Abstract
Analysis of clonotypic isotype class switching (CSR) in Waldenström macroglobulinemia (WM) and IgM monoclonal gammopathy of undetermined significance (MGUS) reveals a normal initial phase of B-cell activation as determined by constitutive and inducible expression of activation-induced cytidine deaminase (AID). Switch μ (Sμ) analysis shows that large deletions are not common in WM or IgM MGUS. In CD40L/IL-4-stimulated WM cultures from 2 patients, we observed clonotypic IgG exhibiting intraclonal homogeneity associated with multiple hybrid Sμ/Sγ junctions. This suggests CSR had occurred within WM cells. Nevertheless, the estimated IgG/IgM-cell frequency was relatively low (1/1600 cells). Thus, for the majority of WM B cells, CSR does not occur even when stimulated in vitro, suggesting that the WM cell is constitutively unable to or being prevented from carrying out CSR. In contrast to WM, the majority of IgM MGUS clones exhibit intraclonal heterogeneity of IgH VDJ. Furthermore, most IgM MGUS accumulate more mutations in the upstream Sμ region than do WM, making them unlikely WM progenitors. These observations suggest that switch sequence analysis may identify the subset of patients with IgM MGUS who are at risk of progression to WM.
Introduction
Waldenström macroglobulinemia (WM) is a lymphoproliferative disorder characterized by pleiomorphic B and plasma cells, with increased levels of monoclonal serum immunoglobulin M (IgM).1 WM, multiple myeloma (MM), and related cancers can evolve from monoclonal gammopathy of undetermined significance (MGUS), a potentially malignant condition increasingly prevalent in older individuals.2 A common feature of these conditions is the presence of monoclonal immunoglobulin, expressed by tumor cells harboring a unique clonotypic rearrangement of immunoglobulin heavy chain (IgH) VDJ associated with a specific immunoglobulin constant region such as IgG, IgA, or IgM. In contrast to IgG or IgA MGUS, which tends to progress to MM, IgM MGUS tends to progress to WM or lymphoma.3 Currently, there is no predictor for progression from IgM MGUS to WM.
WM B cells are characterized as CD20+138-sIgM+sIgD+ cells, some of which express the CD27 memory marker.4-6 Although mainly localized in bone marrow (BM), WM B cells are also present in blood, at clonal frequencies that appear to correlate with serum IgM levels.6 The majority of WM VDJ sequences are from the VH3/JH4 gene families, are hypermutated, and lack intraclonal heterogeneity.6,7 In addition, we have shown that a small proportion of patients with WM have a germline VDJ with few or no mutations.6 In contrast to IgM MM and chronic lymphocytic leukemia (CLL), postswitch clonotypic Ig (IgG or IgA) is undetectable in WM B cells, suggesting that they do not undergo isotype class switching.6-8
Class switch recombination (CSR) juxtaposes the expressed IgH VDJ variable region exon to a downstream set of constant region (CH) exons by recombination events. Activation-induced cytidine deaminase (AID) plays an essential role in CSR, somatic hypermutation (SHM), and gene conversion.9-12 AID mutation in humans is associated with absence of secondary antibody isotypes and SHM in hyper-IgM syndrome type 2.10 Transgenic AID function restores both SHM and CSR.13 Deletional analysis suggests that AID-mediated SHM and CSR may be uncoupled.14 The current CSR model15 proposes that AID initiates U:G mismatches that lead to mutations in the switch (S) region and an increased rate of switch mutation on CSR stimulation,9,16,17 generation of single-strand nicks and double-strand breaks in DNA, and recombination between the Sμ and downstream S regions.15 The S regions consist of highly repetitive 1 to 12 kb with G-rich regions in the nontemplate strand. Deletion of most of the Sμ region severely impairs CSR,18 whereas deletion of the entire Sγ1 region blocks switching to IgG1.19 Recent studies show that this may be more distance-dependent than sequence-specific.20 Noncoding germline transcripts initiated from the exon located upstream of the S region and extending into the corresponding CH gene are also required for CSR. Germline transcripts may form RNA-DNA hybrids to create a single-stranded DNA substrate for AID activity.21 In splenic B cells and in B-cell lines activated to undergo CSR, large deletions of endogenous Sμ occur frequently among cells that remain to produce IgM.22 Downstream S-region deletions in the absence of productive CSR, on the other hand, have rarely been observed.23
In this study, we determine whether CSR is impaired in clonotypic WM and IgM MGUS by analyzing the VDJ-Sμ region, AID expression, and CSR capability ex vivo, in cultures of WM and IgM MGUS B cells stimulated with CD40L/IL-4. Our results show that although the CSR machinery appears normal in both groups, CSR occurs only rarely in WM.
Patients, materials, and methods
Patients
BM aspirates and blood samples were collected from 18 patients with WM and 5 patients with IgM MGUS at the Cross Cancer Institute and University of Alberta Hospital (Edmonton, AB, Canada). WM was identified based on consensus criteria.1 IgM MGUS was defined as serum M-protein concentration less than 3.0 g/dL with normal BM and no evidence of a lymphoproliferative disorder or associated symptoms, signs, or laboratory evidence of disease related to the paraprotein. Clinical data and the concentration of monoclonal proteins of patients with IgM MGUS and WM are summarized in Table 1. The study was approved by the Health Research Board (University of Alberta) and Alberta Cancer Board (Edmonton, AB, Canada). Informed consent was provided in accordance with the Declaration of Helsinki.
Summary of 18 WM and 5 IgM MGUS patients in this study
. | WM . | IgM MGUS . |
|---|---|---|
| Age, y | 72 (59-81) | 77 (64-85) |
| Female/male, no. | 4/14 | 1/4 |
| IgM paraprotein level, g/dL | 4.17 (1.6-6.8) | 0.9 (0.5-1.3) |
| Hemoglobin level, g/L | 116 (84-135) | 122 (115-141) |
| Platelet count, × 109/L | 203 (108-318) | 296 (242-313) |
| Red blood cell count, × 1012/L | 3.65 (2.81-4.15) | 3.92 (3.68-3.96) |
| β2-Microglobulin level, nM | 2.57 (2.01-3.01) | 3.02 (2.39-4.03) |
| Albumin level, g/L | 40 (37-49) | 42 (40-46) |
| BM clonal involvement, % | 55 (30-90) | 0 |
. | WM . | IgM MGUS . |
|---|---|---|
| Age, y | 72 (59-81) | 77 (64-85) |
| Female/male, no. | 4/14 | 1/4 |
| IgM paraprotein level, g/dL | 4.17 (1.6-6.8) | 0.9 (0.5-1.3) |
| Hemoglobin level, g/L | 116 (84-135) | 122 (115-141) |
| Platelet count, × 109/L | 203 (108-318) | 296 (242-313) |
| Red blood cell count, × 1012/L | 3.65 (2.81-4.15) | 3.92 (3.68-3.96) |
| β2-Microglobulin level, nM | 2.57 (2.01-3.01) | 3.02 (2.39-4.03) |
| Albumin level, g/L | 40 (37-49) | 42 (40-46) |
| BM clonal involvement, % | 55 (30-90) | 0 |
Data are presented as median (range).
Molecular identification of clonotypic IgM VDJ sequences
Identification of clonotypic IgM VDJ sequences has been previously described.6 Clonotypic sequences were validated to confirm that the correct clone had been identified by amplification of clonotypic product from sorted CD20+ cells from BM, using reverse transcription-polymerase chain reaction (RT-PCR). Accurately identified clonotypic sequences were detected in 56% ± 24% of these individual cells. Patient-specific primers were designed based on the patient-specific complementarity-determining region 2 (CDR2) and CDR3 and were confirmed to be specific for the target clone, without cross-reactivity to the clonotypic VDJ from other WM clones having the same VH gene family.
RT-PCR
Total RNA was prepared by TRIzol reagent (Invitrogen, Burlington, ON, Canada) according to the manufacturer's instructions. First-strand cDNA was reverse transcribed from 1 μg total RNA using dT15 primer and Superscript RnaseH- reverse transcriptase (Invitrogen) in a total volume of 20 μL. All PCRs were amplified from 1 μL 1:10 cDNA for 30 cycles, using standard buffer and Taq DNA polymerase (Invitrogen) in a total volume of 50 μL. Amplification cycles were as follows: clonotypic CDR2/CDR3, clonotypic isotypes, and β2-microglobulin, 94°C for 30 seconds, 60°C for 30 seconds, and 72°C for 30 seconds; AID, 94°C for 30 seconds, 55°C for 30 seconds, and 72°C for 30 seconds; germline transcripts, 94°C for 30 seconds, 60°C for 30 seconds, and 72°C for 1 minute. Primer sequences are summarized in Table 2.
Summary of PCR primers
RT-PCR and primer . | Sequence . |
|---|---|
| AID | |
| AID 5′ | 5′-GAGGCAAGAAGACACTCTGG-3′ |
| AID 3′ | 5′-GTGACATTCCTGGAAGTTGC-3′ |
| AID 5′int | 5′-CAACCTCAGTCTGAGGATCTTC-3′ |
| AID 3′int | 5′-GAAGATCCTCAGACTGAGGTTG-3′ |
| Clonotypic VDJ, clonotypic isotypes, repertoire analysis | |
| FR1c | 5′-GGTGCAGCTG (G/C) (A/T) G (G/C) AGTC (G/A/T) GG-3′ |
| JHc | 5′-ACCTGAGGAGACGGTGACC (A/G) (G/T) (G/T) GT-3′ |
| FR3 | 5′-CCGAGGACACGGC (T/C) (C/G) TGTATTACTG-3′ |
| Cμ | 5′-GGAATTCTCACAGGAGAC-3′ |
| Cδ | 5′-GTGTCTGCACCCTGATAT-3′ |
| Cγ | 5′-GGGGAAGACCGATGGGCCCT-3′ |
| Cα | 5′-GAGGCTCAGCGGGAAGACCTT-3′ |
| Internal control | |
| β2m 5′ | 5′-CCAGCAGAGAATGGAAAGTC-3′ |
| β2m 5′int | 5′-TGTCTTTCAGCAAGGACTGG-3′ |
| β2m 3′ | 5′-GATGCTGCTTACATGTCTCG-3′ |
| Switch region analysis | |
| Sw15as | 5′-GAAAGCTGGATGGAGTTGTC-3′ |
| SwA | 5′-CCGCAGGCAGCCAATAGAGT-3′ |
| Sγ | 5′-TCCCAGTGTCCTGCATTACTTC-3′ |
| Germline transcripts | |
| slμ | 5′-GTGATTAAGGAGAAACACTTTGAT-3′ |
| slγ1/2 | 5′-GGGCTTCCAAGCCAACAGGGCAGGACA-3′ |
| slγ3 | 5′-AGGTGGGCAGGCTTCAGGCACCGAT-3′ |
| slγ4 | 5′-TTGTCCAGGCCGGCAGCATCACCAGA-3′ |
| slϵ | 5′-GACGGGCCACACCATCC-3′ |
| asCμ | 5′-CCGAATTCAGACGAGGGGGAAAAGGGTT-3′ |
| asCγ? | 5′-GTTTTGTCACAAGATTTGGGCTC-3′ |
| asCγ2 | 5′-GTGGGCACTCGACACAACATTTGCG-3′ |
| asCγ3 | 5′-TTGTGTCACCAAGTGGGGTTTTGAGC-3′ |
| asCγ4 | 5′-ATGGGCATGGGGGACCATTTGGA-3′ |
| asCϵ | 5′-CGGAGGTGGCATTGGAGG-3′ |
RT-PCR and primer . | Sequence . |
|---|---|
| AID | |
| AID 5′ | 5′-GAGGCAAGAAGACACTCTGG-3′ |
| AID 3′ | 5′-GTGACATTCCTGGAAGTTGC-3′ |
| AID 5′int | 5′-CAACCTCAGTCTGAGGATCTTC-3′ |
| AID 3′int | 5′-GAAGATCCTCAGACTGAGGTTG-3′ |
| Clonotypic VDJ, clonotypic isotypes, repertoire analysis | |
| FR1c | 5′-GGTGCAGCTG (G/C) (A/T) G (G/C) AGTC (G/A/T) GG-3′ |
| JHc | 5′-ACCTGAGGAGACGGTGACC (A/G) (G/T) (G/T) GT-3′ |
| FR3 | 5′-CCGAGGACACGGC (T/C) (C/G) TGTATTACTG-3′ |
| Cμ | 5′-GGAATTCTCACAGGAGAC-3′ |
| Cδ | 5′-GTGTCTGCACCCTGATAT-3′ |
| Cγ | 5′-GGGGAAGACCGATGGGCCCT-3′ |
| Cα | 5′-GAGGCTCAGCGGGAAGACCTT-3′ |
| Internal control | |
| β2m 5′ | 5′-CCAGCAGAGAATGGAAAGTC-3′ |
| β2m 5′int | 5′-TGTCTTTCAGCAAGGACTGG-3′ |
| β2m 3′ | 5′-GATGCTGCTTACATGTCTCG-3′ |
| Switch region analysis | |
| Sw15as | 5′-GAAAGCTGGATGGAGTTGTC-3′ |
| SwA | 5′-CCGCAGGCAGCCAATAGAGT-3′ |
| Sγ | 5′-TCCCAGTGTCCTGCATTACTTC-3′ |
| Germline transcripts | |
| slμ | 5′-GTGATTAAGGAGAAACACTTTGAT-3′ |
| slγ1/2 | 5′-GGGCTTCCAAGCCAACAGGGCAGGACA-3′ |
| slγ3 | 5′-AGGTGGGCAGGCTTCAGGCACCGAT-3′ |
| slγ4 | 5′-TTGTCCAGGCCGGCAGCATCACCAGA-3′ |
| slϵ | 5′-GACGGGCCACACCATCC-3′ |
| asCμ | 5′-CCGAATTCAGACGAGGGGGAAAAGGGTT-3′ |
| asCγ? | 5′-GTTTTGTCACAAGATTTGGGCTC-3′ |
| asCγ2 | 5′-GTGGGCACTCGACACAACATTTGCG-3′ |
| asCγ3 | 5′-TTGTGTCACCAAGTGGGGTTTTGAGC-3′ |
| asCγ4 | 5′-ATGGGCATGGGGGACCATTTGGA-3′ |
| asCϵ | 5′-CGGAGGTGGCATTGGAGG-3′ |
β2m indicates β2-microglobulin.
Antibodies, cell sorting, and single-cell RT-PCR
Immunophenotypic analysis of mononuclear cells was by direct immunofluorescence using fluorescein isothiocyanate (FITC)-conjugated anti-CD20 (mouse IgG2a, rituximab24 ) and phycoerythrin (PE)-conjugated anti-IgM (goat Fab′2; Southern Biotechnology, Mississauga, ON, Canada). Isotype-matched antibodies were used as negative controls. Single-cell sorting and single-cell heminested RT-PCR have been previously described.6,25 Primer sequences and combination of primer sets for detection of various gene transcripts are summarized in Tables 2 and 3.
Combination of primer sets for single-cell heminested RT-PCR
. | PCR primers, sense/antisense . | . | |
|---|---|---|---|
| Transcript type . | First-stage PCR . | Second-stage PCR . | |
| AID 5′ | AID 5′/AID 3′ | AID 5′/AID 3′int | |
| AID 3′ | AID 5′/AID 3′ | AID 5′int/AID 3′ | |
| Clonotypic | FRIc/Cμ | CDR2/CDR3 | |
| β2m, control | β2m 5′/β2m 3′ | β2m 5′int/β2m 3′ | |
. | PCR primers, sense/antisense . | . | |
|---|---|---|---|
| Transcript type . | First-stage PCR . | Second-stage PCR . | |
| AID 5′ | AID 5′/AID 3′ | AID 5′/AID 3′int | |
| AID 3′ | AID 5′/AID 3′ | AID 5′int/AID 3′ | |
| Clonotypic | FRIc/Cμ | CDR2/CDR3 | |
| β2m, control | β2m 5′/β2m 3′ | β2m 5′int/β2m 3′ | |
Amplification of genomic sequences of clonotypic Sμ region
gDNA was prepared from mononuclear cells by the TRIzol method (Invitrogen) according to the manufacturer's instructions. Clonotypic Sμ regions were amplified from 20 to 80 ng gDNA template using CDR2 (sense) and Sw15as or SwA (antisense) primers (Table 2) and high-fidelity Taq DNA polymerase (Invitrogen) in a total volume of 50 μL. PCR was run at 94°C for 2 minutes, 30 cycles of amplification at 94°C for 30 seconds, 55°C for 30 seconds, and 68°C for 4 (Sw15as) or 8 (SwA) minutes and final extension at 68°C for 8 minutes. The product was analyzed on 0.5% agarose gel electrophoresis. For WM1-01, PCR product was subcloned into TOPO TA cloning system (Invitrogen) for DNA sequence analysis.
DNA sequence analysis
DNA sequencing was done using Big Dye 1.1 reagent (Applied Biosystems, Foster City, CA) following the manufacturer's instructions. The sequencing reaction was run on an ABI Prism 3100 Genetic Analyzer (Applied Biosystems) using ABI Prism DNA Sequencing Analysis version 5.1 and Seqscape software version 2.1 for data analysis.
Clonal isotype analysis
Clonal transcripts of various isotypes were amplified from cDNA by CDR2 or CDR3R (patient-specific; sense) and CH1 primers (Cα, Cδ, Cγ, or Cμ; antisense) and analyzed on 2% agarose gel electrophoresis. Enhanced PCR protocols involved use of 5 times more cDNA than the standard reaction described in “RT-PCR” and 35 cycles of PCR.
Analysis of IgH VDJ heterogeneity by DNA fragment analysis
Heterogeneity of IgH VDJ expression by populations of cells was analyzed by RT-PCR using 5′-hexachloro-fluorescein phosphoramidite (HEX)-labeled FR3 (sense) and JHc or CH1 (antisense) primers following the standard PCR protocol. The PCR product mixed with formamide and size standard (GeneScan-500 [ROX], Applied Biosystems) was analyzed on an ABI Prism 3100 Genetic Analyzer (Applied Biosystems) according to the manufacturer's instructions. Data analysis was performed using Gene-Mapper software version 3.5.
Activation of B cells by CD40L and IL-4
All tissue culture reagents, unless otherwise stated, were purchased from Invitrogen Life Technologies (Burlington, ON, Canada). CD40L-NIH 3T3 transfectants were a gift of Dr Gordon J Freeman (Dana-Farber Cancer Institute, Boston, MA) and were maintained in 45% DMEM, 45% nutrient mixture F12 ham, 10% fetal bovine serum, and 400 μg/mL geneticin. Stimulator cells were prepared by γ irradiation of CD40L-NIH3T3 cells with 96 Gy. Mononuclear cells isolated from BM aspirate or peripheral blood were cocultured for 6 days with stimulator cells at 6:1 ratio in IMEM supplemented with 5% fetal bovine serum, 10-5 M 2-mercaptoethanol, 3 × 10-5 M ethanolamine (Fisher Scientific, Ottawa, ON, Canada), and 10 ng/mL IL-4 (Sigma-Aldrich, Oakville, ON, Canada). At the end of cultivation, nonadherent cells were harvested for RNA, DNA, and single-cell RT-PCR.
Results
Deletions in the VDJ-S region of patients with WM and IgM MGUS are rare events, localizing to the Sμ tandem repeats
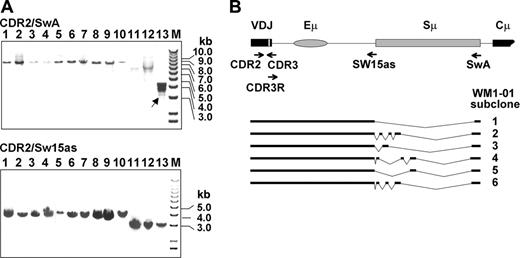
We explored the possibility that WM B cells may have incurred deletions in the IgH VDJ-S region, which severely impair CSR.18 The VDJ-S region is defined here as the DNA sequence downstream of the CDR2 in the rearranged clonotypic V gene, including Sμ switch region. The VDJ-S region of each patient is a fixed size, varying among patients depending on JH usage, with VDJH1-S generating a larger fragment than VDJH6-S. To determine whether deletions had occurred in this region, we amplified the VDJ-S of WM and IgM MGUS patients from gDNA and analyzed the products by agarose gel electrophoresis. Two separate reactions were conducted on each patient sample (Figure 1): one amplifying the VDJ-S region up to the Sμ tandem repeats (patient-specific CDR2 and Sw15as primers; 3-4 kb) and the other amplifying the VDJ-S region including the Sμ tandem repeats (CDR2/SwA; 7-8 kb). Each amplified fragment was validated as clonotypic by directly sequencing the CDR3 region, downstream of CDR2, which represents a patient-specific and tumor-specific sequence. In this analysis, deletions were detectable in 1 of 10 WM and 0 of 3 IgM MGUS patients. In the patient harboring deletions (WM1-01), multiple CDR2-SwA fragments were observed, approximately 3 kb shorter than the expected size. These fragments were not observed in the matching CDR2/Sw15as reaction, suggesting that the deletions originated in Sμ. Sequence analysis of subcloned CDR2/SwA fragments confirmed the presence of clones harboring distinct deletion profiles in the Sμ tandem repeats (Figure 1B). These clones also had a relatively high frequency of point mutations within the rearranged tandem repeats (∼0.5 kb; mutation rate = 1.8 × 10-2 bp, sequence not shown) despite containing an unmutated clonotypic VDJ. It is worth noting that although this patient exhibited deletions that may result from some level of CSR activity, the upstream VDJ sequence is unmutated, suggesting that somatic hypermutation and class-switching activities had become uncoupled.
IgM MGUS and WM patients share similar overall VDJ-S mutation profiles, with key differences defining those IgM MGUSs that are unlikely to be precursors to WM
Several studies have established a general pattern for mutation profiles in the VDJ-S region of B cells during adaptive immune responses.26 This pattern includes a gradient of mutations, higher in the VDJ region from somatic hypermutation, and lower in S region, where mutations may be a key event in initiating CSR. We analyzed gDNA sequences in the CDR2/Sw15as fragment of 12 WM and 4 IgM MGUS patients by direct sequencing to see if these patterns held true and to identify sequence features that may be indicators of relationships between these 2 conditions, or account for their lack of class switching. As shown in Figure 2, all patients, with the exception of WM1-01, exhibited this characteristic gradient of mutations. In addition, WM1-02, who had an unmutated VDJ (1% mutation, below the 2% cut-off), also exhibited a gradient of mutations. We enumerated potential CSR-related mutations in a 1.6-kb region immediately upstream of the Sμ tandem repeats, similar to previous studies on CSR.27 Of 12 WM clones, 4 had an unmutated 1.6-kb segment, 7 had 1 to 2 point mutations, and 1 (WM1-01) had a 24-bp deletion just upstream of Sw15. One of the IgM MGUS patients had a similar profile to the WM patients, with no mutations in the 1.6-kb region, whereas the remaining 3 IgM MGUS patients had a higher mutation rate in this region (4-7 bp). The average mutation frequency in WM and IgM MGUS switch region is calculated to be 0.57 × 10-3 bp and 2.5 × 10-3 bp, respectively (Table 4; P = .001). A closer examination of subcloned VDJ sequences of these 3 IgM MGUS patients revealed intraclonal diversity (Table 5), further distinguishing these IgM MGUS patients from WM patients. It is possible that the IgM MGUS patient with few mutations in upstream Sμ represents a patient group with the potential to progress to WM, whereas those IgM MGUS patients having a mutated upstream Sμ are not likely to give rise to WM having unmutated upstream Sμ.
Switch mutation analyses of WM and IgM MGUS
Group . | No. of sequences . | No. of nucleotides sequenced . | No. of mutations seen . | No. of mutated sequences/total sequences . | Mutation*frequency, × 10-3 bp . |
|---|---|---|---|---|---|
| WM‡ | 11 | 17875 | 10 | 7/11 | 0.56† |
| IgM MGUS | 4 | 6500 | 16 | 3/4 | 2.5† |
Group . | No. of sequences . | No. of nucleotides sequenced . | No. of mutations seen . | No. of mutated sequences/total sequences . | Mutation*frequency, × 10-3 bp . |
|---|---|---|---|---|---|
| WM‡ | 11 | 17875 | 10 | 7/11 | 0.56† |
| IgM MGUS | 4 | 6500 | 16 | 3/4 | 2.5† |
The fidelity of high-fidelity Taq polymerase was determined to be less than 1/10 000 bp. The error rate has been taken into consideration when calculating the significance level of the P value
P = .001; calculated by the Fisher exact test
WM1-01 clone containing 24-bp deletion in the switch region is not included in this analysis
VDJ intraclonal heterogeneity in IgM MGUS
Patient ID . | VH . | JH . | % VDJ mutation* . | VDJ intraclonal diversity . | No. of diversified clones/no. of total clones sequenced . | Switch μ mutation . |
|---|---|---|---|---|---|---|
| MGUS1 | 3 | 4 | 4.1 | + | 5/21 | + |
| MMGUS2 | 3 | 4 | 9.0 | + | 3/7 | + |
| MMGUS4 | 3 | 4 | 7.5 | + | 3/8 | + |
| MMGUS5 | 3 | 4 | 6.1 | + | 6/11 | ND |
| MMGUS3 | 6 | 4 | 5.4 | – | 0/8 | – |
Patient ID . | VH . | JH . | % VDJ mutation* . | VDJ intraclonal diversity . | No. of diversified clones/no. of total clones sequenced . | Switch μ mutation . |
|---|---|---|---|---|---|---|
| MGUS1 | 3 | 4 | 4.1 | + | 5/21 | + |
| MMGUS2 | 3 | 4 | 9.0 | + | 3/7 | + |
| MMGUS4 | 3 | 4 | 7.5 | + | 3/8 | + |
| MMGUS5 | 3 | 4 | 6.1 | + | 6/11 | ND |
| MMGUS3 | 6 | 4 | 5.4 | – | 0/8 | – |
ND indicates not determined.
Mutation rate is calculated based on the consensus VDJ sequences
PCR amplification of WM and IgM MGUS VDJ-S regions reveals rare deletion events. (A) CDR2/SwA and CDR2/Sw15as fragments analyzed by agarose gel electrophoresis reveal multiple VDJ-S fragments in patient WM1-01 (lane 13). Arrow indicates the deleted switch region of WM1-01 clone. Lanes 1-3, IgM MGUS clones using VH3/JH4 gene segments; lanes 4-9, WM clones using VH3/JH4 gene segments; lane 10, WM clone using VH3/JH3 gene segments; lanes 11-13, WM clones using VH3/JH6 gene segments; M indicates marker. Expected sizes of CDR2/SwA fragments are 7.8 (lanes 1-9), 8.1 (lane 10), and 6.8 kb (lanes 11-13). Expected sizes of CDR2/Sw15as fragments are 4 (lanes 1-9), 4.3 (lane 10), and 3 kb (lanes 11-13). (B) Deletions in subcloned WM1-01 VDJ-S fragments. The expected germline configuration is presented at the top, with subcloned fragments indicated by bars. VDJ refers to rearranged variable, diversity, and joining gene segments.
PCR amplification of WM and IgM MGUS VDJ-S regions reveals rare deletion events. (A) CDR2/SwA and CDR2/Sw15as fragments analyzed by agarose gel electrophoresis reveal multiple VDJ-S fragments in patient WM1-01 (lane 13). Arrow indicates the deleted switch region of WM1-01 clone. Lanes 1-3, IgM MGUS clones using VH3/JH4 gene segments; lanes 4-9, WM clones using VH3/JH4 gene segments; lane 10, WM clone using VH3/JH3 gene segments; lanes 11-13, WM clones using VH3/JH6 gene segments; M indicates marker. Expected sizes of CDR2/SwA fragments are 7.8 (lanes 1-9), 8.1 (lane 10), and 6.8 kb (lanes 11-13). Expected sizes of CDR2/Sw15as fragments are 4 (lanes 1-9), 4.3 (lane 10), and 3 kb (lanes 11-13). (B) Deletions in subcloned WM1-01 VDJ-S fragments. The expected germline configuration is presented at the top, with subcloned fragments indicated by bars. VDJ refers to rearranged variable, diversity, and joining gene segments.
WM cells express unmutated full-length AID and AID splice variants, similar to normal B cells
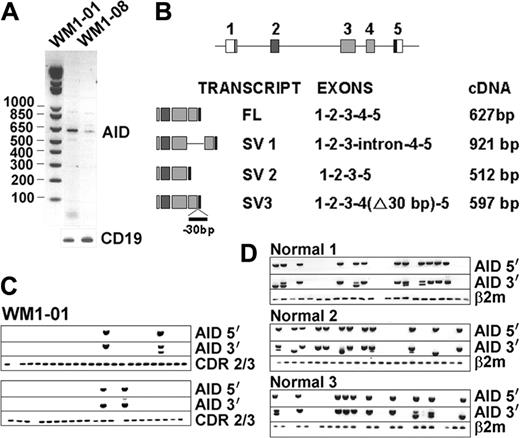
AID, a central enzyme in CSR, was analyzed by RT-PCR in BM and blood of 17 WM and 5 IgM MGUS patients. Two WM BM samples, but not the matching blood samples, tested positive for AID expression (Figure 3A). One sample carried unmutated VDJ (0%), whereas the other was highly mutated (6.2%). Sequence analysis revealed that amplified WM AID transcripts were unmutated. In addition to full-length (FL) transcripts, 3 other splice variants (SVs) were identified as summarized in Figure 3B. Speculating that AID SVs may impair CSR by interfering with the wild-type protein, we analyzed FL and SV coexpression in sorted CD20+ single B cells. Only a small proportion of individual WM cells accounted for AID expression, some of which coexpressed both FL and SV transcripts (Figure 3C). To assess whether certain SVs may be unique to tumor B cells, we determined the AID-expression profile in normal B cells stimulated to undergo CSR using CD40L and IL-4 (Figure 3D). Single-cell analysis of these normal activated CD20+ cells revealed expression of both FL and SV transcripts, similar to tumor B cells. Therefore, in the few patient samples where we could detect AID transcripts, both full-length and splice variants were unmutated and comparable to normal B cells. The causes and consequences of this low-level constitutive AID expression in WM remain to be determined.
CD40L/IL-4 stimulation up-regulates AID and germline transcripts in WM and IgM MGUS B cells independent of postswitch clonal isotypes
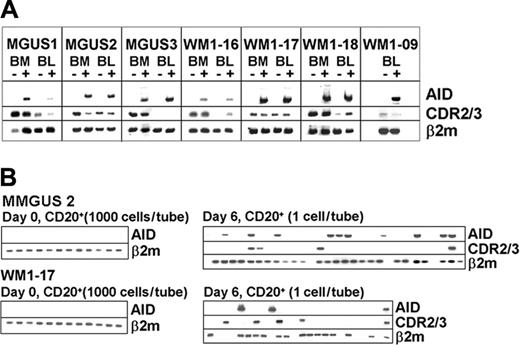
In vitro activation by CD40L and IL-4 up-regulates AID and germline transcripts and induces CSR in normal B cells.28,29 To assess if B cells from WM and IgM MGUS could be induced in the same manner, BM and blood mononuclear cells were cocultured with CD40L-NIH3T3 cells and recombinant IL-4 for 6 days. Expressions of AID, germline transcripts, clonal isotype transcripts, and the repertoire of IgH were compared before and after activation. Studies of 4 WM and 3 IgM MGUS patients showed that AID is inducible in both BM and blood (Figure 4A). Single-cell analysis also showed that both clonotypic and nonclonotypic B cells accounted for AID expression at comparable cell frequency (Figure 4B). Thus, the initial response of clonotypic B cells to the stimulation is comparable to that of autologous nonclonotypic B cells.
Mutation profiles of VDJ-S regions from patients with WM and IgM MGUS. The horizontal bar at the top depicts the DNA segment amplified by CDR2 and Sw15as primers. Patient mutation profiles are aligned directly underneath; vertical lines indicate point mutations, triangles indicate deletions (176 bp in WM1-06 and 24 bp in WM1-01). Total point mutations in the 1.6-kb region (dark gray bar) are summarized to the right.
Mutation profiles of VDJ-S regions from patients with WM and IgM MGUS. The horizontal bar at the top depicts the DNA segment amplified by CDR2 and Sw15as primers. Patient mutation profiles are aligned directly underneath; vertical lines indicate point mutations, triangles indicate deletions (176 bp in WM1-06 and 24 bp in WM1-01). Total point mutations in the 1.6-kb region (dark gray bar) are summarized to the right.
Analysis of clonotypic CSR (Figure 5A) revealed that in general, clonal IgM transcripts and clonal IgD transcripts were present in samples both before and after activation. Using standard PCR conditions with primers specific for CDR2 or CDR3 and IgH constant regions for 30 cycles of PCR, as previously reported,6 postswitch clonal isotypes were undetectable before or after in vitro stimulation of CSR. Some infrequent postswitch clonal isotypes, however, were detectable at day 6 in 2 of 4 WM (WM1-17, WM1-18) and 2 of 3 IgM MGUS (MMGUS1, MMGUS2) clones provided that the amount of cDNA template and cycles of amplification were increased. In the remaining 2 WM (WM1-09 and WM1-16) and 1 IgM MGUS (MMGUS3), clonal class switching remained undetectable. All clonal isotype transcripts were validated by direct sequencing of the entire PCR product to demonstrate that the correct clonotypic sequence was properly joined to the correct constant region. In all cases studied, germline transcripts were up-regulated (Figure 5B), indicating that CSR has been stimulated. It is not possible to distinguish the clonal origin of the germline transcripts because they do not encode VDJ regions.
The B-cell repertoire was further evaluated for IgH VDJ diversity using DNA fragment analysis (Figure 5C). Here, WM1-17 had a single monoclonal peak when amplified by FR3/JHc or FR3/Cμ, suggesting that clonotypic IgM predominates in the WM B-cell compartment. Monoclonal IgD, although present, was at lower intensity. In the unstimulated sample, the repertoire of α and γ chains was polyclonal and at a low expression level because IgG and IgA species were detected only with analysis of 10 times more PCR product. In the stimulated sample, signal intensity of α and γ chains remained unchanged, but diversity profiles shifted toward monoclonality as judged by the profile of DNA fragment peaks that indicate a loss of heterogeneity in CDR3 length. This analysis suggests the outgrowth of clonal postswitch B cells from the polyclonal B-cell population. Nevertheless, the entire B-cell repertoire is dominated by clonotypic IgM B cells.
The frequency of clonotypic IgG and IgM cells was compared. Poststimulated CD20+ cells were sorted and clonal isotype frequencies were analyzed by heminested RT-PCR. In WM1-17, we found that clonal IgG-expressing cells are present at a frequency of approximately 1 in 10 000 B cells (0.01%; Figure 5D) compared to the clonal IgM frequency of 8 in 48 B cells (16%). The estimated ratio of IgG/IgM-expressing cells is calculated to be 1:1600. Thus, although clonotypic IgG transcripts could be demonstrated after in vitro B-cell stimulation, clonal IgG-producing cells are rare.
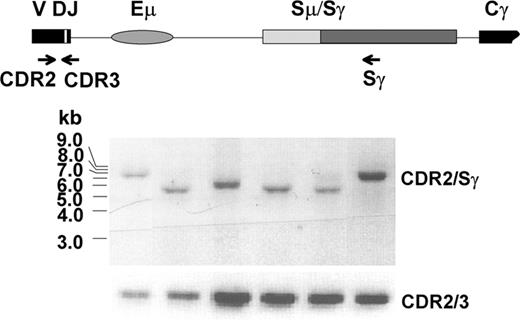
Clonal switch μ/γ junctions can be identified in WM after CD40L/IL-4 activation
To determine whether clonal IgG transcripts are expressed by one or many B clones that have undergone CSR, clonal switch μ/γ junctions were determined. The VDJ-S fragment was amplified by heminested PCR from activated WM1-17 and WM1-18 gDNA using corresponding VH leader (in both cases, VH3) and Sγ primers, followed by second amplification using CDR2 and Sγ primers. A representative result of subclone analysis from WM1-17 is shown in Figure 6. Clonality of each subclone was verified by successive CDR2/CDR3 amplification and DNA sequencing. Agarose gel analysis showing variable-sized CDR2/Sγ fragments and sequence analysis of the switch μ/γ junctions confirmed that each fragment arose from a unique recombination event. Thus, the postswitch clonotypic B cells that are present in 6-day cultures were generated by a series of apparently normal, independent CSR events.
The VDJ of clonotypic IgG exhibits intraclonal homogeneity and is identical to that of clonotypic IgM
If postswitch clonotypic B cells are premalignant remnants of the original parent B-cell clone giving rise to WM, they should exhibit intraclonal heterogeneity. If they arise after transformation of the WM clone, they should exhibit intraclonal homogeneity. To address this issue, we evaluated the extent, if any, of VDJ heterogeneity among clonotypic IgG transcripts by sequencing multiple CDR2/Cγ subclones (Figure 5A) and comparing the WM clonotypic IgM sequence and the CDR2/CDR3 sequence derived from 8 IgG subclones. In the 2 WM samples (WM1-17 and WM1-18) analyzed, we found that there is no intraclonal diversity among IgG subclones and the VDJ sequences were identical to that of their respective WM clonotypic IgM sequence.
AID is constitutively expressed in some WM samples. Single-stage RT-PCR showed positive bands in 2 WM BM samples (A). Various splice variants (SVs) identified in WM are summarized in panel B. Single-cell analysis of WM1-01 CD20+ cells (C) and activated CD20+ cells of healthy controls (D) were analyzed for clonotypic CDR2/CDR3 and AID expression.
AID is constitutively expressed in some WM samples. Single-stage RT-PCR showed positive bands in 2 WM BM samples (A). Various splice variants (SVs) identified in WM are summarized in panel B. Single-cell analysis of WM1-01 CD20+ cells (C) and activated CD20+ cells of healthy controls (D) were analyzed for clonotypic CDR2/CDR3 and AID expression.
Overall, analysis of in vitro stimulation of WM B cells suggests that the initial phase of CSR in WM and IgM MGUS is comparable to that occurring in normal B cells, as determined by AID induction and up-regulation of germline transcripts. Clonal IgG transcripts are detectable in some WM and IgM MGUS when stimulated with CD40L and IL-4. Clonal IgG-producing cells are relatively infrequent. Nevertheless, they are derived from several independent CSR events. Lack of heterogeneity in the CDR2/CDR3 sequence of clonal IgG and the homology to clonal IgM sequences suggested that IgG-producing clonotypic B cells are derived from WM B cells rather than the expansion of clones that preexist neoplastic transformation. We are unable to distinguish whether B cells expressing clonotypic IgG continuously arise in vivo and undergo clonal expansion in vitro, or alternatively, if they are generated from multiple independent CSR events occurring during in vitro stimulation.
Discussion
In the present report, clonal Sμ region, AID expression, and clonal CSR were analyzed in WM and IgM MGUS. Analysis of clonotypic switch regions from 10 WM and 3 IgM MGUS samples reveals that large deletions able to hinder CSR are not commonly found in WM or IgM MGUS. Sequence analysis showed that WM clones have fewer point mutations within the Sμ upstream region than do IgM MGUS clones. Unlike WM, the clonotypic B cells of IgM MGUS exhibit intraclonal heterogeneity of VDJ sequences and harbor frequent mutations in upstream Sμ. Constitutive expression of AID is rare in WM and undetectable in IgM MGUS but is inducible by CD40L/IL-4 activation in both conditions. AID is found in some, but not all, clonotypic B cells. Expression of AID splice variants is not unique to WM B cells because they are also found in stimulated B cells from healthy donors. Although undetectable using standard PCR conditions, with an enhanced PCR amplification strategy, clonotypic transcripts encoding post-switch isotypes can be detected in cells cultured with CD40L/IL-4 in some WM and IgM MGUS. As predicted, clonotypic IgG-producing cells are infrequent. Sequence analysis revealed that multiple, independent recombination events can be identified among clonotypic IgG-producing cells. Moreover, clonal IgG transcripts show intraclonal homogeneity, with a VDJ sequence identical to that of the autologous, clonotypic IgM.
The switch region located within J-C intron of immunoglobulin genes is known to be essential for CSR. Deletions within the Sμ sequence severely impair CSR.18 In CLL, large internal deletions were found in 34% of cases but these deletions did not correlate with VDJ mutational status or clinical outcome.30 A recent study of normal B cells suggests that endogenous Sμ deletions might result from recombination in the absence of appropriate downstream acceptor sites.15 Our finding that WM and IgM MGUS do not generally delete regions of endogenous Sμ suggests that the failure of CSR in WM involves abnormalities that are different from those seen in CLL. For one case of WM that has Sμ deletions, the multiple and heterogeneous deletions appear to differ from single deletion reported in CLL.30 This may suggest that switch deletion is a pretransformation event in CLL, whereas ongoing somatic alteration in the switch region may occur in WM.
Switch sequence analysis revealed striking differences between WM and IgM MGUS. We showed that switch sequence of WM exhibits low rates of mutation (0-2 bp/1.6 kb, 0.56 × 10-3 bp, n = 11) whereas the majority of IgM MGUSs harbor frequently mutated Sμ (0-7 bp/1.6 kb, 2.5 × 10-3 bp, n = 4). Thus, switch sequence analysis may help predict the likelihood of tumor progression. It is unlikely that a parent clone having multiple mutations in a given region will give rise to a malignant cell having unmutated sequence in that same region. Thus, IgM MGUS clones with frequently mutated switch sequences are less likely part of the precursor pool from which WM arises. Alternatively, those IgM MGUSs having intraclonal homogeneity of IgH VDJ clonotypic sequences and an unmutated switch region (such as MMGUS3) have molecular characteristics that are more consistent to WM clones. In combination with our knowledge of biased VH3 gene usage in WM (17 of 18 WM patients studied so far), these parameters may help set informative criteria for identifying those patients with IgM MGUS at risk of transformation to WM. Future studies with a larger cohort of IgM MGUS patients along with the clinical follow-up of these IgM MGUS individuals are required to evaluate this hypothesis.
AID is inducible in both WM and IgM MGUS. (A) AID expression is compared before and after CD40L/IL-4 stimulation by bulk RT-PCR. (B) Aliquots of 1000 cells/tube from unstimulated CD20+ B cells were analyzed for AID transcripts (left side of each row) or single CD20+ cells from poststimulated samples were sorted and analyzed for clonotypic and AID transcripts (right side of each row).
AID is inducible in both WM and IgM MGUS. (A) AID expression is compared before and after CD40L/IL-4 stimulation by bulk RT-PCR. (B) Aliquots of 1000 cells/tube from unstimulated CD20+ B cells were analyzed for AID transcripts (left side of each row) or single CD20+ cells from poststimulated samples were sorted and analyzed for clonotypic and AID transcripts (right side of each row).
CD40L/IL-4 activated clonal isotype expression in healthy donor, WM, and IgM MGUS cells. Clonal isotype transcripts were determined by RT-PCR using standard (Std) or enhanced (Enh) amplification protocol (A). Day 0, unstimulated; day 6, after stimulation; CDR2/CDR3 (lane 1), CDR2/Cμ (lane 2), CDR2/Cδ (lane 3), CDR2/Cα (lane 4), and CDR2/Cγ (lane 5). In normal control, lanes 1 and 3 are not determined and CDR2 is replaced by FR1c. In WM1-09, CDR2 is replaced by CDR3R. Representative results of germline transcript up-regulation after CD40L/IL-4 activation are shown in panel B. The primers (sense/antisense) and expected size of PCR products are as follows: M, sIμ/asCμ, 537 bp; G1, sIγ1/2/asCγ1, 603 bp; G2, sIγ1/2/asCγ2, 597 bp; G3, sIγ3/asCγ3, 670 bp; G4, sIγ4/asCγ4, 411 bp; E, sIϵ/asCϵ, 125 bp. CDR3 repertoire analysis of WM1-17 was conducted for B cells expressing each isotype (FR3/Cμ, FR3/Cδ, FR3/Cα, and FR3/Cγ) and total B cells (FR3/JHc) (C). IgM (CDR2/Cμ), IgG (CDR2/Cγ), and total clonal frequency (CDR2/CDR3) was analyzed in poststimulated CD20+ cells of WM1-17 (D).
CD40L/IL-4 activated clonal isotype expression in healthy donor, WM, and IgM MGUS cells. Clonal isotype transcripts were determined by RT-PCR using standard (Std) or enhanced (Enh) amplification protocol (A). Day 0, unstimulated; day 6, after stimulation; CDR2/CDR3 (lane 1), CDR2/Cμ (lane 2), CDR2/Cδ (lane 3), CDR2/Cα (lane 4), and CDR2/Cγ (lane 5). In normal control, lanes 1 and 3 are not determined and CDR2 is replaced by FR1c. In WM1-09, CDR2 is replaced by CDR3R. Representative results of germline transcript up-regulation after CD40L/IL-4 activation are shown in panel B. The primers (sense/antisense) and expected size of PCR products are as follows: M, sIμ/asCμ, 537 bp; G1, sIγ1/2/asCγ1, 603 bp; G2, sIγ1/2/asCγ2, 597 bp; G3, sIγ3/asCγ3, 670 bp; G4, sIγ4/asCγ4, 411 bp; E, sIϵ/asCϵ, 125 bp. CDR3 repertoire analysis of WM1-17 was conducted for B cells expressing each isotype (FR3/Cμ, FR3/Cδ, FR3/Cα, and FR3/Cγ) and total B cells (FR3/JHc) (C). IgM (CDR2/Cμ), IgG (CDR2/Cγ), and total clonal frequency (CDR2/CDR3) was analyzed in poststimulated CD20+ cells of WM1-17 (D).
In an attempt to identify normal B-cell counterparts and to evaluate mutational status of immunoglobulin genes, expression of AID has been studied in various lymphoproliferative disorders. High AID expressors found in follicular lymphoma (FL), Burkitt lymphoma (BL), and diffuse large B-cell lymphoma (DLBCL) are comparable to germinal center B cells.31 SHM of VH genes coincides with AID expression in FL and DLBCL.32 Alternatively, AID expression appears to be absent from malignant cells in multiple myeloma, as expected of late-stage B/plasma cells.33 Interestingly, some types of CLL B cells constitutively express AID, predominantly among those with unmutated IgH VDJ genes.15,33,34 In our studies, most WM and IgM MGUS patients lack detectable AID expression, a phenotype characteristic of late-stage B cells. In WM1-01, AID expression persists among 2 BM samples collected 3 years apart (data not shown) but a very small subpopulation of tumor cells, in fact, accounted for AID expression (Figure 3). AID SVs are colocalized with full-length transcripts in some individual cells from WM and IgM MGUS patients and healthy donors. The mechanisms by which SHM is suppressed in a WM clone expressing AID but having unmutated IgH VDJ remain to be explored.
Several independent recombinations took place in clonal class switching of WM. Subclones of CDR2/Sγ fragments amplified from CD40L/IL-4 activated B cells of WM1-17 (Figure 5D) were shown to have variable size. Each one was confirmed clonotypic by CDR2/CDR3 analysis and direct sequencing.
Several independent recombinations took place in clonal class switching of WM. Subclones of CDR2/Sγ fragments amplified from CD40L/IL-4 activated B cells of WM1-17 (Figure 5D) were shown to have variable size. Each one was confirmed clonotypic by CDR2/CDR3 analysis and direct sequencing.
Our identification of multiple hybrid Sμ/Sγ junctions among clonotypic IgG transcripts suggests that clonal CSR had occurred within WM cells. Clonotypic IgG transcripts do not exhibit intraclonal heterogeneity, thus ruling out an origin from circulating premalignant, clonally related B cells. It is difficult to distinguish experimentally whether the clonotypic IgG B cells preexist in vivo and have undergone clonal expansion during the 6-day culture or clonotypic IgM B cells have undergone CSR in vitro. First, preexisting IgG clones, if present, are undetectable in the prestimulated sample. Moreover, a search for recombination products such as switch circles, although possible, cannot identify recombination products from events involving clonotypic IgH VDJ in a population that also includes some polyclonal B cells, because they do not include the clonotypic signature. Regardless of their origin, post-switch clonotypic B cells have an extremely low frequency in WM (IgG/IgM-cell frequency = 1/1600). Thus, for the majority of WM B cells, CSR does not occur even when stimulated in vitro, suggesting that the WM cell is constitutively unable to or being prevented from carrying out CSR.
In conclusion, WM and IgM MGUS share several common characteristics including paraproteinemia, VH gene usage, hypermutated VDJ, lack of constitutive AID expression, and normal AID inducibility as well as similar pattern of postswitch clonal isotype responsiveness to CD40L/IL-4 stimulation. Despite these similarities, they are different in terms of switch mutation and intraclonal diversity of VDJ. Overall, our studies suggest that CSR is a rare event in WM cells. Postswitch clonal isotype expression occurs at very low cell frequencies, with clonotypic IgG B cells deriving from multiple CSR events in transformed B cells. Finally, analysis of switch sequence and clonotypic VDJ may help in predicting those patients with IgM MGUS who are at risk of progression to WM.
Prepublished online as Blood First Edition Paper, November 29, 2005; DOI 10.1182/blood-2005-09-3613.
Supported by a Smokler Research Grant from the Research Fund for Waldenstrom Ltd.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal