGACAGAGAGGG) at an annealing temperature of 60°C.
DNA sequences were analyzed by using an ABI Prism 377 DNA Sequencer and the DNA Sequencing Analysis software version 3.4 (PE Applied Biosystems) and Sequencer version 3.4.1 software (Gene Codes Corporation, Ann Arbor, MI).
FACS, Western blotting, and immunofluorescence
Immune assessment, including immunophenotyping of peripheral blood mononuclear cells, and T-cell proliferation studies followed standard procedures. Peripheral blood mononuclear cells were isolated by Ficoll Paque (Amersham Biosciences, Freiburg, Germany) density gradient from 2 AP-3-deficient patients, their healthy siblings, and unrelated healthy donors. For immunophenotyping and natural killer (NK) T-cell analysis, the following antibodies were used: anti-CD3-APC (clone UCHT1), anti-CD4-PerCP (clone SK3), anti-CD25-PE (clone M-A251), anti-CD56-PE (clone B159), anti-6B11-PE (all from BD Biosciences, Heidelberg, Germany); anti-CD8-PE (clone B9.11), anti-CD16-FITC (clone 3G8), anti-CD19-FITC (clone J4.119), anti-Vα24-FITC (clone C15), anti-Vβ11-PE (clone C21; all from Beckman Coulter, Krefeld, Germany), and the respective isotype-matched controls. NKT cells were detected as Vα24+Vβ11+CD3+ cells or as 6B11+CD3+ cells.
To detect CD63 at the plasma membrane, fibroblasts of the AP-3-deficient patient no. 30 and of 2 unrelated donors were incubated stained with FITC-conjugated anti-CD63 (clone H5C6) and a corresponding isotype control antibody (BD Biosciences).
Flow cytometry was performed using a FACSCalibur flow cytometer (BD Biosciences). Subsequent data analysis was performed using WinMDI 2.8 software (The Scripps Research Institute, La Jolla, CA).
For Western blotting and immunofluorescence studies, the following antibodies were used: anti-β3A (“Marlene,” 1:100; kind gift from Margaret Robinson, Cambridge, United Kingdom), polyclonal anti-δ3A (1:1000; kindly provided by Andrew Peden, Genentech, San Francisco, CA), and anti-σ3A (1:250; BD Biosciences). AP-3-δ and -β were detected on a 7% gel; AP-3-σ was detected on a 10% gel.
The following antibodies were used to detect neutrophil proteins: anti-MPO (1:1000; Abcam, Hiddenhausen, Germany), anti-PR3 (1:300; Santa Cruz, Heidelberg, Germany), anti-HNE (1:300; Merck Biosciences Calbiochem; Schwalbach/TS, Germany), anti-MMP9 (1:300; Acris, Hiddenhausen, Germany), and anti-GAPDH (1:1000; Santa Cruz Biotechnology, Santa Cruz, CA).
Cell extracts were separated in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), blotted, and analyzed with Kodak Image-Station 440CF (Kodak, Rochester, NY). For immunofluorescence, fibroblasts were fixed with 4% paraformaldehyde (Fluka, Buchs, Switzerland) in phosphate-buffered saline (PBS) and permeabilized with 0.1% Triton-X100 (Sigma, St Louis, MO). The staining pattern was documented with an Axiovert 200 (Zeiss, Oberkochen, Germany) using a 63×/1.4 numeric aperture Plan Apochromat objective, and was computer analyzed using Axiovision 4.2.
Electron microscopy
Skin biopsies from patient no. 30 and from 2 unrelated healthy donors were fixed with 2.5% glutaraldehyde (Polysciences, Warrington, PA) in 0.1 M sodium cacodylate, pH 7.3 (Merck-Schuchardt, Hohenbrunn, Germany), and postfixed with 2% osmium tetroxide (Polysciences, Eppelheim, Germany) in the same buffer. After dehydration the specimens were embedded in Epon resin. Thin sections were stained with uranyl acetate and lead citrate.
For electron microscopic studies of granulocytes, 1 × 106 neutrophils were incubated with 3 × 107 TB Pseudomonas bacteria (kind gift from B. Tümmler, Department of Pediatric Pulmonology, Hannover Medical School, Germany) and incubated for 0, 7.5, and 20 minutes at 37°C in RPMI 1640 (PAA Laboratories, Pasching, Austria) containing 10% human AB serum (PAA Laboratories) and 10 mM HEPES buffer pH 7.4 (Gibco/Invitrogen, Karlsruhe, Germany), pelleted by centrifugation, and fixed in 2.5% glutaraldehyde in 0.1 M sodium cacodylate, pH 7.3. In addition, primary granulocytes were analyzed directly after hypotonic erythrocyte lysis. Electron microscopy was performed using a Philips electron microscope 301 (Philips Electronic Instruments, Eindhoven, The Netherlands) at an acceleration voltage of 80 kV and an objective with a focal length of 1.6 mm and an aperture of 50 μm. Images on electron microscope film 4489 (Eastman Kodak, Rochester, NY) were scanned using Agfa Duoscan and the image acquisition software Agfa Fotolook PS 3.03 (Agfa-Gevaert, Mortsel, Germany). The images were then processed using Adobe Photoshop 5.0 (Adobe Systems, San Jose, CA).
Neutrophil studies
For neutrophil studies, freshly purified granulocytes from patient no. 30 and from 2 healthy individuals were used. In some experiments, granulocytes were differentiated in vitro from purified CD34+ cells in the presence of recombinant human G-CSF and GM-CSF (AMGEN, Thousand Oaks, CA), following a previously published protocol.28 Primary granulocytes were purified by gradient centrifugation over Ficoll Paque and consecutive hypotonic erythrocyte lysis, yielding a purity of greater than 95%.
For chemotaxis experiments, neutrophils were first starved in RPMI 1640 supplemented with 1% BSA (Carl Roth, Karlsruhe, Germany) at pH 7.0 for 4 hours. Chemotaxis was assessed in duplicates in fibronectin-coated 0.3-μm pore size transwell migration plates (Corning/Sigma-Aldrich, Munich, Germany) as previously described.29 The number of migrated cells with respect to input cells was analyzed by flow cytometry after the addition of 50 000 uniform-dyed microspheres (Bangs Laboratories, Fishers, IN) to assure volume-independent acquisition. The inwells were flushed and controlled for the absence of residual adherent cells.
Phagocytosis was evaluated using FITC-labeled K12 Escherichia coli bacteria (Molecular Probes/Invitrogen, Karlsruhe, Germany). Neutrophils (5 × 105) were exposed to inactivated K12 E coli at a ratio of 1:50 in 100 μL RPMI 1640 supplemented with 10% FCS and 10 mM HEPES buffer pH 7.4 and cultured at a temperature of 37°C. Uptake of bacteria was analyzed by flow cytometry after quenching extracellularly attached bacteria with 0.4% trypan blue solution (Gibco/Invitrogen) at 4°C.
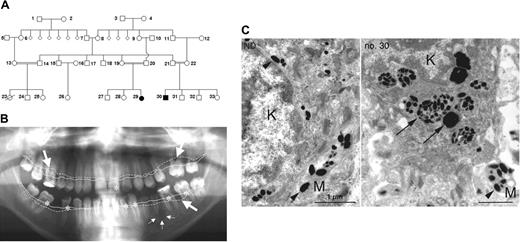
Pedigree and clinical manifestation. (A) Pedigree showing 2 affected individuals (no. 29 and no. 30). (B) Panoramic radiograph revealing aggressive periodontitis as clinical manifestation of congenital neutropenia in patient no. 30. Coronal carious decay is marked by asterisks; periradicular radiolucency around the apex of the second lower left premolar is indicated by small arrows. Bold arrows indicate generalized horizontal alveolar bone loss with vertical drop. Note the difference between expected alveolar bone level (continuous line) and actual alveolar bone margin (dotted line). (C) Transmission electron microscopy of the basal keratinocyte (K) and a melanocyte process (M) from a healthy individual (left) and from patient no. 30 (right). In a healthy individual, mature melanosomes (arrowheads) are translocated from melanocytes to keratinocytes. In AP-3 deficiency, keratinocytes contain large phagosomes of densely packed immature melanosomes (arrows).
Pedigree and clinical manifestation. (A) Pedigree showing 2 affected individuals (no. 29 and no. 30). (B) Panoramic radiograph revealing aggressive periodontitis as clinical manifestation of congenital neutropenia in patient no. 30. Coronal carious decay is marked by asterisks; periradicular radiolucency around the apex of the second lower left premolar is indicated by small arrows. Bold arrows indicate generalized horizontal alveolar bone loss with vertical drop. Note the difference between expected alveolar bone level (continuous line) and actual alveolar bone margin (dotted line). (C) Transmission electron microscopy of the basal keratinocyte (K) and a melanocyte process (M) from a healthy individual (left) and from patient no. 30 (right). In a healthy individual, mature melanosomes (arrowheads) are translocated from melanocytes to keratinocytes. In AP-3 deficiency, keratinocytes contain large phagosomes of densely packed immature melanosomes (arrows).
E coli lysis was assessed according to a previously described protocol.30 In this assay, lysis of a mutant strain of E coli (E coli ML-35; kindly provided by Reinhard Seger, Children's Hospital Zürich, Switzerland) can be measured based on β-galactosidase activity. The E coli mutant has no lactose permease but expresses β-galactosidase. On perforation of the bacterial envelope, β-galactosidase cleaves the artificial substrate ortho-nitrophenyl-β-d-galactopyranoside.
Results
Clinical phenotype
We investigated the molecular etiology of congenital neutropenia in 2 patients (aged 15 and 21 years), offspring of a consanguineous Turkish family (Figure 1A). Their complex phenotype, associating congenital neutropenia, thrombocytopenia, mild developmental delay, coarse facial features, splenomegaly, and hypopigmentation have been reported previously.23 Despite remarkably low neutrophil counts (ranging from 0 to 0.5 × 109/L [0 to 500/μL]), both patients had a relatively benign clinical course and did not have life-threatening infections. The most prominent finding was severe dental decay and aggressive periodontitis (Figure 1B). Microbiologic analysis revealed the presence of Porphyromonas gingivalis, Tannerella forsythensis, and Treponema denticola, a microbial spectrum usually associated with severe periodontitis. Patient no. 29 had recurrent skin abscesses; patient no. 30 has had 2 episodes of pneumonia to date. Temporary administration of G-CSF enhanced the peripheral neutrophil counts to normal levels, an effect that was not sustained because of poor compliance.
There was no evidence of prolonged bleeding. Patient no. 30 tolerated circumcision without hemorrhagic complications. Both patients showed splenomegaly and thrombocytopenia ranging from 50 000 to 150 000 platelets/μL. The ultrastructure of peripheral blood thrombocytes revealed immature dense granules (not shown). A bone marrow aspirate and biopsy on patient no. 30 revealed normal cellularity and complete maturation of all lineages without dysplastic changes (not shown).
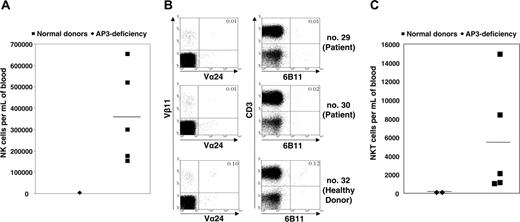
Immunophenotypic analysis. (A) Absolute numbers of peripheral blood CD3-CD56+ NK cells in patient no. 30, as compared with healthy family donors. (B) For fluorescence-activated cell sorting (FACS) analysis of NKT cells, 100 000 events were recorded for each sample. NKT cells were detected as Vα24+Vβ11+CD3+ or 6b11+CD3+ cells. A marked reduction in the numbers of NKT cells was found in AP-3-deficient patients (no. 29 and no. 30) as compared with a homozygous healthy family donor (no. 32). (C) Absolute numbers of NKT cells in peripheral blood of AP-3-deficient patients as compared with healthy family donors (2-sided Student t test for unequal variances using logarithmic values, P < .002). (A, C) Horizontal bars indicate median values.
Immunophenotypic analysis. (A) Absolute numbers of peripheral blood CD3-CD56+ NK cells in patient no. 30, as compared with healthy family donors. (B) For fluorescence-activated cell sorting (FACS) analysis of NKT cells, 100 000 events were recorded for each sample. NKT cells were detected as Vα24+Vβ11+CD3+ or 6b11+CD3+ cells. A marked reduction in the numbers of NKT cells was found in AP-3-deficient patients (no. 29 and no. 30) as compared with a homozygous healthy family donor (no. 32). (C) Absolute numbers of NKT cells in peripheral blood of AP-3-deficient patients as compared with healthy family donors (2-sided Student t test for unequal variances using logarithmic values, P < .002). (A, C) Horizontal bars indicate median values.
Both patients showed decreased visual acuity, photophobia, and nystagmus associated with decreased pigmentation of the eye fundus (not shown). Because of aberrant melanosome maturation in melanocytes, immature melanosomes were transported to keratinocytes and completely degraded in large phagosomes inside the basal layer of the epidermis (Figure 1C), as was evidenced in transmission electron microscopy (TEM) studies.
Immunologic phenotype
Immunologic investigations revealed normal serum levels of IgG, IgM, and IgA. Both patients mounted specific antibodies in response to tetanus and polio vaccines but failed to generate antibodies against mumps and rubella. Immunophenotyping of peripheral blood cells showed normal numbers of B and T cells (Table S1, available on the Blood website; see the Supplemental Materials link at the top of the online article). However, CD3-CD56+ NK cells were reduced in numbers (Figure 2A). The decrease in CD56+ cells prompted us to also assess the rare subpopulation of NKT cells. We stained lymphocytes from patients no. 29 and no. 30 with monoclonal antibodies against Vβ11 and Vα24 or CD3 and 6B11, respectively, and determined the absolute number of NKT cells in peripheral blood. Interestingly, compared with healthy family members, the numbers of NKT cells were significantly reduced in patients no. 29 and no. 30 (Figure 2B-C).
Genomewide linkage analysis
In light of the atypical clinical presentation of this family, we carried out a genomewide screen to localize the genetic cause. The only place in the genome where multiple adjacent markers had LOD scores greater than 1 was on chromosome 5q (see Table 1). The positively scoring markers from D5S1501 (at 78.5 Mb [megabase] from 5ptel) through D5S1505 (119.1 Mb from 5ptel) indicated a huge interval in which the affected individuals are homozygous and the markers segregate perfectly with the disease. However, AP3B1 is located at approximately 77.3 Mb, which is between the unlinked marker D5S2500 and the first linked marker D5S1501 (Figure 3A). Therefore, we genotyped 2 additional markers, D5S424 (76.2 Mb) (Figure 3A) and D5S2041 (76.7 Mb) to show that AP3B1 is positioned inside the interval of perfect segregation. The peak multipoint LOD score using the 6 positively scoring markers in Table 1 is 3.30.
Markers in the linkage region on chromosome 5 and single-marker LOD scores under a fully penetrant recessive model
. | . | . | Recombination fraction . | . | . | . | . | . | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Marker . | Genetic, cM . | Sequence, Mb . | 0.00 . | 0.01 . | 0.03 . | 0.05 . | 0.10 . | 0.15 . | |||||
| D5S2500 | 69.2 | 58.7 | −∞ | −2.83 | −1.94 | −1.53 | −0.97 | −0.66 | |||||
| D5S424* | 82.0 | 76.2 | 2.41 | 2.35 | 2.24 | 2.13 | 1.85 | 1.57 | |||||
| D5S2041* | 82.0 | 76.7 | 1.20 | 1.17 | 1.11 | 1.05 | 0.90 | 0.75 | |||||
| D5S1501 | 85.2 | 78.5 | 2.20 | 2.15 | 2.04 | 1.93 | 1.66 | 1.39 | |||||
| D5S644 | 104.8 | 95.8 | 2.27 | 2.23 | 2.15 | 2.06 | 1.83 | 1.60 | |||||
| D5S2501 | 117.0 | 110.0 | 1.57 | 1.54 | 1.47 | 1.41 | 1.24 | 1.07 | |||||
| D5S1505 | 129.8 | 119.1 | 1.32 | 1.29 | 1.22 | 1.16 | 1.00 | 0.84 | |||||
| D5S1480 | 147.5 | 144.1 | −∞ | −3.66 | −2.34 | −1.73 | −0.94 | −0.54 | |||||
. | . | . | Recombination fraction . | . | . | . | . | . | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Marker . | Genetic, cM . | Sequence, Mb . | 0.00 . | 0.01 . | 0.03 . | 0.05 . | 0.10 . | 0.15 . | |||||
| D5S2500 | 69.2 | 58.7 | −∞ | −2.83 | −1.94 | −1.53 | −0.97 | −0.66 | |||||
| D5S424* | 82.0 | 76.2 | 2.41 | 2.35 | 2.24 | 2.13 | 1.85 | 1.57 | |||||
| D5S2041* | 82.0 | 76.7 | 1.20 | 1.17 | 1.11 | 1.05 | 0.90 | 0.75 | |||||
| D5S1501 | 85.2 | 78.5 | 2.20 | 2.15 | 2.04 | 1.93 | 1.66 | 1.39 | |||||
| D5S644 | 104.8 | 95.8 | 2.27 | 2.23 | 2.15 | 2.06 | 1.83 | 1.60 | |||||
| D5S2501 | 117.0 | 110.0 | 1.57 | 1.54 | 1.47 | 1.41 | 1.24 | 1.07 | |||||
| D5S1505 | 129.8 | 119.1 | 1.32 | 1.29 | 1.22 | 1.16 | 1.00 | 0.84 | |||||
| D5S1480 | 147.5 | 144.1 | −∞ | −3.66 | −2.34 | −1.73 | −0.94 | −0.54 | |||||
The 6 rightmost column headings are recombination fractions between the putative disease gene and a marker. Genetic locations are from the Marshfield map.41
Markers were genotyped to prove that AP3B1 (at 77.3 Mb) lay inside the linkage interval.
Mutation detection and DNA analysis
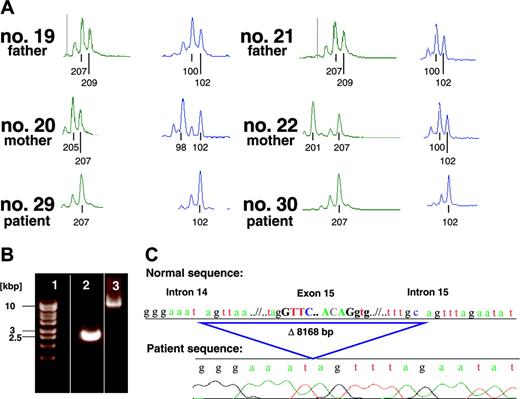
We first sequenced the AP3B1 cDNA with pairs of primers for several intervals. The PCR product representing exons 11 to 16 of the patient was shortened by approximately 150 bp when compared with the healthy control of the expected length of 664 bp (not shown). Exon by exon analysis of the patient's cDNA showed a complete deletion of exon 15. Exon skipping is usually caused either by a splice-site mutation or by a genomic deletion. To distinguish between these 2 possibilities, we carried out a genomic Long-Range PCR using DNA of patient no. 30 and DNA of his homozygous healthy cousin as a control. The healthy cousin's amplification product had the expected length of 11 018 bp. In contrast, the amplification product of the affected individual displayed a length of approximately 3000 bp (Figure 3B), suggesting a homozygous genomic deletion of approximately 8000 bp. To determine the exact length and boundaries of the deletion, the PCR product was sequenced. The deleted interval spanned 8168 bp, including a big part of intron 14, the complete exon 15, and a small part of intron 15 (Figure 3C). Loss of exon 15 results in the absence of the amino acids 491 to 550 while preserving the reading frame. Therefore, it is expected that an intact, albeit defective, AP3B1 protein is translated in the patient's cells.
Molecular identification of AP3B1 mutation. (A) Genotypes of the marker D5S424 (76.2 Mb) and D5S1501 (78.5 Mb) of the affected homozygous individuals (no. 29 and no. 30) and the heterozygous parents of the affected individuals (nos. 19-22). (B) Genomic Long-Range PCR of the affected individual no. 30 (lane 2) and the homozygous healthy cousin as a control (lane 3). The PCR product of lane 2 displays a length of only approximately 3000 bp (base pair) in contrast to the expected length of 11 018 bp (lane 3), suggesting a genomic deletion of approximately 8000 bp. Lane 1, size marker. (C) Sequence analysis of the Long-Range (LR) PCR product of the affected individual no. 30, revealing a deletion of 8168 bp.
Molecular identification of AP3B1 mutation. (A) Genotypes of the marker D5S424 (76.2 Mb) and D5S1501 (78.5 Mb) of the affected homozygous individuals (no. 29 and no. 30) and the heterozygous parents of the affected individuals (nos. 19-22). (B) Genomic Long-Range PCR of the affected individual no. 30 (lane 2) and the homozygous healthy cousin as a control (lane 3). The PCR product of lane 2 displays a length of only approximately 3000 bp (base pair) in contrast to the expected length of 11 018 bp (lane 3), suggesting a genomic deletion of approximately 8000 bp. Lane 1, size marker. (C) Sequence analysis of the Long-Range (LR) PCR product of the affected individual no. 30, revealing a deletion of 8168 bp.
Protein analysis
According to the GenBank annotation of AP3B1 (NP_003655), the functional adaptin domain, which is shared by multiple proteins in different adaptor complexes, spans amino acids 39 to 560, ending in exon 16. Therefore, sequence analysis suggests that skipping exon 15 at positions 491 to 550 is likely to have deleterious consequences.
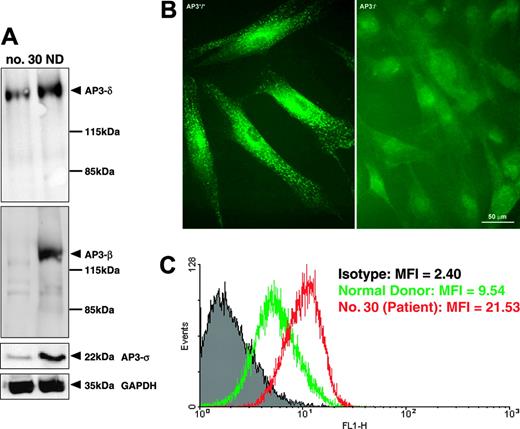
To assess the biologic significance of an exon 15 deletion in AP3B1, we analyzed the expression of AP3B1 on the RNA and protein levels. Using quantitative Lightcycler reverse transcriptase (RT)-PCR analysis, we documented that the levels of AP3B1 mRNA did not differ between patients and controls (not shown). However, intact assembly of the AP-3 complex was severely disturbed. Protein extracts from fibroblasts obtained from patient no. 30 showed decreased concentrations of the AP-3 β, δ, and σ subunits when compared with fibroblasts from a healthy donor (Figure 4A). Furthermore, AP3B1-deficient fibroblasts did not react with anti-δ polyclonal antibodies in immunofluorescence studies (Figure 4B), suggesting that the deletion of AP3B1 exon 15 prevents proper assembly of the AP-3 complex.
Deficient assembly and function of AP-3 complex. (A) Western blot analysis of fibroblast protein extracts showing decreased concentrations of AP-3 β, δ, and σ subunits. (B) Immunofluorescence analysis demonstrating deficiency of the δ subunit of the AP-3 complex in fibroblasts from patient no. 30 (right) compared with fibroblasts from a healthy control (left). (C) FACS analysis of fibroblasts stained for cell-surface expression of CD63 (LAMP3) revealed increased expression of CD63 on fibroblasts from patient no. 30 (red line) compared with control (green line). The mean fluorescence intensity (MFI) is indicated for the isotype control (gray), healthy donor, and patient no. 30.
Deficient assembly and function of AP-3 complex. (A) Western blot analysis of fibroblast protein extracts showing decreased concentrations of AP-3 β, δ, and σ subunits. (B) Immunofluorescence analysis demonstrating deficiency of the δ subunit of the AP-3 complex in fibroblasts from patient no. 30 (right) compared with fibroblasts from a healthy control (left). (C) FACS analysis of fibroblasts stained for cell-surface expression of CD63 (LAMP3) revealed increased expression of CD63 on fibroblasts from patient no. 30 (red line) compared with control (green line). The mean fluorescence intensity (MFI) is indicated for the isotype control (gray), healthy donor, and patient no. 30.
It has been previously shown that the AP-3 complex interacts with the tyrosine-based signals of the tetraspanin molecule CD63 and that in the absence of AP3B1 lysosomal missorting leads to increased cell-surface expression of CD63.12 In line with previous studies, we documented that CD63 expression at the cell surface was enhanced (Figure 4C), thus strengthening the concept that a deletion of exon 15 of AP3B1 functionally compromises the AP-3 complex. Taken together, these data suggest that exon 15 of AP3B1 is critically involved in the stabilization and function of the AP-3 complex.
Neutrophil function
We reasoned that a defective lysosomal compartment might contribute to neutropenia and potentially neutrophil dysfunction. We performed transmission electron microscopy on purified neutrophils from our patients and healthy donors. Compared with healthy donors, AP-3-deficient neutrophil granulocytes showed uniformly marked alterations in azurophilic and specific granules. In contrast to a distinct compartmentalization of specific and azurophilic granules in normal neutrophils, AP-3-deficient neutrophils showed a disorganized pattern characterized by distorted shapes and a less well-defined density spectrum of their granules (Figure 5A-B). To assess the function of neutrophil granulocytes, we focused our investigations on chemotaxis, phagocytosis, and killing of ingested bacteria. First, we isolated peripheral blood neutrophils from patient no. 30 and from 2 healthy control donors by gradient centrifugation and subjected the cells to a transwell migration assay. After 30 and 60 minutes of incubation time, cells in the lower well were harvested, and the percentage of cells migrating toward an fMLP gradient was determined by FACS. Although somewhat delayed, AP-3-deficient neutrophils migrated as efficiently toward the chemotactic stimulus as neutrophils obtained from the 2 healthy donors (Figure 5F).
Ultrastructural and functional analysis of neutrophil granulocytes. (A-B) Transmission electron microscopy (TEM) of representative native neutrophil granulocytes from a healthy individual (ND; A) and patient no. 30 (B). The neutrophils of the patient (B) contained higher amounts of granules, demonstrating high degrees of variability in size, shape, and electron density. Immature granules are indicated by arrows. (C-D) TEM of in vitro-differentiated neutrophil granulocytes on exposure to Pseudomonas bacteria. No difference is seen in phagolysosome formation and bacterial degradation in granulocytes from the healthy individual (C) compared with patient (no. 30; D). (E) Western blot analysis of peripheral blood neutrophils obtained from 2 healthy control individuals with and without prior G-CSF treatment and patient no. 30. Complete blots are shown in Figure S1. (F) Chemotaxis assay of peripheral blood neutrophils from 2 healthy control donors (ND 1 and ND 2) and patient no. 30. Cells either migrated toward medium without chemoattractant (negative control) or toward medium supplemented with 10-7 M fMLP. (G) Uptake of FITC-labeled K12 E coli by in vitro-differentiated neutrophils from 2 healthy control donors and patient no. 30. (H) Killing of lactose permease-deficient E coli in peripheral blood neutrophils from 2 control donors (ND 1 and ND 2) and patient no. 30.
Ultrastructural and functional analysis of neutrophil granulocytes. (A-B) Transmission electron microscopy (TEM) of representative native neutrophil granulocytes from a healthy individual (ND; A) and patient no. 30 (B). The neutrophils of the patient (B) contained higher amounts of granules, demonstrating high degrees of variability in size, shape, and electron density. Immature granules are indicated by arrows. (C-D) TEM of in vitro-differentiated neutrophil granulocytes on exposure to Pseudomonas bacteria. No difference is seen in phagolysosome formation and bacterial degradation in granulocytes from the healthy individual (C) compared with patient (no. 30; D). (E) Western blot analysis of peripheral blood neutrophils obtained from 2 healthy control individuals with and without prior G-CSF treatment and patient no. 30. Complete blots are shown in Figure S1. (F) Chemotaxis assay of peripheral blood neutrophils from 2 healthy control donors (ND 1 and ND 2) and patient no. 30. Cells either migrated toward medium without chemoattractant (negative control) or toward medium supplemented with 10-7 M fMLP. (G) Uptake of FITC-labeled K12 E coli by in vitro-differentiated neutrophils from 2 healthy control donors and patient no. 30. (H) Killing of lactose permease-deficient E coli in peripheral blood neutrophils from 2 control donors (ND 1 and ND 2) and patient no. 30.
Next, we evaluated the capacity to ingest bacteria. Neutrophil granulocytes from patient no. 30 and from 2 healthy control individuals were incubated with FITC-labeled Fc-conjugated E coli bacteria. Phagocytosis was measured after 20, 40, and 60 minutes by FACS. As shown in Figure 5G, the patient's neutrophils showed similar phagocytic activity compared with the control cells, suggesting that the Fc-mediated phagocytosis is not perturbed in the absence of the AP-3 complex. To determine the killing activity of AP-3-deficient neutrophils, we used a fluorometric assay, allowing us to visualize β-galactosidase activity on fusion of the phagolysosome. However, AP-3-deficient neutrophils were as effective as control neutrophils in lysing E coli bacteria (Figure 5H). Similar results were seen when the phagolysosomes on ingestion of pathogenic Pseudomonas bacteria were compared between normal and AP-3-deficient neutrophils by means of electron microscopy (Figure 5C-D). Thus, despite marked alterations in the lysosomal organelles, no defective neutrophil function could be documented.
In neutropenic dogs, mutations in the AP3B1 gene cause perturbed intracellular trafficking and reduced abundance of neutrophil elastase.3 To assess the global expression pattern of neutrophil granule contents, we performed a series of Western blots, comparing cell extracts from 1 patient (no. 30) and from 2 healthy controls, respectively. As shown in Figure 5E, the patient's cells showed comparable levels of myeloperoxidase and proteinase-3 levels, while abundance of neutrophil elastase and matrix metalloproteinase 9 (gelatinase) was reduced.
Discussion
We here describe 2 novel patients with HPS2 and determine, for the first time, a homozygous genomic exon deletion as the molecular cause for this rare disorder. All previously reported human patients as well as the affected dogs have nonsense mutations. Furthermore, in contrast to the previously published 4 cases, our patients show novel clinical features such as splenomegaly, thrombocytopenia, and developmental delay, suggesting that the clinical spectrum of AP-3 deficiency might be more variable than previously described. Another interesting novel feature of our patients is the paucity of NK and NKT cells. Finally, we provide evidence that neutrophil function appears to be intact despite disorganized lysosomal compartments.
The deletion of exon 15 completely abrogates proper assembly and/or stability of the AP-3 complex. It has previously been shown in mice that complete deficiency of β3A results in complete lack of μ3, the AP-3 subunit to which β3A binds directly,5 and in reduced levels of the δ and σ subunits.10 Similarly, human patients with HPS2 characterized by compound heterozygous mutations resulting in residual AP3B1 expression or in complete β3A deficiency showed decreased levels of σ3 and δ3,13 suggesting that normal β3A chain is critically important to stabilize the heterotetrameric AP-3 complex. Peden et al7 have recently undertaken a study to delineate functional domains controlling protein configurations in the AP-3 complex by using a complementation assay in β3A-deficient cells obtained from the pearl mouse. Whereas the adaptin domain is important for assembly into heterotetrameric complexes, the C-terminal hinge and ear domains mediate the function of AP-3.7
Therefore, the μ chain could not be detected by Western blotting, whereas the σ and δ subunits appeared reduced. Thus, our patients provide an “in vivo” model of an adaptin domain deletion rendering the AP-3 β chain incapable of forming the AP-3 complex.
AP-3 is critically involved in the biogenesis of specialized endosomal organelles referred to as lysosome-related organelles.31,32 Previous in vitro experiments have shown that in AP-3-deficient melanosomes, tyrosinase accumulates inappropriately in vacuolar and multivesicular endosomes, thus preventing the full maturation of melanosomes.33,34 Here, we provide evidence that the incomplete maturation of melanosomes is seen in humans in vivo. Immature melanosomes are degraded in large phagolysosomes of basal keratinocytes, resulting in hypopigmented skin and retina.
Neutrophil granulocytes represent a cellular compartment that contains a specialized lysosomal compartment. On engulfing exogenous particles, phagosomes are destined to fuse with lysosomes to degrade phagocytosed material. So far, the role of AP-3 in the formation of phagolysosomes has not been addressed. Interestingly, AP-3 deficiency leads to neutropenia in humans (congenital neutropenia)12 and dogs (cyclic neutropenia)3 but not in mice.10 The reason for this species specificity remains unknown. Furthermore, the exact pathophysiology of neutropenia in AP-3 deficiency is only beginning to be unraveled. Benson3 has presented an elegant hypothesis that offers an interesting interpretation of the puzzling question of why AP-3-deficient individuals might have neutropenia. According to this model, neutrophil elastase is missorted to the plasma membrane, thereby mediating deleterious effects. Interestingly, analysis of the AP-3-deficient neutrophils revealed decreased abundance of neutrophil elastase and gelatinase, a protein usually found in specific granules, whereas other granule proteins such as proteinase 3 and MPO were unaffected. These findings suggest that distinct granule proteins might be involved in the pathogenesis of neutropenia in AP-3-deficient patients. Further studies are needed to characterize their role in greater detail.
Although we provide ultramorphologic evidence of disordered vesicular transport, we were unable to define unequivocally any functional compromise in the 2 lysosomal compartments in neutrophil granulocytes, using both in vitro-generated neutrophils and primary peripheral blood neutrophils. These observations are consistent with the relatively mild clinical course of our patients. In contrast to other groups of patients with (severe) congenital neutropenia, AP-3-deficient patients have not had life-threatening infections, suggesting that the function of residual neutrophils could partially compensate for innate immunity.
A surprising finding was the persistent paucity of NK and NKT cells in our patients. Studies in the mutant pearl mouse have shown that, as a consequence of AP-3 deficiency, the generation of NKT cells is severely affected.35,36 This can be explained by perturbed CD1d-mediated antigen presentation,37 an immunologic mechanism evolved to present the universe of lipid antigens to T cells. In humans, however, AP-3 binds to CD1b38 but does not bind to CD1d.36 Thus, it is puzzling to explain deficient CD1d-dependent NKT-cell development. We propose that secondary mechanisms might account for decreased NKT-cell numbers in our patients. To date, the only other molecularly defined human disease in which NKT cells are missing is SAP deficiency,39,40 also known as X-linked lymphoproliferative syndrome (XLP; OMIM catalog no. 308240). Both knockout mice and human patients with XLP have an almost complete absence of detectable NKT cells.39,40 Whereas SAP-deficient patients show a propensity to fatal EBV infections, the 4 previously published patients with HPS2 and the 2 novel cases in our study were not particularly prone to viral infections, suggesting that NKT cells may not play a critical role in antiviral immune defense.
In summary, our report extends the clinical and molecular phenotype of HPS2 and provides further insights into the role of the AP-3 complex for the innate immune system.
Prepublished online as Blood First Edition Paper, March 14, 2006; DOI 10.1182/blood-2005-11-4377.
Supported by the Deutsche Forschungsgemeinschaft (grants KFO110 and GR1617/3), the Deutsche Jose-Carreras Stiftung, the Bundesministerium für Bildung und Forschung, and the Intramural Research Program at the National Institutes of Health, National Library of Medicine.
J.J., G.B., A.A., B.G., and C.K. contributed equally to this study.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Dr A. Jobke for outstanding support; Dr Martin Zimmermann for statistical advice; E. Mallon, A. Hundt, and G. Preiss for their technical support; and the family for their collaboration.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal