Loss of major histocompatibility class II (MHC II) expression in diffuse large B-cell lymphoma (DLBCL) correlates with worse outcome, possibly from decreased immunosurveillance. Primary mediastinal B-cell lymphoma (PMBCL) is a subtype of DLBCL which reportedly has frequent loss of MHC II proteins; however, PM-BCL has better survival than DLBCL. To investigate this paradox, we used geneexpression profiling (GEP) data and immunohistochemistry to study expression of MHC II and its regulatory genes and to determine their relationship to PMBCL survival. We found that GEP levels correlated between MHC II genes and the transcriptional regulator MHC2TA but not other adjacent genes, implying that transcriptional regulation of MHC II in PMBCL was intact and that MHC II gene deletion was unlikely. MHC II average expression was lower than in certain subtypes of DLBCL; however, only 12% had complete loss of MHC II expression. Poor patient survival in PMBCL correlated with incremental decreases in MHC II expression. Although overall survival was better, survival of the lowest 10% of MHC II expressers was similarly poor in DLBCL and PMBCL. MHC II expression may define a therapeutic target in both these diseases. (Blood. 2006;108:311-318)
Introduction
Primary mediastinal large B-cell lymphoma (PMBCL) is considered to be a subset of diffuse large B-cell lymphoma (DLBCL), characterized by predominant mediastinal involvement, large B cells with clear cytoplasm, convoluted or centroblastic nuclei, and thin bands of compartmentalizing sclerosis.1 Immunophenotypically, PMBCL is unusual among mature B-cell neoplasms in that, despite expression of B-cell surface antigens, including CD20 and CD79a, and IgH rearrangements and mutations, PMBCL cells usually lack surface IgG. According to the World Health Organization's classification, they also exhibit loss of expression of the major histocompatibility class II (MHC II) proteins.1,2 Unlike DLBCLs, PMBCLs frequently overexpress MAL, an integral membrane protein usually found in lipid rafts, which is also expressed in certain normal thymic medullary B cells, from which PMBCLs may be derived.3 Cases of PMBCL also tend to exhibit genetic abnormalities involving chromosomal gains at 9p, associated with increased expression of JAK2 at 9p24, and 2p, associated with the REL locus at 2p16.4,5 Unlike DLBCLs, PMBCLs seldom exhibit BCL2 or BCL6 rearrangements or translocations.6
Patients with PMBCL are typically younger than patients with other subtypes of DLBCL and may show female predominance. The disease is aggressive and can involve multiple thoracic structures (lung, pericardium, breast) but is less likely than other DLBCLs to extend to extrathoracic regions.7 As a subset of DLBCL, PMBCL is currently treated similarly, with adriamycincontaining combination chemotherapy regimens and monoclonal antibody therapy. Involved-field radiation therapy is often added to debulk localized disease.8 Survival and responses to treatment of PMBCL have been controversial, possibly because of confusion of this disease with other subtypes of DLBCL arising in the mediastinal area. Some studies have reported substantially better outcome for PMBCL (82% at 3 years),9 and some have reported survivals that are very similar to or worse than other forms of DLBCL (approximately 50% at 5 years).10
Two studies, one at Dana-Farber Cancer Institute,11 and one at the NCI by the Leukemia and Lymphoma Molecular Profiling Project (LLMPP),7 have reported results of microarray geneexpression profiling showing PMBCL to be a unique entity, separable from other subgroups of DLBCL. Although in many ways resembling the germinal centerlike subtype of DLBCL, PMBCL also had increased expression of a number of genes characteristically expressed in Hodgkin lymphoma. The DanaFarber study also noted lower expression of specific MHC II genes in PMBCL, in agreement with previous immunohistochemical studies.11
MHC II genes are a family of genes mostly expressed from a single locus on chromosome 6p.12 These genes are constitutively expressed in antigen-presenting cells, such as B cells, dendritic cells, monocytes, and macrophages, with inducible expression in many other cell types. The proteins deriving from these genes present peptide antigens derived from exogenous proteins to CD4+ T cells. MHC II proteins are divided into 2 types, classic and nonclassic. Classic proteins consist of HLA-DR, -DP, and -DQ, each of which are heterodimers of an alpha and beta protein, encoded by A and B genes.13 Classic proteins are directly responsible for displaying peptide antigens on the cell surface. Nonclassic proteins assist in and modulate the function of the classic proteins and consist of HLA-DM, -DO, and invariant chain (Ii, CD74). HLA-DM and -DO are also heterodimers similar to the classic proteins. Invariant chain, unlike the other MHC II proteins, is expressed from a gene on chromosome 5q.
A large study of DLBCL by the LLMPP has determined that specific MHC II genes constitute 1 of 4 prognosis-related gene signatures.14 The MHC class genes all have similar sites in their regulatory regions for the cooperative binding of a transcriptosome complex of RFX, NF-Y, and CREB. This complex in turn creates a protein-binding site for CIITA, which does not bind DNA directly.13 CIITA (encoded by the MHC2TA gene) serves as a master transactivator of the MHC II genes. CIITA expression is usually highly correlated with MHC II expression in most normal cell types where it is expressed. Loss of functional expression of any of the 3 genes encoding the subunits of the heterotrimer RFX or of the MHC2TA gene encoding CIITA leads to elimination of all MHC II expression, as evidenced in the congenital immunodeficiency diseases known as the MHC II deficiency syndromes.15
Loss of expression of MHC II genes is reportedly a common event in DLBCL involving immune-privileged sites like testes, ovary, eye, or brain.16 Immune-privileged DLBCL (IP-DLBCL) is relatively rare, is different clinically and phenotypically from other DLBCLs, and occurs most commonly in patients who are immunodeficient. It has been reported that IP-DLBCL of brain or testis shows loss of expression of MHC II genes in more than half the cases, and that this loss of expression is often accompanied by large homozygous or heterozygous deletions of the MHC II locus.16,17 In the same reports it was found that non-IP-DLBCL lost MHC II expression in 5% of cases without accompanying deletions of the 6p locus. In agreement with these findings, we have recently reported that complete loss of MHC II expression occurs in only about 10% of DLCBL in a data set of 240 de novo DLBCL and found evidence for MHC II locus deletions in less than 1%, although all patients with MHC II expression below the median had a poorer outcome.18,19 Because the mechanism for loss of MHC II expression in DLBCL does not usually appear to involve loss of the genes themselves, but probably genetic or epigenetic regulation, it may be possible to restore the expression of these genes.
Another possible mechanism of loss of MHC II expression may be specific to PMBCL. Loss of functional expression of the signaling molecule SOCS1, a gene located on chromosome 16p13, in the near vicinity of MHC2TA, was shown in 9 of 20 primary PMBCL samples and 2 of 2 PMBCL cell lines.20,21 Although these losses of SOCS function were shown in all but 1 case to be due to small insertions or deletions within the SOCS gene, 1 PMBCL cell line, Karpas 1106, had a biallelic deletion of approximately 700 Kb (kilobase), in the region including SOCS1.21 Although this specific deletion did not include MHC2TA, deletions of this size involving SOCS1 certainly could, because the MHC2TA gene is within 400 Kb of SOCS1.
The purpose of the present work was to study the patterns of expression of MHC II and regulatory genes in PMBCL, relate these patterns to protein expression and possible deregulation with neoplasia, and determine how these patterns are related to patient survival.
Patients, materials, and methods
PMBCL and DLBCL clinical, gene-expression profiling, and comparative genomic hybridization data
In 2003, the LLMPP published a report on the development of a molecular PMBCL predictor for distinguishing PMBCL from other types of DLBCL.7 This paper used a training set of 35 cases and a validation set of 274 cases of DLBCL, for which gene expression data from the Lymphochip microarray were available. Lymphochip type II microarray data consist of relative gene expression derived from the ratio of fluorescence of sample mRNA to a reference mRNA made from multiple pooled cell lines. The Lymphochip probes consist of more than 12 000 cDNA expressed sequence tags (ESTs) derived from a number of libraries enriched for genes related to lymphoid cells or believed to play a role in cancer or immune function.22 We used a subset of this data consisting of de novo untreated cases only, for which clinical information was available. Forty-two cases were classified as PMBCL by the molecular predictor (23 from the training set and 19 from the validation set) and 221 cases were classified as other types of DLBCL. Thirty of the 42 cases classified as PMBCL were originally clinically considered to be PMBCL, with mediastinal presentation, and the rest were originally considered DLBCL, but were classified as PMBCL by geneexpression predictor score. These included 82 cases of activated B-cell-like (ABC) DLBCL, 95 cases of germinal center B-cell-like (GCB) DLBCL, and 44 cases of DLBCL that could not be identified with either group (unclassified). In addition, there were 9 samples of different types of normal B cells. For the purposes of this paper, our definition of PMBCL is that of the molecular predictor.
The cases from the training and validation sets in the LLMPP PMBCL paper7 had been compared with different reference pools. Therefore, it was first necessary to adjust for the differences between the reference pools. The 2 sets originally consisted of log2 transformed fluorescence ratios between each sample and reference, representing different ESTs, normalized so that the median of the well-measured spots was set to 0. To normalize the test set to the validation set, 25 DLBCLs from the validation set were rearrayed using the new reference. For each gene, we considered the pairwise difference between the log2 ratios of these samples with the old and new references. We then subtracted the median of these differences from all of the log2 ratios of that gene on all of the training set data.
ESTs representing the MHC II family genes, MHC2TA, RFX, and genes located near or within the MHC II locus on chromosome 6p21 and the MHC2TA location on chromosome 16p13 were sought in the microarray database. Table 1 is a list of these genes and their location relative to the MHC II genes or to MHC2TA. Expression data for all the subunits of RFX and HLA-DO were not found, so these genes were excluded from further analysis. Multiple ESTs representing many of the genes were found in the data. In the case of multiple representations of a single gene, all of the ESTs for that gene were averaged to a single gene expression value. Overall, average MHC II expression was defined as the average of the expressions of HLA-DRA, -DRB, -DPA, -DPB, -DQA, -DQB, -DMA, -DMB, and CD74. Data for Figure 4 was median-centered.
Genes in the vicinity of the MHC class II or MHC2TA loci represented in GEP data
Gene . | Position relative to locus . |
|---|---|
| MHC class II locus | |
| HSPA1L | −597 Kb from HLA-DRA |
| HSPA1A | −594 Kb from HLA-DRA |
| BAT8 | −532 Kb from HLA-DRA |
| RDBP | −453 Kb from HLA-DRA |
| CREBL 1 | −317 Kb from HLA-DRA |
| PBX2 | −255 Kb from HLA-DRA |
| TAP2 | Between HLA-DQA and DMB |
| TAP1 | Between HLA-DQA and DMB |
| PSMB9 | Between HLA-DQA and DMB |
| BRD2 | Between HLA-DMA and DPA |
| RXRB | +77 Kb from HLA-DPB |
| RING1 | +85 Kb from HLA-DPB |
| RPS18 | +148 Kb from HLA-DPB |
| TAPBP | +186 Kb from HLA-DPB |
| DAXX | +195 Kb from HLA-DPB |
| BAK1 | +452 Kb from HLA-DPB |
| MHC2TA locus | |
| ABAT | −2203 Kb from MHC2TA |
| USP7 | −1914 Kb from MHC2TA |
| PRO0149 | −1786 Kb from MHC2TA |
| MHC2TA | 0 Kb from MHC2TA |
| SOCS1 | +379 Kb from MHC2TA |
| LITAF | +709 Kb from MHC2TA |
| GSPT1 | +1039 Kb from MHC2TA |
| TNFRSF17 | +1088 Kb from MHC2TA |
Gene . | Position relative to locus . |
|---|---|
| MHC class II locus | |
| HSPA1L | −597 Kb from HLA-DRA |
| HSPA1A | −594 Kb from HLA-DRA |
| BAT8 | −532 Kb from HLA-DRA |
| RDBP | −453 Kb from HLA-DRA |
| CREBL 1 | −317 Kb from HLA-DRA |
| PBX2 | −255 Kb from HLA-DRA |
| TAP2 | Between HLA-DQA and DMB |
| TAP1 | Between HLA-DQA and DMB |
| PSMB9 | Between HLA-DQA and DMB |
| BRD2 | Between HLA-DMA and DPA |
| RXRB | +77 Kb from HLA-DPB |
| RING1 | +85 Kb from HLA-DPB |
| RPS18 | +148 Kb from HLA-DPB |
| TAPBP | +186 Kb from HLA-DPB |
| DAXX | +195 Kb from HLA-DPB |
| BAK1 | +452 Kb from HLA-DPB |
| MHC2TA locus | |
| ABAT | −2203 Kb from MHC2TA |
| USP7 | −1914 Kb from MHC2TA |
| PRO0149 | −1786 Kb from MHC2TA |
| MHC2TA | 0 Kb from MHC2TA |
| SOCS1 | +379 Kb from MHC2TA |
| LITAF | +709 Kb from MHC2TA |
| GSPT1 | +1039 Kb from MHC2TA |
| TNFRSF17 | +1088 Kb from MHC2TA |
Positional differences calculated from the 5′ ends of the genes compared to the 5′ end of the nearest MHC class II gene or MHC2TA; positive numbers indicate centromeric direction.
Chromosome comparative genomic hybridization (CGH) data from 16 of the cases23 were down-loaded from http://www.ncbi.nlm.gov/projects/sky/.
Tissue immunohistochemistry
Tissue sections or tissue microarray cores from paraffin-embedded tissues were available from 17 PMBCL samples from our data set, for our use in validating the gene expression results. Approval was obtained from the University of Arizona Institutional Review Board for these studies. Informed consent was provided in accordance with the Declaration of Helsinki. Because the material was limited, we chose to use a single, representative MHC II molecule for staining. HLA-DR was chosen because it has been specifically related to survival in previous publications,24 because it is by far the most highly expressed of the MHC II family, and because it has a readily available antibody (immunoglobulin G2b [IgG2b]; Novocastra, Newcastle upon Tyne, United Kingdom). The antibody, at a 1:50 dilution, was optimized by epitope-recovery methods on an automated immunostainer (Ventana Benchmark System; Ventana Medical Systems, Tucson, AZ) with a biotin-avidin-diaminobenzidine-based detection system. Samples were counterstained with hematoxylin.
Immunohistochemistry results were initially examined by 2 hematopathologists (L.M.R. and M.A.J.) and a consensus counting methodology agreed on after an initial training session using a multiheaded microscope. Samples were then counted by 1 of the 2 pathologists. As in our previous studies, an area of minimum intensity staining within the tumor was chosen, with the rationale that areas of tumor which have lost MHC II expression can escape immunosurveillance and are likely to contribute to poor prognosis.19,25 Approximately 200 tumor cells were counted per case, and each cell was scored as negative or positive. The numbers of cells scored as positive were divided by the total cells counted and multiplied by 100 for a percentage positive score. Pathologists performing cell counts were blinded to the GEP results for the samples. Photomicrographs of stained slides were taken on a labophot-2 mocroscope using a 10× eyepiece and a 40×/0.65 objective lens (Nikon, Melville, NY). A SPOT-RT 2.2.0 color camera and SPOT Advanced 4.0.9 software (Diagnostic Instruments, Sterling Heights, MI) were used to capture and digitally acquire images, which were then inserted into PowerPoint 10 (Microsoft, Redman, WA) for processing.
Data analysis
Overall survival was estimated by the method of Kaplan and Meier. Tests of differences in survival between groups were by the log-rank test. These were done on Statview software (version 4.5; Abacus Concepts, Berkeley, CA). Pearson correlations, t tests for determining significance of differences in gene expression, and other statistical and mathematic manipulations of the data were done with Excel software (version 10; Microsoft).
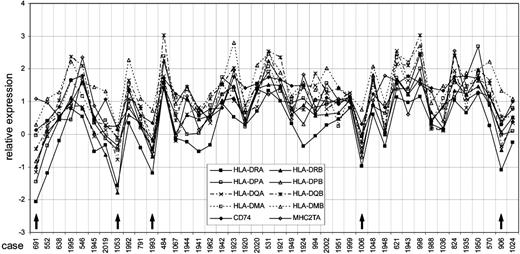
Coordinated expression of the MHC class II genes and CIITA in 42 individual cases of PMBCL. The x-axis shows individual cases by study ID number, and the y-axis shows relative expression values. Cases with substantial, coordinated loss of MHC class II expression are marked with ↑. Cases are sorted as follows: from the left, patients who did not survive are plotted sorted by survival time, then patients who survived are plotted by follow-up time. The first surviving case is 1941.
Coordinated expression of the MHC class II genes and CIITA in 42 individual cases of PMBCL. The x-axis shows individual cases by study ID number, and the y-axis shows relative expression values. Cases with substantial, coordinated loss of MHC class II expression are marked with ↑. Cases are sorted as follows: from the left, patients who did not survive are plotted sorted by survival time, then patients who survived are plotted by follow-up time. The first surviving case is 1941.
Results
Coordination of MHC II gene and protein expression
First, we plotted the relative gene expressions of the individual MHC II genes and MHC2TA (Figure 1). By visual inspection, the expression of the genes was coordinated between the MHC II genes and MHC2TA for cases with both high and low relative expression. In this study 5 of 42 (12%) PMBCL cases have substantial, coordinate loss of expression of all of the MHC II genes, which is similar to what we previously reported in DLBCL.19 In all these cases of loss, except possibly case 691, coordination of MHC2TA expression with the other genes implies that transcriptional regulation may still be functioning even in low-expressing cases.
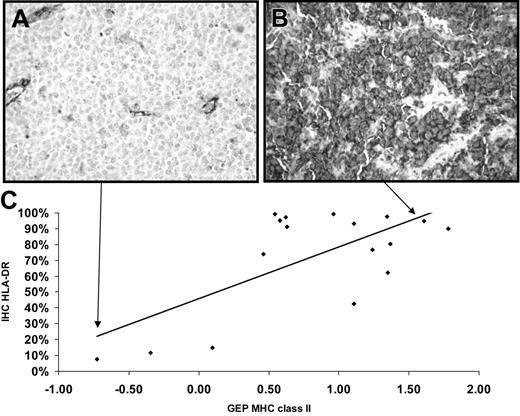
To show that the microarray results were related to cellular display of the proteins, IHC was performed on the subset of PMBCL samples available to us (Figure 2A-B), and results related to the average MHC II expression GEP values (Figure 2C). The Pearson correlation (R) between the percentage of cells positive for protein and gene expression data was 0.68, indicating a reasonably good agreement between microarray expression values and protein surface expression on PMBCL cells.
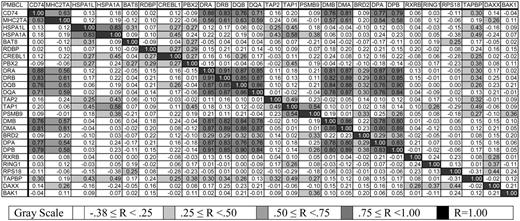
Then, to demonstrate the specificity of coordination of the expression of the MHC II genes and their transcriptional activator MHC2TA, we calculated Pearson correlation coefficients between the MHC II genes and their neighbors on chromosome 6p, CD74/Ii on chromosome 5, and MHC2TA on chromosome 16 (Figure 3). In the figure, highly correlated expression is shown in dark gray, and this color is seen at the intersections of the MHC II genes with each other and with MHC2TA. This analysis demonstrated high correlation coefficients for the classic and nonclassic MHC II genes within the class II locus (range, 0.76-DPA versus DQB or DQA to 0.91-DRA versus DRB or DPB) and between CD74 and MHC II genes (range, 0.71 versus DQA to 0.88 versus DRA). However, correlations between MHC II genes (including CD74) and genes adjacent to or within the MHC II locus were lower and did not overlap (range, -0.12 -DMA versus RING1 to 0.25 -DRB versus DAXX). In addition, correlation between the transactivator gene MHC2TA and any of the MHC II genes, including CD74, was higher (range, 0.54 versus DPA to 0.63 versus DQB or CD74) than between MHC2TA and the genes adjacent to or within the MHC II locus (range, -0.12 versus BAT8 to 0.26 versus DAXX). These results implied that there was no larger global regulation of the MHC II gene locus (eg, extended locus methylation) but that the MHC II genes are specifically regulated in our PMBCL data set.
Possibility of large deletions of DNA as explanation of MHC II loss
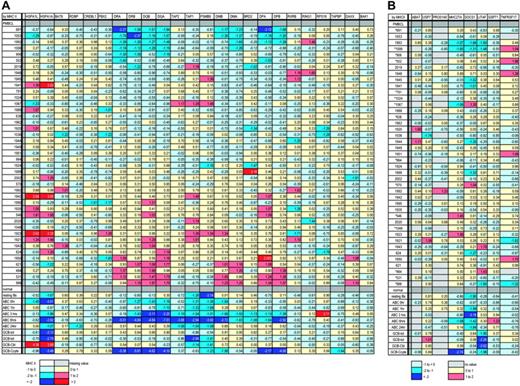
Because large genetic deletions in the MHC II locus have been implicated in the loss of MHC II expression in IP-DLBCL, we made a positional expression profile26 of the expression of genes located near or within about 500 Kb of the MHC II locus, for which we had GEP data (Figure 4A). Relative locations of these genes are listed in Table 1. Our rationale was that if genes located near or between the MHC II genes which have been down-regulated were still being expressed at levels similar to or higher than those in normal cells in the individual cases, large deletions probably did not exist within the locus in those cases.18 The data show that in the 5 cases with lowest MHC II expression as well as in the other cases the expression of genes between and around the MHC II genes was not decreased, implying that the MHC II genes are not down-regulated by large deletions in our PMBCL cases.
Immunohistochemical detection of HLA-DR and relationship to gene expression. (A) Photomicrograph demonstrating a low HLA-DR-expressing PMBCL case, case 691. Staining of endothelial cells and some histiocytes serves as an internal positive control. (B) Photomicrograph demonstrating a high HLA-DR-expressing PMBCL case, case 824. Original magnification for both × 100. (C) Graph of the relationship between results of immunohistochemistry (IHC) of HLA-DR in percentage of positive tumor cells (y-axis) and average MHC class II gene expression from microarray results (x-axis). Pearson correlation R = 0.68. A trend line is shown to help judge the extent of correlation. Arrows show where the cases pictured above are located in the graph.
Immunohistochemical detection of HLA-DR and relationship to gene expression. (A) Photomicrograph demonstrating a low HLA-DR-expressing PMBCL case, case 691. Staining of endothelial cells and some histiocytes serves as an internal positive control. (B) Photomicrograph demonstrating a high HLA-DR-expressing PMBCL case, case 824. Original magnification for both × 100. (C) Graph of the relationship between results of immunohistochemistry (IHC) of HLA-DR in percentage of positive tumor cells (y-axis) and average MHC class II gene expression from microarray results (x-axis). Pearson correlation R = 0.68. A trend line is shown to help judge the extent of correlation. Arrows show where the cases pictured above are located in the graph.
Pearson correlation coefficients between average gene expression of MHC class II genes, adjacent and intervening genes, MHC2TA, and CD74. In no case is R less than -.38.
Pearson correlation coefficients between average gene expression of MHC class II genes, adjacent and intervening genes, MHC2TA, and CD74. In no case is R less than -.38.
Another way that the MHC II genes could be coordinately down-regulated would be by deletion of the MHC2TA gene, which is the master transactivator for all MHC II expression. A recent report of a homozygous deletion found in the vicinity of the MHC2TA gene at the SOCS1 locus21 prompted us to make a positional expression profile for the genes near the MHC2TA locus as well. Genes on chromosome 16p13 near MHC2TA were much sparser in the microarray data than genes in the MHC II locus, but those found were mapped in Figure 4B. Of the lowest expressing cases of PMBCL, shown at the top of the map, only cases 1006 and 1993 show substantial down-regulation of both SOCS1 and MHC2TA, indicating the possibility of a large deletion, including MHC2TA. It is difficult to extrapolate the possibility of an MHC2TA deletion from the low expression of the SOCS1 gene, because it is expressed at very low levels in most of the normal B cells as well as in the PMBCL cases. However, most of the samples do not show coordinate down-regulation of MHC2TA and adjacent genes, minimizing the possibility of large deletions in the MHC2TA locus in most of the PMBCL cases.
Sixteen of the PMBCL cases (Figure 4) had published CGH data available, and no large deletions in either the MHC II at 6p or the MHC2TA locus at 16q were found by this technique, including for case 1006.
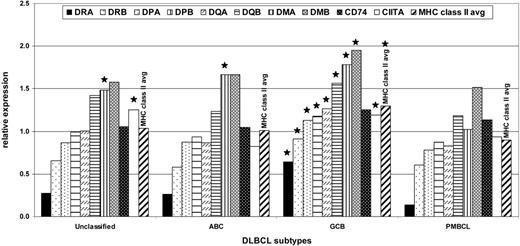
Expression of MHC II genes in DLBCL subtypes
Because it has been reported that PMBCL shows frequent loss of MHC II expression compared with other types of DLBCL,11 we analyzed the average relative expression of MHC II genes in PMBCL compared with the average relative expression of these genes in other types of DLBCL (Figure 5). PMBCL had significantly lower expression of all of the individual MHC II genes except CD74, and lower expression of the MHC II average, than the GCB subtype, which is the most common subtype of DLBCL. Expression of most of the MHC II genes was not significantly different in PMBCL than in the ABC subtype and unclassified DLBCL cases. Only HLA-DMA expression in PMBCL was significantly lower in all the subtypes. The average MHC II gene expression of the normal cells was extremely variable (see Figure 4A, bottom), and there were few or no replicates of the normal cell types, so it was not possible to say with confidence how the expression of the MHC II genes in any of the DLBCL subtypes compared with normal cells.
Positional expression profiling of the MHC class II and MHC2TA loci. (A) Positional expression profiling map showing relative expression of MHC class II genes and those genes physically located telomeric, within, and centromeric to the MHC class II locus. (B) Positional expression profiling map showing relative expression of MHC2TA and those genes physically located near the MHC2TA locus. For both sections, asterisks indicate cases with CGH data available, and color coding is used to indicate the relative expression of genes. Each row indicates one of the PMBCL cases, by individual patient study number, sorted by average MHC class II expression, from low to high. The last 9 rows are activated B cells (ABC, with hours of activation indicated) and germinal center B cells (GCB). Tot indicates total from a normal lymph node; Cbl, centroblastic; and Ccyte, centrocytes from healthy individuals. MHC class II genes are shown with dots. Gene expression (e) is colored as follows: dark blue, e <-2; medium blue, -2 ≤ e <-1; light blue, -1 ≤ e < 0; yellow, 0 ≤ e < 1; orange, 1 ≤ e < 2; red, e ≥ 2; light gray squares pattern, no value.
Positional expression profiling of the MHC class II and MHC2TA loci. (A) Positional expression profiling map showing relative expression of MHC class II genes and those genes physically located telomeric, within, and centromeric to the MHC class II locus. (B) Positional expression profiling map showing relative expression of MHC2TA and those genes physically located near the MHC2TA locus. For both sections, asterisks indicate cases with CGH data available, and color coding is used to indicate the relative expression of genes. Each row indicates one of the PMBCL cases, by individual patient study number, sorted by average MHC class II expression, from low to high. The last 9 rows are activated B cells (ABC, with hours of activation indicated) and germinal center B cells (GCB). Tot indicates total from a normal lymph node; Cbl, centroblastic; and Ccyte, centrocytes from healthy individuals. MHC class II genes are shown with dots. Gene expression (e) is colored as follows: dark blue, e <-2; medium blue, -2 ≤ e <-1; light blue, -1 ≤ e < 0; yellow, 0 ≤ e < 1; orange, 1 ≤ e < 2; red, e ≥ 2; light gray squares pattern, no value.
Graph of the relative expression of the MHC class II genes, MHC2TA, and the average MHC class II gene expression in the different types of DLBCL. Bars topped with a star indicate that the value represented is significantly different (P < .05) from the corresponding value for PBMCL.
Graph of the relative expression of the MHC class II genes, MHC2TA, and the average MHC class II gene expression in the different types of DLBCL. Bars topped with a star indicate that the value represented is significantly different (P < .05) from the corresponding value for PBMCL.
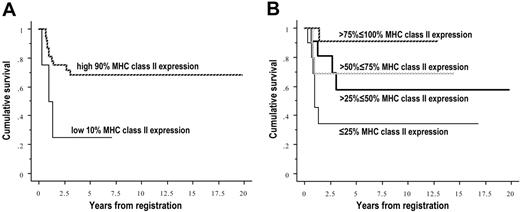
MHC II gene expression and survival and clinical features
Figure 6 shows Kaplan-Meier plots of cumulative survival versus MHC II average gene expression. The left side shows the low 10% of MHC II gene expressers versus the rest of the cases. This division was used in order to be able to compare the low 10% of PMBCL cases with our previous data from the low 10% of other DLBCLs.27 The right side shows 4 approximately equal groups of increasing gene expression, to show that survival increases significantly and incrementally with increasing gene expression. The 5-year survival estimates for plot A on the left were 68% for those patients in the greater than 10% to 100% expression level versus 25% for patients in the lowest 10% expression levels. The 5-year estimates for survival for plot B on the right were 91% in the greater than 75% to 100% group, 69% in the greater than 50% to 75% group, 58% in the greater than 25% to 50% group, and 34% for those patients in the lowest 25% of expression.
Kaplan-Meier plots of survival by average MHC class II gene expression in PMBCL. (A) Cases were divided into the low 10% expressers (n = 4) versus the rest of the cases (n = 38). (B) Cases were divided into 4 approximately equal groups by expression (n = 10, 11, 10, 11 for increasing expression). Survival increases significantly and incrementally with increasing expression, P = .041 for the left graph, P = .039 for the right.
Kaplan-Meier plots of survival by average MHC class II gene expression in PMBCL. (A) Cases were divided into the low 10% expressers (n = 4) versus the rest of the cases (n = 38). (B) Cases were divided into 4 approximately equal groups by expression (n = 10, 11, 10, 11 for increasing expression). Survival increases significantly and incrementally with increasing expression, P = .041 for the left graph, P = .039 for the right.
Because there was insufficient clinical data collected for the majority of the samples to calculate the International Prognostic Index (IPI), it was not possible to compare the prognostic value of the MHC II genes with the IPI to determine whether MHC II expression was an independent survival predictor. The IPI is composed of 5 risk factors which each can be given a score of 0 or 1, the lower values corresponding to 0 in all cases; the risk factor scores are summed to equal the IPI.28 The risk factors are stage (1 or 2 versus 3 or 4), age (≤ 60 versus > 60), LDH (lactate dehydrogenase) levels (≤ normal versus > normal), performance status (0 or 1 versus ≥ 2), and number of extranodal sites (≤ 1 versus > 1). Information was available for all of the risk factors except the number of extranodal sites. We therefore calculated the values for all of the individual risk factors possible and found that these were not individually significantly predictive of survival (data not shown). Then we calculated a partial IPI score, consisting of the sum of the 4 risk factor scores for which we had clinical information, and this was not significantly predictive of survival either (data not shown), whether divided into halves (0 or 1 versus 2 or 3) or into the 4 individual numbers.
We found no clinical differences between the lower and higher MHC II expressers except survival, using the limited clinical data. There were no significant differences with decreasing MHC II expression in location (North America versus Europe), performance status, LDH level, sex, or stage. Among the 4 patients in the lowest 10% of MHC II expression, there were 2 women and 2 men. Two were diagnosed in the third decade of life, one in the fourth, and one in the sixth. One patient was in stage II, 1 in stage III, and 2 in stage IV. The serum LDH level was above the upper limit of normal for 2 patients and within normal limits in the other 2 patients. Two patients each were from the United States and Europe.
Discussion
In this study, we have demonstrated that loss of MHC II molecules is not characteristic of PMBCL and is similar to non-GCB subtypes of DLBCL. However, when loss of expression does occur, those cases have the same extremely poor clinical outcome as MHC II-negative DLBCL. Using positional expression profiling and CGH, we did not find evidence of large deletions at the MHC II or CIITA loci, implying that the genes are intact and may be targets for therapeutic manipulation.
Similar to what we previously reported in DLBCL, about 10% of the cases of PMBCL appeared to have substantial loss of expression of all the MHC II genes by GEP.27 This was in contrast to previous IHC studies which had shown loss of MHC II expression in PBMCL on a much larger percentage of cases, sometimes in a majority of those studied,9,29 but similar to a study showing only 20% of samples losing HLA-DR staining.30 Previous studies of HLA-DR expression on different sets of archival non-PMBCL DLBCL samples, which had undergone different preservation and storage conditions, reported that about 5% and 40% do not stain for this MHC II protein.16,27,31 In fact, the WHO classification of lymphoid neoplasms describes the characteristic immunophenotype of PMBCL as negative for MHC II.1 Our speculation is that GEP was a more sensitive indicator of the presence of lowered levels of MHC II expression than IHC, which appears to have a variable threshold for detection of HLA-DR and tends to overestimate the number of non-MHC II-expressing cases.
The average expression of the MHC II genes in PMBCL cases appeared to be slightly lower than that in the most common subtype of DLBCL, the GCB subtype, and similar to the average expressions of the genes in the ABC subtype, and in the unclassified cases, from the LLMPP data. This was unexpected because the PMBCL subtype most closely resembles the GCB subtype in other GEP respects. In fact, the 19 PMBCL cases from the validation set were all classified as GCB by the original DLBCL predictor, prior to the development of the molecular predictor for PMBCL. Because the differences between average expression of the MHC II genes may be due to differences in MHC II gene expression in the B-cell subsets from which the tumors were derived, it would be interesting to compare the DLBCL data with normal B-cell expression data. However, our data set was limited because it had so few and such variable and highly processed samples of normal B-cell populations to compare our tumor samples with. MHC II gene expression varies during differentiation of the B-cell lineage. MHC II genes are not expressed in pro-B-cell populations and are highly expressed by mature B cells, and their expression is extinguished by down-regulation of expression of CIITA protein during maturation into plasma cells.32 GCB DLBCL cell lines have a different reaction to IL-4 signaling than ABC DLBCL. In GCB DLBCL cell lines, IL-4 induces expression of its known STAT-induced target genes, whereas in ABC DLBCL lines it does not.33 Because MHC II genes are IL-4 target genes in B cells,13 if this difference also occurs in vivo, it could explain the higher expression of MHC II genes in GCB DLBCL cases. It is also possible that the differences in expression between the different DLBCL subtypes are due to a different local environment, although this is unlikely because 5 GCB and 3 ABC samples of DLBCL from mediastinal locations were not significantly different in MHC II expression compared with GCB or ABC DLBCL samples from other sites (data not shown).
Interestingly, HLA-DM appeared to be expressed at a somewhat lower level in the PMBCL subtype than in the other DLBCL subtypes. HLA-DM is involved with modulation of the antigenloading of the classic MHC II proteins, especially the replacement of CLIP (the degradation product of invariant chain which serves to block loading of endogenous antigens on the MHC II classic molecules).13 It would therefore be interesting to know whether the antigens displayed on classic MHC II molecules on the surface of PMBCLs are different from those on other DLBCLs—for instance, if they are more frequently endogenous rather than exogenous antigens.
PMBCL also seemed to resemble other DLBCL subtypes in the relationship of loss of MHC II expression to survival. There were survival increases with increasing expression of MHC II genes across both DLBCL and PMBCL cases. Although the average expression of MHC II genes was lower in PMBCL than in some other types of DLBCL, and the overall survival generally higher, the estimated survival at 5 years of the lowest 10% MHC II expressers was very similar in both the DLBCL set (24%), from our previous work,27 and the PMBCL (25%) subset in this publication. As a caveat, there were 3 samples from our validation set that were present in both the PMBCL lowest 10% expressers subset (n = 4) and the DLBCL 10% lowest expressers subset (n = 24). This does show, however, that the extent of substantial loss was similar in all the subtypes of DLBCL in our data sets.
The mechanism of lowered MHC II expression in PMBCL remains unclear. Our positional expression profiling analysis demonstrated PMBCL was unlikely to down-regulate the expression of the MHC II genes by large genetic deletions in the MHC II locus, as had been previously described in IP-DLBCL.16 PMBCL also did not appear in most cases of MHC II loss to have genetic deletions of the MHC2TA gene, which encodes CIITA, the major transactivator of the MHC II family, even though deletion of the SOCS1 gene, which is located near the MHC2TA gene, had been seen in a PMBCL cell line.21
In summary, our data seemed to show that there could be similar mechanisms of substantial and partial loss of MHC II gene expression across the PMBCL, ABC, GCB, and unclassified subtypes of DLBCL. Further investigations may extend our study of MHC II expression to other types of non-Hodgkin lymphoma (NHL), such as follicular, Burkitt, and mantle-cell lymphoma, with the hypothesis that other types of NHL also have a range of MHC II expression related to survival, with a small subset showing substantial loss with extremely poor outcome. We are also investigating the mechanisms of down-regulation of the MHC II genes, including transcriptional or epigenetic regulation, as well as testing in vitro treatments for increasing MHC II expression in DLBCL. Our long-term goal is to therapeutically modify MHC II expression in patient tumors in the hope of improving patient outcome.
Prepublished online as Blood First Edition Paper, March 16, 2006; DOI 10.1182/blood-2005-11-4742.
Supported by the Warmer Foundation and Director's Challenge Grant NIH/NCI U01-CA84967.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank the additional members of the LLMPP Research Group for helping produce the large and well-annotated data sets that are providing new insights into the molecular pathology of NHL: James O. Armitage, Rita Braziel, Elias Campo, Joseph M. Connors, Jan Delabie, Richard I. Fisher, Timothy C. Greiner, Elaine S. Jaffe, Julie Vose, Dennis D. Weisenburger, and Wyndham H. Wilson. In particular Dr Campo's laboratory performed the CGH referenced in this work.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal