The introduction in 1998 of imatinib mesylate (IM) revolutionized management of patients with chronic myeloid leukemia (CML) and the second generation of tyrosine kinase inhibitors may prove superior to IM. Real-time quantitative polymerase chain reaction (RQ-PCR) provides an accurate measure of the total leukemiacell mass and the degree to which BCR-ABL transcripts are reduced by therapy correlates with progression-free survival. Because a rising level of BCR-ABL is an early indication of loss of response and thus the need to reassess therapeutic strategy, regular molecular monitoring of individual patients is clearly desirable. Here we summarize the results of a consensus meeting that took place at the National Institutes of Health (NIH) in Bethesda in October 2005. We make suggestions for (1) harmonizing the differing methodologies for measuring BCR-ABL transcripts in patients with CML undergoing treatment and using a conversion factor whereby individual laboratories can express BCR-ABL transcript levels on an internationally agreed scale; (2) using serial RQ-PCR results rather than bone marrow cytogenetics or fluorescence in situ hybridization (FISH) for the BCR-ABL gene to monitor individual patients responding to treatment; and (3) detecting and reporting Philadelphia (Ph) chromosome-positive subpopulations bearing BCR-ABL kinase domain mutations. We recognize that our recommendations are provisional and will require revision as new evidence emerges. (Blood. 2006;108:28-37)
Introduction
Although chronic myeloid leukemia (CML) was recognized as a distinct form of leukemia in the first half of the 19th century, it was not until advances in technology for characterizing human chromosomes in the late 1950s led to the discovery in 1960 that the leukemia cells harbored a consistent abnormality that came to be known as the Philadelphia (Ph1 or now Ph) chromosome. During the subsequent 30 years identification and quantification of Phpositive metaphases in the bone marrow proved valuable for confirming the diagnosis and monitoring the response to therapy. In the last 15 years the introduction of techniques for identifying and measuring BCR-ABL transcripts has enabled more precise assessment of response to specific therapies for CML, notably the use of allogeneic stem cell transplantation, interferon-α, and tyrosine kinase (TK) inhibitors. With each of these therapeutic approaches, serial monitoring of individual patients can predict those at higher risk of “disease progression.” The use of real-time quantitative polymerase chain reaction (RQ-PCR) has also been used with advantage to monitor other types of leukemia. Thus, in general, serial measurement of leukemia-specific transcripts is a valuable approach to monitoring individual patients and in some cases to indicating the need to reassess therapy.
The methodology used for identifying BCR-ABL transcripts has evolved over the years. Initially it was possible only to identify the presence or absence of BCR-ABL transcripts by either single-step amplification or a 2-step “nested” amplification with internal primers to increase the sensitivity.1-3 In 1993 Cross and colleagues introduced a competitive technique that allowed transcript numbers to be expressed per microgram of leukocyte RNA4 or as a ratio of BCR-ABL/ABL on a log scale.5 This method of expressing results was adapted for real-time PCR when this technology became available.6-11 An alternative method for expressing results of novel and effective treatment for CML was introduced by Hughes and colleagues in 2003, who monitored the response to imatinib in previously untreated patients with CML entered in the International Randomized Study of Interferon versus STI571 (IRIS study)12 ;in order to normalize results of measuring reductions in BCR-ABL transcripts in 3 geographically dispersed laboratories, the investigators introduced the concept of log10 reduction from a standardized baseline for untreated patients. Some clinicians have found this a more “user-friendly” unit of measurement than the ratio expressed as a percentage.
In 2003 a Europe Against Cancer (EAC) Program established standardized protocols for fusion transcript quantitation in multiple centers using Taqman methodology.13 The choice and stability of candidate control genes were evaluated with various specific recommendations.14,15 Two years later, however, there is still considerable diversity in the way in which RQ-PCR for BCR-ABL is carried out and the results reported in different laboratories. Although there is some level of consensus about suitable control genes, methods have not been standardized across all laboratories, and guidelines for acceptable levels of reproducibility and sensitivity are lacking. In addition, certified international reference and control materials are not yet available, although efforts to standardize methods and to develop guidelines for data analysis and for reporting levels of minimal residual disease (MRD) are in progress.13,16,17 For these reasons, a number of investigators with an interest in these techniques participated in a meeting that took place at the National Institutes of Health (NIH) in Bethesda in October 2005. The issues discussed included appropriate RNA quality, PCR methodology, reverse transcription and PCR amplification efficiency, appropriate control genes for normalization, standards, sensitivity and reporting of PCR negative results, quality assurance of the assay, international reference and control material, and the expression of data on an international scale. Some of these issues are addressed in this paper. More precise methodologic details will be provided in a separate publication which will include key recommendations for generating reliable quantitative data for BCR-ABL transcripts (Hughes and Branford,18 and Branford et al, manuscript in preparation).
Some of the patients with CML who receive imatinib and respond initially but then lose their response prove to have 1 or sometimes more than 1 Ph-positive subclone characterized by the presence of 1 or other of a range of mutations in the BCR-ABL kinase domain (KD) that code for a specific amino-acid substitution.19-22 Cell lines transfected with these various mutant forms of BCR-ABL are variably resistant to imatinib in vitro.19,20,23 The position of the mutation within the KD may be clinically relevant, inasmuch as patients with mutations in the P-loop (usually defined as spanning amino acids 248-256) of the KD may have survival inferior to those with mutations at other KD sites or those without mutations,24,25 a conclusion that is still controversial. In practice, it may be more important to characterize a specific mutation within or outside the P-loop because each mutation may be associated with a specific clinical prognosis. For these reasons a case can be made for searching for KD mutations in selected patients and indeed for quantitating such mutant clones when they are identified. Thus, this paper also addresses the issue of how best to identify and quantitate mutant subclones. It makes recommendations for monitoring individual patients in order to recognize the presence of subclones that may prove clinically relevant.
RQ-PCR for BCR-ABL transcripts
Control gene for standardization
Choosing an appropriate control gene is important for generating reliable and reproducible data. Comparison with control gene results helps to identify RNA samples of unacceptable quality.13,26,27 In samples deemed of acceptable RNA quality the use of a control gene compensates for variations in transcript levels due to sample degradation after collection, helps to adjust for differences in the efficiency of the reverse transcription (RT) step and variations in the amount of RNA, and aids in assessing the sensitivity of each sample measurement.13,16-18,28 The control gene should satisfy the following criteria: (1) it should have an expression level broadly similar to that of BCR-ABL at diagnosis of CML; (2) it should have stability similar to BCR-ABL; and (3) primers for the gene should be proven not to amplify sequences from genomic DNA such as pseudogenes. Similar levels of RNA stability are essential since delays in sample processing are common and substantial changes in expression can occur very rapidly after blood collection.28,29 Therefore, differential stability of the target and control may lead to inaccurate RQ-PCR data.
The 3 control genes that have been studied extensively and appear most suitable for BCR-ABL quantitation are BCR, ABL, and β-glucuronidase (GUSB). ABL has been used by many investigators in studies evaluating MRD in patients treated by allogeneic stem cell transplantation, interferon-α, and more recently with imatinib.26,30-36 Further investigation of ABL as the control gene has revealed comparable mean stability of ABL and BCR-ABL, but substantial differences between individual samples were observed upon storage.16 At high transcript levels (ie, in patients with CML at diagnosis or CML still predominantly Ph positive), the fact that the ABL control also measures BCR-ABL gives a spuriously high result and may therefore underestimate the BCR-ABL/ABL ratio.13 At lower levels of disease this distortion is small and irrelevant. BCR was initially investigated as a control gene since it has a similar expression level and stability to that of BCR-ABL.37 Subsequent experiments did indeed confirm that BCR degrades at the same rate as BCR-ABL.18 BCR was the control gene selected for the IRIS study.12 The EAC, which did not test BCR among the 17 control genes evaluated, recommended ABL as the control of choice for RQ-PCR diagnostics and MRD detection in patients with leukemia. GUSB and β2-microglobulin were also considered suitable.13,14,38 Wang and colleagues have argued in favor of GUSB, which, in contrast to BCR and ABL, has the theoretic advantage that it is not rearranged in the leukemia-cell population.39 Thus, though ABL is currently the most widely used control gene, BCR and GUSB are equally suitable. However, many of the numerous other genes used as controls do not satisfy one or other of the criteria specified above.
Optimization of RQ-PCR methodology
For valid RQ-PCR data, it is imperative to consider and optimize each stage of the procedure, including sample collection, RNA extraction, RT, and the quantitative PCR. The quality of the RNA is extremely important for reproducible data, and consistency in sample collection, tissue type, transportation, and storage conditions will maximize the accuracy and reliability of analysis. Sufficient cDNA should be seeded into each amplification reaction to enable sensitive detection of MRD. Therefore, appropriate validation of all procedures should be undertaken before an assay is deemed suitable. Once a laboratory achieves the acceptable reproducibility limits and produces acceptable data in quality control assessments, meaningful results can be reported on an international scale. There have now been a number of investigator meetings on both sides of the Atlantic designed to establish the best methodology for performing and reporting RQ-PCR data for BCR-ABL, and the following are the consensus recommendations agreed most recently at the National Institutes of Health (NIH) meeting (Bethesda, MD). The reasoning and evidence for these recommendations with supporting data will be published separately (S.B., N.C.P.C., A.H., J.R., G.S., J.K., J.G., T.H.; “Harmonizing current methodology for detecting BCR-ABL transcripts by real-time quantitative PCR: specific recommendations and rationale” manuscript in preparation).
Key recommendations for optimization
Appropriate sample for analysis and RNA extraction. Peripheral blood (PB) is suitable for analysis of BCR-ABL transcripts in chronic-phase CML. A minimum of 5 mL should be collected and most investigators recommend 10 mL. In practice, the real criterion should be a minimum number of nucleated cells (eg, at least 1-2 × 107) and not the volume of blood, which means that a larger quantity of blood may be appropriate if the leukocyte count is low. The use of Ficoll is not recommended because use of fractionated leukocytes is less sensitive than analyses based on total leukocytes following red-cell lysis.
Although the differences in low levels of leukemia measured in peripheral blood and bone marrow are small, the serial use of RQ-PCR values based on interchangeable use of both sources can lead to misinterpretation of results.40 Since blood samples clearly correlate with clinical response and are easy to collect on a regular basis, peripheral blood should routinely be used for monitoring patients in chronic phase (in advanced-phase disease the use of bone marrow may sometimes give results significantly higher than those obtained with use of peripheral blood).
EDTA anticoagulant is appropriate for PCR analysis. Some investigators believe that heparin inhibits the PCR reaction, although this may not be a problem if cells are washed adequately prior to RNA extraction.
To prevent significant degradation of transcripts, samples should be processed within 36 hours of collection, although ideally samples should be processed within 24 hours for most sensitive measurement of MRD. The use of commercial preparations designed to prevent nuclease degradation and thus “stabilize” RNA needs further assessment.
Reverse transcription. Random primers are recommended for the reverse transcription reaction and a final concentration of at least 25 μM improves the sensitivity.13
The choice of RT can influence the yield and efficiency of the RT. Moloney murine leukemia virus (MMLV) at a concentration of 4to8U/μL of reaction and Superscript (supplied at 200 U/μL) are both suitable, but Superscript (Invitrogen, Carlsbad, CA) is favored by some. Other sources of the enzyme are also acceptable.
To maximize reproducibility and to enhance detection of low-level residual disease, the ideal might be to divide the sample into 2 aliquots and to assay both separately for BCR-ABL, including the RNA extraction stage. It would be simpler, however, to assay a patient sample in duplicate just from the start of the RT stage. Duplicates starting with a single preparation of cDNA are not appropriate because they do not detect variability in the RT reaction.
RQ-PCR assay design. Technologies based on hydrolysis and hybridization probes are both suitable and can give comparable results41,42 provided that standardized and validated procedures are followed, as recommended by the EAC.13
Assay design should take account of the polymorphic site in BCR exon 13,7,43,44 and also the fact that the small intron between ABL exons 2 and 3 can be amplified efficiently from genomic DNA, which can thereby distort results and decrease sensitivity. The probes and primers should be RNA specific and tested with genomic DNA.
Contamination prevention. Strict precautions should be undertaken to limit the possibility of contamination, including the physical separation of areas for (1) the preparation of plasmid dilutions; (2) the extraction of RNA and for the addition of cDNA to tubes containing master mix before PCR amplification; (3) the preparation of PCR master mix; and (4) PCR amplification and product detection. PCR products, undiluted BCR-ABL, and control plasmids should never be introduced into the sample preparation area.
Gowns and gloves should be worn in the work areas, and gowns should be dedicated to each area.
The use of aerosol-resistant pipette tips or positive displacement pipettes is essential to minimize the risk of contamination.
Work surfaces and equipment should be wiped regularly with a decontaminating agent such as 2% to 10% hypochlorite.
Replacing thymidine with uracil and use of uracil-N-glycosylase in the single-round PCR mix may help to control for contamination with previously amplified product, but the preventive methods detailed are more important.
Appropriate standards and the influence of PCR amplification efficiency. RNA or DNA standards can be used for RQ-PCR but DNA standards may have superior stability. Plasmids that contain both target and control sequences will limit variability.
When using RNA standards it is necessary to ensure that the long-term degradation rates of the control do not differ from the target, thus avoiding inaccurate ratios.
In the case of assays that measure more than 1 transcript with a common plasmid, the amplification efficiency of the PCR reactions for each target should be comparable.
The correlation coefficient of the standard curve should be 0.98 or greater to certify the linearity of the assay.
Ten-fold dilutions of standards that span the dynamic range of the assay should be included in each set of runs conducted during the day using the same primer/probe mix to compensate for inherent variations of the assay produced by probe degradation or variation between batches of reagents.
Measuring the reliability of the RQ-PCR assay. The correct interpretation of sequential BCR-ABL values relies on the measurement reliability of an individual assay. All associated variables of the procedure should be included when determining the reproducibility, including RNA extraction, RT, the quantitative PCR, and operators.
The use of Ct values (defining the cycle number when sample fluorescence reaches a predetermined threshold) to determine the coefficient of variation (CV) is inappropriate since these values are logarithmic units and result in a misleading representation of reproducibility.
Performance characteristics for each laboratory's BCR-ABL assay should be determined including accuracy, precision, analytic sensitivity and specificity, and reportable range. This is still relevant for laboratories using the standardized methods.
Different levels of acceptable reproducibility will be required over the dynamic range to account for the wider variability at very low levels of BCR-ABL.
Quality control samples of at least a low and high level of BCR-ABL are necessary to monitor the performance of the assay, including the maintenance of linearity, the stability of the standards, and the success of the RT and quantitative PCR steps.
Acceptable sensitivity and undetectable BCR-ABL. The determination of the sensitivity of detection should cover the quality of a particular RNA sample and the efficiency of the RT step, as well as the detection limit of the quantitative PCR. Using a standardized baseline or median diagnostic BCR-ABL value may be appropriate when calculating the sensitivity of a sample, but the formula for calculating sensitivity should be determined separately for each control gene.
For a sample to be classified as “transcripts undetectable,” all parallel reactions (ie, Taqman microtiter plate wells or Lightcycler capillaries) should be negative. Reporting of undetectable BCR-ABL values should incorporate a sensitivity level of the individual sample to avoid misleading interpretation of negative values. If only 1 of the reactions is positive, the test should be repeated or the result confirmed with a nested primer PCR. In practice, equivocal results need to be considered in the clinical context.
Cut-off values for the determination of positivity need to be decided. The cut-off value will represent the limit of detection and is usually the lowest BCR-ABL plasmid dilution that can be amplified reliably (ie, 5-10 molecules). Thus, the overall sensitivity of the assay depends on the number of cells in the primary lysate, the level of control gene transcripts, and the performance of the PCR, and it should be calculated for each individual assay.
Quality assurance of the RQ-PCR assay
Quality control samples of at least a low and high level of BCR-ABL are necessary to monitor the performance of the assay.
All assays should include appropriate negative controls that are subject to the whole test process, including the extraction step.
Current methods for reporting results
There are currently various different methods in use for reporting results of RQ-PCR data on individual patients. One approach is to report BCR-ABL copies per microgram of RNA, but this is associated with a number of shortcomings since measurement of RNA concentration is inherently unreliable, and variations in RNA quality and efficiency of the RT step are not taken into account. More commonly, BCR-ABL copy numbers are expressed as a ratio to copy numbers of the control gene, or as this ratio expressed as a percentage whereby equal target gene and control copy numbers at diagnosis would be expressed as 100%. Another approach introduced more recently is to report the reduction of transcript numbers on a log10 scale from a standardized baseline for untreated patients established in the laboratory where the assay is being performed.12
Proposal for expressing results on an international scale
It is highly desirable that a standardized international scale for measuring BCR-ABL transcripts should be established. Theoretically, one might aim to adopt a single RQ-PCR assay at all PCR centers with every detail of the process specified, controlled, and optimized. This would indeed remove many of the variables that cause interlaboratory differences in results, but a major disadvantage would be the lack of flexibility necessary to improve the assay in individual laboratories.
In practice, a number of different RQ-PCR methods are valid for monitoring patients with CML, so the alternative to a single “global” protocol would be: (1) to select a limited number of RQ-PCR assays that are already established and widely adopted; (2) to establish a set of agreed principles that would always be applied when making adjustments or technical improvements to these assays (as listed in “Key recommendations for optimization”); and (3) to convert local laboratory results to an international scale once the approved assays have been selected and consensus principles accepted, as suggested previously by the EAC group.14
The molecular monitoring component of the IRIS study referred to above established for the first time a low level of BCR-ABL transcripts, called a major molecular response (MMR), that correlated with excellent progression-free survival.12 The standardized baseline was calculated by measuring the level of BCR-ABL/BCR in 30 patients with chronic-phase CML from blood collected before any treatment was started. The same 30 samples were assayed in the 3 laboratories. For example, if the median BCR-ABL/BCR ratio for these 30 samples in laboratory A was 50%, then patients with BCR-ABL/BCR levels of 0.05% or below in that laboratory were assessed as having achieved a “3-log or more reduction from the standardized baseline,” which is equivalent to a MMR. By this method the BCR-ABL level for MMR was independent of the actual baseline level in a particular patient.
The advantage of defining the molecular response according to the reduction from a standardized baseline is that once a laboratory has established the BCR-ABL transcript level that is equivalent to a MMR as determined in the IRIS trial, results can be expressed on a common scale between participating laboratories.12 However, the 30 pretreatment samples used for the IRIS standardization are no longer available for more widespread standardization. An alternative strategy to determine the value of MMR in another laboratory was demonstrated by the study from the laboratory of Hochhaus and coworkers in Mannheim, which involved exchange of over 50 RNA samples between the German and Australian groups.45 A BCR-ABL/ABL ratio of 0.12% in the German laboratory was shown to be equivalent to an MMR as defined in the IRIS trial. However, it is impractical to exchange large numbers of samples for the standardization of multiple international laboratories.
In practice, the use of the “log reduction” terminology has led to confusion since it implies that the value is a relative one, although the way the “3-log or more reduction” level was established makes it an absolute value that could theoretically be defined as a specific number of BCR-ABL transcripts. Nevertheless, it is probably preferable to move away from the term “log reduction” in routine clinical practice, and express all results on a standardized international numeric scale, but the international scale still needs to be anchored to some absolute values. We propose that the international scale should be anchored to 2 values that have already been defined. The standardized “baseline,” as established in the IRIS trial, is taken to represent 100% on the international scale and a 3-log reduction from the standardized baseline (MMR) is fixed at 0.10%.
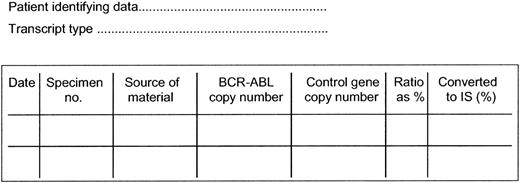
In order to determine the international scale conversion factor for each laboratory the RQ-PCR values must be referenced to a set of verified samples of known value, which may be plasmids, lyophilized cells, cell extracts, or in vitro “stabilized” RNA. Reference standards have already been prepared at several centers and are being distributed to other laboratories. The use of prepared material will eventually allow each laboratory to determine a BCR-ABL/control gene ratio as a percentage equivalent to a MMR as established in the IRIS trial. The conversion factor (CF) for each laboratory is then linked to this value. The BCR-ABL values are multiplied by the CF to convert to the international scale and expressed as BCR-ABL values on the international scale. The process for conversion is illustrated in Table 1 using the MMR values that have already been established in 3 laboratories, and the suggested format for reporting is shown in Figure 1. It should be noted that when ABL is the control gene the use of the conversion factor at high BCR-ABL transcripts levels may be less satisfactory owing to the nonlinearity of the ratios.
Example of the use of a conversion factor to convert BCR-ABL values obtained in a given laboratory to the international scale
Laboratory . | MMREq, % . | 0.1%MMREq (%) = conversion factor . | Formula for conversion of a given result to the international scale (BCR-ABLL × CF = BCR-ABLIS) . |
|---|---|---|---|
| Adelaide | 0.08 | 0.1/0.08 = 1.25 | BCR-ABLL × 1.25 |
| Mannheim | 0.12 | 0.1/0.12 = 0.83 | BCR-ABLL × 0.83 |
| London | 0.045 | 0.1/0.045 = 2.22 | BCR-ABLL × 2.22 |
Laboratory . | MMREq, % . | 0.1%MMREq (%) = conversion factor . | Formula for conversion of a given result to the international scale (BCR-ABLL × CF = BCR-ABLIS) . |
|---|---|---|---|
| Adelaide | 0.08 | 0.1/0.08 = 1.25 | BCR-ABLL × 1.25 |
| Mannheim | 0.12 | 0.1/0.12 = 0.83 | BCR-ABLL × 0.83 |
| London | 0.045 | 0.1/0.045 = 2.22 | BCR-ABLL × 2.22 |
BCR-ABLL = BCR-ABL/control ratio expressed as a percentage in a given laboratory. MMREq = BCR-ABLL that is equivalent to a MMR as extablished in the IRIS trial.12 In order to convert a given local result to the international scale, it is necessary to use a conversion factor (CF). This is calculated as follows: CF = 0.1% divided by MMREq (since 0.1% is the agreed value for MMR on the international scale). Once a laboratory-specific conversion factor has been derived, it can be used to convert all local values to the international scale. (This calculation will be invalid if the reproducibility or linearity of the assay is poor, in which case the methodology will need to be optimized.)
In the near future it will be more appropriate for a commercial or national reference laboratory to prepare on a large scale stable quantitative control material that is then validated by an appropriate international agency and made available to clinical laboratories for determining the conversion factor. Follow-up quality control samples of different BCR-ABL levels will be necessary for continued assessment and validation of the methods. In the more distant future it would be desirable if commercial laboratories were able to produce kits comprising a complete set of standard reagents, as has for example been achieved for monitoring levels of hepatitis C virus and HIV.46,47
Suggested method for sequential reporting of results of RQ-PCR assays. Interpretation (assuming adequate quality RNA): (1) BCR-ABL detectable at a level greater than 1.0%, which suggests that the patient has some or 100% Ph-positive marrow metaphases (the patient may still be responding to therapy or may be relapsing from a Ph-negative status); (2) BCR-ABL detectable at a level greater than 0.1%, which suggests that the patient has not achieved or has lost a major molecular response; (3) BCR-ABL detectable at or below the level of 0.1% indicates achievement of a major molecular response (as defined by the IRIS study); and (4) BCR-ABL is not detectable, meaning that the BCR-ABL level is below the level of sensitivity of the assay, which should be at least 0.01% on the international scale, a value equivalent to a 4-log reduction below baseline. The laboratory value for a given result can be converted to a value on the international scale by use of a conversion factor. This factor is based on the relationship of the laboratory specific value for an MMR to the value equivalent to an MMR as established in the IRIS trial, namely a 3-log reduction below an internationally agreed standardized baseline. The conversion factor will be specific for each laboratory but may be affected by any change in the technical aspects of the assay. If the quality of the RNA is poor, no useful conclusion can be drawn from the results of the test.
Suggested method for sequential reporting of results of RQ-PCR assays. Interpretation (assuming adequate quality RNA): (1) BCR-ABL detectable at a level greater than 1.0%, which suggests that the patient has some or 100% Ph-positive marrow metaphases (the patient may still be responding to therapy or may be relapsing from a Ph-negative status); (2) BCR-ABL detectable at a level greater than 0.1%, which suggests that the patient has not achieved or has lost a major molecular response; (3) BCR-ABL detectable at or below the level of 0.1% indicates achievement of a major molecular response (as defined by the IRIS study); and (4) BCR-ABL is not detectable, meaning that the BCR-ABL level is below the level of sensitivity of the assay, which should be at least 0.01% on the international scale, a value equivalent to a 4-log reduction below baseline. The laboratory value for a given result can be converted to a value on the international scale by use of a conversion factor. This factor is based on the relationship of the laboratory specific value for an MMR to the value equivalent to an MMR as established in the IRIS trial, namely a 3-log reduction below an internationally agreed standardized baseline. The conversion factor will be specific for each laboratory but may be affected by any change in the technical aspects of the assay. If the quality of the RNA is poor, no useful conclusion can be drawn from the results of the test.
Summary of key recommendations for international standardization
(1) International standardization needs to be achieved by an exchange of reference standards with values carefully established in reference laboratories; (2) the reference standards will assess the reproducibility of each method and indicate when the techniques are inappropriate for accurate assessment of BCR-ABL levels; (3) reference and quality control samples prepared on a large scale by a commercial company or nationally supported laboratory will be incorporated when available; (4) to convert laboratory BCR-ABL values to the international scale a conversion factor for each laboratory must be determined. The conversion factor is derived from the value that is equivalent to the MMR value as established in the IRIS trial; (5) proposed formula for individual conversion of laboratory BCR-ABL/control percentage values to the international scale: BCR-ABL (local value) × conversion factor = BCR-ABL (international scale); (6) the conversion should be formally tested with a series of quality control samples, with values established in reference laboratories; and (7) the significance of a BCR-ABL value should always be considered in the context of treatment and with regard to sequential BCR-ABL values.
Detection of kinase domain mutations
It is now accepted that the expansion of a Ph-positive clone carrying an Abl KD mutation may be associated with resistance to imatinib,19-22,48-51 and in some cases precedes or accompanies progression to advanced-phase disease.24,25 This means that the KD mutations above a certain level should probably be identified as early as possible because they may indicate the need to reconsider the therapeutic strategy. Conversely, the probability of finding a mutant clone is very low in a patient who has a stable or declining level of BCR-ABL transcripts51 ; the incidence of mutations in imatinib-naive chronic-phase patients and patients in complete cytogenetic remission (CCyR) is usually also low.52-54 Moreover, mutant clones at low level may not necessarily have the same clinical significance as clones that are detected in the context of a rising disease burden.53,55
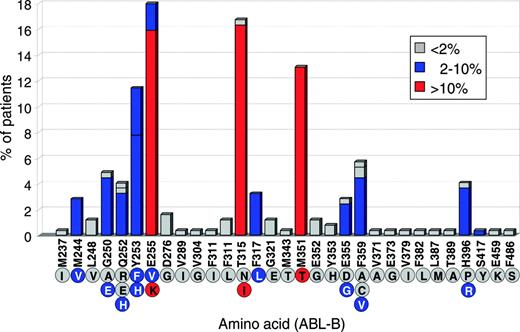
There is currently no universally accepted consensus when patients should be screened for KD mutations, which technique should be used, and how the data should be reported. As new ABL inhibitors become available for imatinib-resistant disease it will be important to define diagnostic standards and guidelines. Mutations have been detected in more than 40 different positions; some of the more common ones are shown in Figure 2.
Method of detection
The available technologies are summarized in Table 2. Direct sequencing has a sensitivity of about 20% and is probably the optimal method for routine use at present. BCR-ABL rather than ABL should be amplified. Forward and reverse strands of BCR-ABL amplicons should be sequenced to confirm the mutation. It is possible that other more sensitive methods like denaturing highperformance liquid chromatography (D-HPLC; transgenomic wave) may replace direct sequencing in the future. Newly identified mutations should be confirmed by amplifying the normal ABL alleles to exclude polymorphisms.
Technologies available for identifying and quantifying BCR-ABL KD mutations
Technology . | Sensitivity, % . | Specificity . | Bias* . | Availability . | Reference . |
|---|---|---|---|---|---|
| Direct sequencing | 15-25 | ++ | No | +++ | 19,21,22,24,59 |
| Subcloning and sequencing | 9 | +++ | No | ++ | 20 |
| Denaturing high-performance liquid chromatography (D-HPLC) | 0.1-10 | ++ | No | ++ | 66-68 |
| Pyrosequencing | 5 | ++ | No | + | 55,69 |
| Double-gradient denaturing electrophoresis | 5 | ++ | No | + | 57 |
| Fluorescence PCR and PNA clamping | 0.2 | ++ | Yes | + | 58 |
| Allele-specific oligonucleotide PCR (ASO-PCR) | 0.01 | ++ | Yes | + | 53,59,60 |
Technology . | Sensitivity, % . | Specificity . | Bias* . | Availability . | Reference . |
|---|---|---|---|---|---|
| Direct sequencing | 15-25 | ++ | No | +++ | 19,21,22,24,59 |
| Subcloning and sequencing | 9 | +++ | No | ++ | 20 |
| Denaturing high-performance liquid chromatography (D-HPLC) | 0.1-10 | ++ | No | ++ | 66-68 |
| Pyrosequencing | 5 | ++ | No | + | 55,69 |
| Double-gradient denaturing electrophoresis | 5 | ++ | No | + | 57 |
| Fluorescence PCR and PNA clamping | 0.2 | ++ | Yes | + | 58 |
| Allele-specific oligonucleotide PCR (ASO-PCR) | 0.01 | ++ | Yes | + | 53,59,60 |
PNA indicates peptide nucleic acid.
Bias indicates that the test is designed to detect specific mutations.
The relative frequency of BCR-ABL kinase domain mutations detected at 31 different positions in clinical specimens from 245 patients in whom mutations were detected (219 with CML and 26 with Ph-positive acute lymphoblastic leukemia). The numbering of amino acids is based on the Abl protein variant B (which includes ABL exon 1b but not exon 1a). The letters inside the circles denote the amino acid encoded by the corresponding mutated nucleotide. At some positions 2 or 3 possible mutant nucleotides encode different amino acids. The percentage of patients in the series with each mutation specified on the y axis is color-coded as shown in the box. Data collated from 20 published papers.19-22,24,25,48,52,56-66
The relative frequency of BCR-ABL kinase domain mutations detected at 31 different positions in clinical specimens from 245 patients in whom mutations were detected (219 with CML and 26 with Ph-positive acute lymphoblastic leukemia). The numbering of amino acids is based on the Abl protein variant B (which includes ABL exon 1b but not exon 1a). The letters inside the circles denote the amino acid encoded by the corresponding mutated nucleotide. At some positions 2 or 3 possible mutant nucleotides encode different amino acids. The percentage of patients in the series with each mutation specified on the y axis is color-coded as shown in the box. Data collated from 20 published papers.19-22,24,25,48,52,56-66
How should mutations be reported?
It is suggested that individual mutations should be reported with terminology that indicates both the amino-acid exchange and the nucleotide exchange. It would be desirable if the quantity of mutant transcripts relative to nonmutated BCR-ABL could be reported. As most of the methods in use, other than pyrosequencing, are at best semiquantitative, this will naturally be an estimate rather than a precise quantification. Some indication of the sensitivity of mutant clones to available TK inhibitors may be useful. The results could be expressed as shown in Figure 3.
Recommendations for monitoring individual patients receiving treatment with tyrosine kinase inhibitors
Indications for cytogenetics and RQ-PCR
At diagnosis of chronic-phase disease. In any new patient whose blood count suggests the diagnosis of a chronic myeloproliferative disorder, the detection of BCR-ABL transcripts in a blood specimen is probably the best way to confirm the diagnosis of CML. It is suggested therefore that circulating BCR-ABL transcript numbers should be measured, and bone marrow (BM) cytogenetics studied in every new patient with CML before initiation of treatment. If collection of BM is not feasible, fluorescence in situ hybridization (FISH) on a PB specimen using dual probes for the BCR and ABL genes is a valuable but secondary method of confirming the diagnosis; it has the advantage that it may detect cytogenetically “silent” BCR-ABL rearrangements and also deletions in the derivative 9q+, which may or may not have prognostic significance.70-72 It may therefore be performed in conjunction with marrow cytogenetics and RQ-PCR for BCR-ABL transcripts, but it is considerably less sensitive than RQ-PCR and should not replace either test. Marrow cytogenetics is essential to identify any unusual translocations or additional cytogenetic abnormalities and RQ-PCR for BCR-ABL at diagnosis will identify whether the commonly observed e13a2 (b2a2) or e14a2 (b3a2) transcripts are present, or 1 of the less common fusion transcripts that are not amplified by the standard primer sets. This will prevent confusion when a patient on therapy has undetectable BCR-ABL transcripts because their transcripts were not amplified in the standard assay.
Suggested headings for expressing results of Abl KD analysis. For example, resistance may be classified as Fully Sensitive (FS), Partially Resistant (PR), Fully Resistant (FR), or Unknown (UKN).
Suggested headings for expressing results of Abl KD analysis. For example, resistance may be classified as Fully Sensitive (FS), Partially Resistant (PR), Fully Resistant (FR), or Unknown (UKN).
While a patient appears to be responding to treatment. Once a patient has started treatment with a tyrosine kinase inhibitor, it is suggested that BCR-ABL transcript levels should be measured at 3-month intervals or more frequently if convenient. Although it has hitherto been customary to examine the bone marrow cytogenetics at 3-month intervals, in a patient who appears to be responding to treatment a reasonable alternative would be to omit the examination at 3 months and simply perform a single examination at 6 months to confirm Ph negativity.
When a patient reaches CCyR. When a patient has achieved CCyR, it is suggested that BCR-ABL transcript numbers should be measured at intervals not longer than 3 months. It may be clinically useful to characterize the level of response at each subsequent clinic visit as (1) transcripts continuing to decline; (2) transcripts undetectable; (3) transcripts at a stable level (or plateau); or (4) transcript numbers rising.12,73 In the last case it is suggested that monitoring should continue at intervals of shorter than 3 months. For a patient in CCyR it may be reasonable to reduce the frequency of routine examination of marrow cytogenetics to every 12 months. If such examinations are abandoned completely, the possible onset of dysplasia or clonal changes in Ph-negative cells will not be detected.74,75
The use of FISH techniques performed on peripheral blood is appreciably less sensitive than RQ-PCR. FISH cannot reliably distinguish patients in CCyR who have achieved an MMR from those with lesser degrees of transcript reduction, and will not identify patients with low but rising transcript levels. It is therefore not the appropriate methodology for monitoring patients in CCyR.
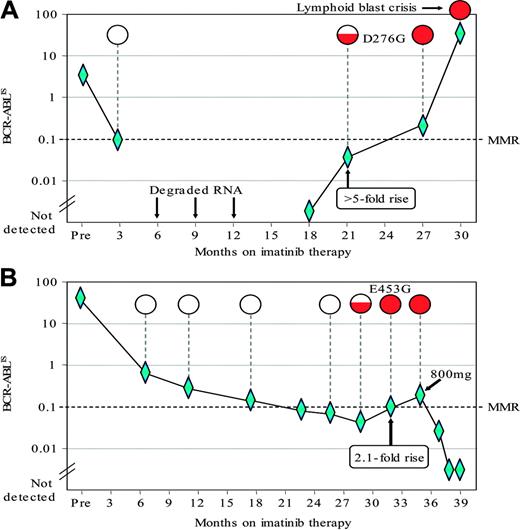
The detection of KD mutations associated with a rise in the BCR-ABL level. The graphs plot the BCR-ABL levels as measured by RQ-PCR76 in 2 late-chronic-phase patients treated in Australia who achieved a CCyR on imatinib 400 mg/daily. The BCR-ABL levels were calculated according to the proposed international scale (IS). The mutation analysis was performed using a direct sequencing technique.77 The mutation results are depicted as open circles when wild-type BCR-ABL was detected; the amount of shading within the circles indicates the relative size of the mutant sub-clone. Diamonds indicate datapoints. (A) After 18 months on imatinib, the patient had undetectable BCR-ABL that was followed by a rise of at least 5-fold. At that time the D276G mutation was detected. The patient remained on 400 mg imatinib and a CCyR was maintained at 27 months. Thereafter the patient progressed rapidly to lymphoid blast crisis. (B) This patient had a rise of 2.1-fold and the E453G mutation was detected prior to the rise. On the basis of the rising BCR-ABL level and the detection of the mutation the dose of imatinib was increased from 400 to 800 mg per day. The BCR-ABL level subsequently decreased and the CCyR was maintained.
The detection of KD mutations associated with a rise in the BCR-ABL level. The graphs plot the BCR-ABL levels as measured by RQ-PCR76 in 2 late-chronic-phase patients treated in Australia who achieved a CCyR on imatinib 400 mg/daily. The BCR-ABL levels were calculated according to the proposed international scale (IS). The mutation analysis was performed using a direct sequencing technique.77 The mutation results are depicted as open circles when wild-type BCR-ABL was detected; the amount of shading within the circles indicates the relative size of the mutant sub-clone. Diamonds indicate datapoints. (A) After 18 months on imatinib, the patient had undetectable BCR-ABL that was followed by a rise of at least 5-fold. At that time the D276G mutation was detected. The patient remained on 400 mg imatinib and a CCyR was maintained at 27 months. Thereafter the patient progressed rapidly to lymphoid blast crisis. (B) This patient had a rise of 2.1-fold and the E453G mutation was detected prior to the rise. On the basis of the rising BCR-ABL level and the detection of the mutation the dose of imatinib was increased from 400 to 800 mg per day. The BCR-ABL level subsequently decreased and the CCyR was maintained.
When a patient appears to have a rising level of BCR-ABL transcripts. If a patient is found to have a rising level of BCR-ABL transcripts, the frequency of measurement should be increased. The definition of “rising level” is not yet universally agreed. The Adelaide group found a strong association between a greater than 2-fold increase in BCR-ABL levels and detection of mutations (Figure 4).51 This analysis was performed using an RQ-PCR technique that was optimized to limit the variability of BCR-ABL quantitative measurement. Others suggest that a more realistic criterion to trigger mutation analysis using less precise RQ-PCR methods would be a rise of at least 5-fold in the BCR-ABL/control gene ratio, which should be confirmed by more than 1 test. In practice, increasing transcript levels are more likely to be associated with KD mutations in patients who never reached a MMR. The Hammersmith group has defined criteria for molecular relapse after allogeneic stem cell transplantation78 ; this may or may not be relevant to patients who achieved molecular negativity on treatment with TK inhibitors, since molecular negativity seems more durable after allogeneic stem cell transplantation than after “successful” treatment with imatinib.79
Indications for mutation analysis
Advanced-phase patients. KD mutations should be sought in any patient presenting in advanced-phase disease. The search could be repeated in such patients if they fail achieve to respond to a TK inhibitor or if having responded, they subsequently have rising numbers of BCR-ABL transcripts.
Chronic-phase patients. There is currently no clear evidence that a chronic-phase patient defined as high risk by Sokal or Hasford criteria is also at high risk for developing KD mutations. However, for chronic-phase patients who start treatment with TK inhibitors, mutation screening is indicated if there is inadequate initial response or any sign of loss of response. At a minimum this would include any patient who has failed to achieve complete hematologic response at 3 months, minimal cytogenetic response at 6 months, or major cytogenetic response at 12 months. A case can be made for screening any patient who has failed to achieve a major cytogenetic remission by 6 months.24 Loss of response is defined provisionally as hematologic relapse, relapse from CCyR to Ph positivity, or an increase in BCR-ABL transcript ratio of 1 log or greater.
Discussion
A qualitative RT-PCR for assessing the presence or absence of BCR-ABL transcripts in patients treated by allogeneic stem cell transplantation was introduced in the late 1980s but was superseded subsequently by a competitive PCR that gave much more precise information about the level of residual disease.4 This technology was in turn replaced by the introduction of real-time RT-PCR techniques, which are now used to monitor response to therapy in CML and a range of other hematologic malignancies. The methodology is demanding, however, and requires considerable attention to detail to ensure reproducible results. Moreover, a variety of different methods and approaches to expressing results are in operation at the national and international levels.12,14,15 The meeting that took place in October 2005 attempted to review some of these different approaches and to arrive at recommendations that might in due course lead to standardized procedures for monitoring BCR-ABL transcripts in CML. For example, there was emphasis on the need to ensure that the patient specimen arrived in the laboratory as rapidly as possible to minimize the risk of RNA degradation and the methods for extraction and reverse transcription needed careful monitoring. It was accepted that 3 specific control genes, ABL, BCR or GUSB, were all suitable, and 1 of these should be selected by laboratories establishing the assay ab initio. It was very important to ensure that standard curves were reproducible over time and achieved a high correlation coefficient. The results in an individual patient should ideally be expressed as a percentage reflecting the ratio of BCR-ABL transcript copies to control copies. Of great importance was the suggestion that individual laboratories could derive a conversion factor that would enable them to convert their own results to an international standard. This could then form the basis for a report form that would very readily interpretable to clinicians worldwide. This approach would not preclude simultaneous reporting of data as a log10 reduction from a baseline standardized in a given laboratory, but in general, the former method of expressing results should take precedence.
The measurement of viral load in patients infected with HIV or hepatitis C gives information about the efficacy of treatment of these 2 infections. For both of these diseases, PCR techniques were developed in individual centers to measure viral copy numbers.46,80 These methods were in due course complemented by the introduction of standard reference materials validated by the World Health Organization (WHO). Thereafter, a series of commercial kits became available, and these are now in widespread use for monitoring the clinical response to treatment for these 2 viral disorders. It seems desirable that the use of RQ-PCR for monitoring response to treatment for leukemia should follow the same pattern. This would mean that the next stage of development would be the introduction of standard control and reference reagents, such as plasmids containing BCR-ABL13,14 or BCR-ABL plus a control gene, a lyophilized preparation of a CML cell line, such as K562,81 or an “armored” RNA as has been used extensively in RT-PCR used to detect various viruses.82 Thereafter, 1 or more of these standard reagents could accompany in vitro diagnostic tests that satisfy criteria for clinical use as required by the appropriate national and international regulatory agencies.
It is clear that tests designed to detect and quantitate relatively low levels of MRD need to attain optimum levels of reliability and sensitivity, since they are becoming critical to directing treatment approaches in individual patients. The experience gained in resolving these issues in the context of CML will prove invaluable for other leukemias and the other hematologic malignancies in which molecular diagnostics and assessment of MRD will become essential tools in the effort to “individualize” patient management.
Although originally it was thought that the detection of any KD mutation in a given patient was the immediate cause of imatinib resistance, and also predicted for disease progression, it subsequently became clear that different mutations were associated with different degrees of resistance, some of which could be overcome by escalating the dose of imatinib or by use of 1 of the second-generation TK inhibitors. Thus far, only the T315I mutant has proved totally resistant to all clinically available BCR-ABL inhibitors. Conversely, some small clones detected in imatinibnaive patients and other small clones in patients receiving imatinib may not necessarily be clinically significant.53,55 Therefore, it may not be necessary on a routine basis to detect mutant clones present at less than 20% of total Ph-positive transcripts. For this purpose direct sequencing is sufficiently sensitive, and at the present time may be the technology of choice. More sensitive assays may prove superior if validated in interventional studies. In due course it may also be possible to define a finite number of BCR-ABL KD mutations that should be identified at lower levels, as they may be associated with a poor prognosis.
Finally, can one really make valid recommendations for the frequency with which bone marrow cytogenetics, RQ-PCR for BCR-ABL transcripts, and the screen for ABL KD mutations are carried out? If this is indeed attempted, as we have done here, it must be with the understanding that any such recommendations are based on incomplete clinical data that emanate from a very rapidly evolving field. They will necessarily be subject to frequent review as the field evolves.
Appendix
The persons attending the meeting included: Adam Bagg, Philadelphia, PA; John Barrett, Bethesda, MD; Susan Branford, Adelaide, Australia; Jorge Cortes, Houston, TX; Michael Deininger, Portland, OR; Brian Druker, Portland, OR; Jean Gabert, Marseille, France; John Goldman, Bethesda, MD; David Grimwade, London, United Kingdom; Margaret Gulley, Chapel Hill, NC; Rudiger Hehlmann, Mannheim, Germany; Andreas Hochhaus, Mannheim, Germany; Eric Hsi, Cleveland, OH; Tim Hughes, Adelaide, Australia; Dan Jones, Houston, TX; Jaspal Kaeda, London, United Kingdom; Suzanne Kamel-Reid, Toronto, ON, Canada; Roger Kurlander, Bethesda, MD; Pierre Laneuville, Montreal, QC, Canada; Janina Longtine, Boston, MA; Jeff Lipton, Toronto, ON, Canada; Giovanni Martinelli, Bologna, Italy; Peter Maslak, New York, NY; Rebecca McClure, Rochester, MN; Claude Preudhomme, Lille, France; Jerry Radich, Seattle, WA; Giuseppe Saglio, Torino, Italy; Simona Soverini, Bologna, Italy; Wendy Stock, Chicago, IL; Richard Stone, Boston, MA; John Thorson, Ann Arbor, MI; Lynn Wang, New York, NY; and Neal Young, Bethesda, MD.
Blood First Edition Paper, prepublished online March 7, 2006; DOI 10.1182/blood-2006-01-0092.
Supported by unrestricted educational grants from Novartis Pharma (Basel, Switzerland) and Bristol-Myers Squibb (Wallingford, CT) to the Foundation for the National Institutes of Health. A.H., J.K., M.B., N.C.P.C., J.G., D.G., R.H., G.M., G.S., and S.S. were supported by the European Leukemia Net within the Sixth European Community Framework Programme for Research and Technological Development.
T.H and S.B. provided the initial draft of the section of methodology for RQ-PCR; M.D. provided the initial draft for the section on KD mutations; and R.H. and M.B. contributed drafts for the clinical monitoring section. All other named authors attended at least 1 meeting and in most cases 2 separate meetings to discuss the content of the manuscript, and all authors read and made constructive comments and corrections on at least 2 and more often 3 to 4 versions of the manuscript during its prepration. J.M.G. prepared a series of drafts leading ultimately to the final manuscript.
This meeting took place under the sponsorship of the National Heart, Lung and Blood Institute, and the Fogarty International Center of the National Institutes of Health, Bethesda, MD. We thank those participants in the European Leukemia-Net who made available to us results of experimental work and discussions supported by the European Union.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal