Abstract
The membrane-spanning 4A (MS4A) family of proteins includes CD20, FcϵRIβ, and HTm4, whose genes are grouped in a chromosomal location that is associated with increased susceptibility to allergy and atopic asthma. One family member, Chandra/MS4a4B, was reported to be expressed in T helper 1 (Th1) T cells but not Th2 T cells. In the present study, Ms4a4b was isolated in a screen of genes differentially expressed during thymocyte development. MS4a4B was detected in immature CD4-CD8-CD44+CD25- thymocytes, turned off during further stages of thymocyte development and reexpressed in mature single-positive thymocytes. MS4a4B expression was found in naive CD8+ and CD4+ peripheral T cells and natural killer (NK) cells but not in B cells. MS4a4B is expressed at the cell surface with its C-terminus located in the cytoplasm. When expressed in a T-cell hybridoma by retroviral vector, MS4a4B protein constitutively associated with lipid raft microdomains, whereas in primary T cells endogenous MS4a4B protein became enriched in rafts after T-cell activation. Overexpression of MS4a4B in primary CD4+ T-cell blasts enhanced T-cell receptor (TCR)-induced Th1 cytokine production. These results suggest that MS4a4B expression is tightly regulated during T-cell development and that MS4a4B expression promotes Th1 function and/or differentiation. (Blood. 2006;107:2400-2408)
Introduction
MS4A (membrane-spanning 4-domain family, subfamily A) is a large family of proteins that includes at least 26 members in mouse and humans.1,2 Each protein is predicted to traverse the membrane bilayer 4 times, and the greatest sequence conservation among family members is found in the transmembrane domains. Although similar in overall structure to the tetraspanin family (CD81, etc) the MS4A family does not show sequence homology to the tetraspanins.
The genes for the MS4A family are linked on chromosome 11q12-q13.1 in human and syntenic mouse chromosome 19, strongly suggesting a common evolutionary precursor.3 Genetic variations at chromosome 11q12-q13 have been implicated in the pathogenesis of allergic diseases in humans.4-11 One gene that is present in this locus is FcϵRIβ, a signal amplifier of the high-affinity IgE receptor FcϵRI and the low-affinity IgG receptor FcγRIII (reviewed in Dombrowicz et al12 ), and therefore a likely candidate susceptibility gene for atopy. FcϵRIβ possesses an immunoreceptor tyrosine activation motif (ITAM) in the cytoplasmic tail. This ITAM motif has been shown to be important for amplification of signal transduction induced by IgE binding.12,13 However, some known polymorphisms in FcϵRIβ have so far been excluded as susceptibility determinants for allergy,14 and other studies provide evidence for genetic linkage only.9 Therefore, it is possible that other genes encoded in the locus may contribute to allergy susceptibility.
The most well-known MS4A family member is CD20, a B-cell-specific molecule. Monoclonal antibodies against CD20 are among the most successful passive immunotherapeutic drugs in the treatment of B-cell lymphomas and autoimmune disease.15 The mechanism of action of anti-CD20 immunotherapy is complex. Although antibody-dependent and complement-mediated killing is clearly part of the mechanism of passive immunotherapy,16-18 CD20 crosslinking by antibodies can also induce growth arrest and apoptosis in lymphoma cell lines.19-22 These results have prompted investigations into the potential signaling function of CD20. Although CD20 is a major target for serine/threonine phosphorylation on mitogen stimulation of B cells (reviewed in Riley and Sliwkowski23 ), it is not known whether its phosphorylation or associations with known signaling molecules are important for function. CD20 is known to physically associate with many proteins in raft detergent-resistant lipid microdomains.24,25 Several studies show that CD20 promotes calcium entry in B cells and in other cells when expressed ectopically26-28 and that lipid raft localization contributes to its calcium entry function.29,30
Ms4a4b was first described as a T helper 1 (Th1)-specific transcript and protein that was repressed by T-cell differentiation under Th2 conditions.31 In this report, we show that Ms4a4b is differentially expressed during immature thymic development. Further analysis of its expression showed that MS4a4B is expressed not only in naive CD4+ but also in CD8+ T cells, natural killer (NK) cells, and in thymocytes at precommitment and mature developmental stages. MS4a4B protein was found on the surface of T cells, and its membrane topology is similar to that of CD20. Using Western blot and a flow cytometric assay, we show that MS4a4B is present in detergent-resistant membranes and that it is localized to lipid rafts on activation of naive primary T cells. We further demonstrate that retroviral vector-mediated overexpression of MS4a4B enhanced IL-2 and IFN-γ production but not IL-4, IL-10, or TNF-α production in activated primary T cells. These results suggest that MS4a4B may play a regulatory role in cytokine production or differentiation of T cells.
Materials and methods
Mice and cells
C57BL/6J mice (The Jackson Laboratory; Bar Harbor, ME) were used as donors for tissues and primary cells. The use of laboratory animals conformed to the Guide for the Care and Use of Laboratory Animals32 and was reviewed by an internal committee for the care and use of animals in research.
NIH 3T3 fibroblast cells, WEHI-231 B cells, MEL-201 erythoroleukemia, 70Z/3 pre-B cells, CTLL-2, and HT-2 T cells were from American Type Culture Collection (ATCC; Manassas, VA) and maintained according to instructions. T-cell clones T32 and 5cc7 were obtained from D. Scott (University of Maryland School of Medicine, Baltimore, MD) and L. J. Berg (University of Massachusetts Medical School, Boston, MA), respectively. The 58α-β (58α) T-hybridoma line was obtained from D. L. Wiest (Fox Chase Cancer Center, Philadelphia, PA), and the α28 and α3A3 T-cell hybridoma lines were described previously.33 Th2 cells (gift of A. Keegan, University of Maryland School of Medicine) were derived by expansion from DO11.10/rag-2-/- lymph node T cells in the presence of antigen, irradiated antigen-presenting cells, IL-4 cytokine, and anti-IL-12 neutralizing antibody. Primary macrophages were produced from adherent spleen cells from C57BL/6 mice that were cultured for 2 weeks in RPMI complete medium containing 10% FCS and 5% monocyte colony stimulating factor (MCSF) containing supernatants from L929 cell culture. The resulting cells were typically greater than 97% CD11b+ cells.
Gene cloning and construction of Ms4a4b retroviral vectors
A cDNA library was generated by polymerase chain reaction (PCR) subtraction (representation difference analysis) cloning using mRNA isolated by fluorescence-activated cell sorting (FACS) in CD44+CD25-CD4-CD8- and CD44-CD25+CD4-CD8- thymocytes of RAG-1-/- mice. The library was screened using probes generated by subtraction of CD44-CD25+ thymocyte RNA from CD44+CD25- thymocyte RNA, and the Ms4a4b sequence was 1 of 150 unique clones isolated based on its differential expression in the CD44+CD25- subpopulation. The full-length sequence of Ms4a4b was obtained by 3′ rapid amplification of cDNA ends (RACE) extension. The full-length clone was amplified from C57Bl/6 mouse thymus cDNA by PCR with Ms4a4b primers (sense, CACGAGGACTAGTAGATCTACCATGCAAGGACAGGAACAGAC; antisense, GGACTGCGGCCGCGTTACCTCAAAAGCCAGTGCAATGAAGTGT), and cloned in PCR2.1 TA-cloning vector (Invitrogen, Carlsbad, CA) and confirmed by sequencing. The sequence obtained fully matched that deposited in the NCBI GenBank NM_021718. A hemagluttinin (HA) epitope tag (YPYDVPDYA) was inserted into the MS4a4B protein between the third and fourth transmembrane domains of MS4a4B in the MIGR retroviral vector by using the QuikChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA). The mutated sequence was confirmed by PCR sequencing. The coding sequence of Ms4a4b was inserted into a MIGR I retroviral vector (from W. Pear, University of Pennsylvania, Philadelphia, PA) by blunt-end ligation. Ms4a4b vectors were selected to transfect a packaging cell line (Phoenix cells; G. Nolan, Stanford University, Stanford, CA), and titers of retrovirus supernatants were determined by infection of NIH 3T3 cells.
Analysis of Ms4a4b mRNA transcription by Northern blot and RT-PCR
Total RNA was isolated from tissues or cultured cells with TriZol (Life Technologies, Gaithersburg, MD). Northern blots were hybridized by 32 P-labeled Ms4a4b probe or G3PDH cDNA probe (Clontech, San Diego, CA) and detected by autoradiography. For reverse transcriptase (RT)-PCR, 2 μg RNA was transcribed into cDNA. Samples were normalized by HPRT PCR (primers: sense, GTAATGATCAGTCAACGGGGGAC; antisense, CCAGCAAGCTTGCAACCTTAACCA; conditions: denaturation, 94°C for 2 minutes; PCR, 94°C for 1 minute, 60°C for 1 minute, 72°C for 1 minute for 30 cycles; extension, 72°C for 8 minutes) and then were amplified with the primers (sense, ACCATGCAAGGACAGGAACAGAC; antisense, CCTCAAAAGCCAGTGCAATGAAGTGT) specific for MS4a4B (denaturation, 94°C for 2 minutes 30 seconds; PCR 94°C for 30 minutes, 57°C for 1 minute, 72°C for 1 minute, 32 cycles; extension, 72°C for 8 minutes). All PCR primers were synthesized by MWG (High Point, NC). PCR products were electrophoresed on 1.8% agarose gel in the presence of ethidium bromide and were photographed.
For quantitative real-time PCR, specific primers were generated as follows: Ms4a4b (sense, ACCATGCAAGGACAGGAACAGAC; antisense, CCTCAAAAGCCAGTGCAATGAAGTGT), Ms4a4c (sense, CATTGCTGTGTCCATCTCTGCTT; antisense, CACACACACACACACACACACAC), and Ms4a4d (sense, TAGAAGTCAACCAGCCATCGCTA; antisense, TGCACACACACACTCTCACACATGA) transcripts using the Roche (Indianapolis, IN) LightCycler and Sybr Green detection chemistry. PCR reactions (94°C, 0.1 or fewer seconds; 58°C, 10 seconds; 72°C, 13 seconds; 40 cycles, followed by melting curve verification) were performed using coupled reverse-transcription/PCR kits from Roche or QIAGEN (Valencia, CA). Transcript quantitation was relative to HPRT and 18S ribosomal RNA standards (Applied Biosystems, Foster City, CA.) Reaction conditions and primer sequences were optimized for specificity and sensitivity. The 4B, 4C, and 4D products were determined by gel electrophoresis to be single bands of expected size. When an Ms4a4b-containing plasmid was used a control target, the 4B primers, and only the 4B primers, amplified a product. Thus, the 4C and 4D primers did not crossreact with Ms4a4b. The melting curves of the 4B product amplified from cloned target and from spleen cDNA were identical. This shows that the Ms4a4c message (present in spleen) cannot be amplified using 4B primers. Finally, the 4B primers were unable to amplify a product from lung, which was strongly positive using the 4D primers, or from brain, which was strongly positive using 4C primers. These data show that the 4B primers are unable to amplify a product using either 4C or 4D gene products as templates. For semiquantitative RT-PCT, the products were verified by DNA sequencing. All results were within the linear response of the assay.
Immunostaining, flow cytometric analysis, and cell sorting
Antibodies used for immunostaining mouse cells were obtained from BD-Biosciences (San Diego, CA). Biotinylated anti-HA was from Roche. For intracellular staining for MS4a4B, cells were first surface stained, then fixed and permeabilized using CytoFix&Perm (BD-Biosciences) before incubating with biotinylated MS4a4B or Ig control reagents. Flow cytometric analysis was performed using a FACSCalibur (BD-Biosciences) and FlowJo software (Tree Star, San Carlos, CA). Cell sorting was performed in the Flow Cytometry Core of the Holland Lab (defunct), American Red Cross.
Preparation of anti-MS4a4B antibody
A 16-amino acid cytoplasmic terminal peptide (TNVPGNVYKNHPGEIV) was synthesized and coupled to KLH as an immunogen. Custom antibody to MS4a4B-peptide was prepared by BioSource International (Hopkinton, MA) using a standard protocol for immunization of rabbits. Antibody titers were determined by enzyme-linked immunosorbent assay (ELISA). Anti-MS4a4B antibody in serum from the final bleeding was purified by affinity chromatography with peptide-coupled Sepharose 4B column and further characterized as described under “Results.”
Western blot and immunoprecipitation
Isolation of lipid rafts by sucrose gradient fractionation
Lipid rafts were isolated from cell lysates by discontinuous sucrose gradient ultracentrifugation as described.36 Rafts and nonraft fractions were identified by Western blot with Cholera Toxin B-HRP conjugate (Sigma, St Louis, MO), rabbit anti-LAT antibody (Upstate Biotechnology, Lake Placid, NY), and rabbit anti-CD71 (transferrin receptor) (Santa Cruz Biotechnology, Santa Cruz, CA). Flow cytometric analysis of association with detergent-resistant membranes was performed as described.37 In some cases, cells were preincubated at 37°C for 30 minutes with methyl-beta-cyclodextrin (Sigma) or at 37°C for 5 minutes with sphingomyelinase (Sigma) prior to staining.
Retroviral infection of cell lines
Retroviral infection of cell lines or primary CD4+ T cells35,38 was performed as previously described. For T-cell hybridomas, GFP+ cells were isolated by FACS sorting (FACSVantage; BD-Biosciences) after retroviral infection and then stimulated with antigen as previously described.35 Cell supernatants were harvested and assayed for IL-2 content by CTLL-2 bioassay and by ELISA (OPTIEIA; BD-Biosciences) as described previously.35
After infection, primary T cells were stimulated with PMA (20 ng/mL) and ionomycin (1 μg/mL) in the presence of monensin (2 μM) for 4 hours before harvest. Washed cells were permeabilized and fixed with CytoFix&Perm reagents (BD-Biosciences), and then stained with anti-cytokine antibodies (BD-Biosciences). The percentage of cytokine-positive cells was determined by flow cytometry, analyzing viable GFP+ cells.
Results
Expression of MS4a4B mRNA
To isolate genes whose expression is regulated during T-lineage commitment, we performed a representation difference analysis (RDA) experiment using mRNA from RAG-1-deficient thymocytes just prior to and just after T commitment. These stages were identified by the expression of cell-surface markers CD25 and CD44 on double-negative thymocytes. Precommitted CD44+CD25-CD4-CD8- thymocytes and the postcommitted CD25+CD44-CD4-CD8- thymocyte populations were isolated simultaneously by cell sorting. About 600 cDNA clones isolated from the screen were sequenced, and about 100 were retested for differential expression using independently amplified, RDA subtracted probes. A synopsis of the general classes of genes and their distribution between the precommitted CD44+CD25-CD4-CD8- thymocytes and the postcommitted CD25+CD44-CD4-CD8- thymocyte populations are shown in Table S1 on the Blood website; see the Supplemental Table link at the top of the online article. As expected, the CD44+CD25- library of sequences was enriched for genes expressed in non-T lineages such as NK, whereas the complementary CD44-CD25+ library was enriched for T-cell-specific transcripts. Interestingly, the CD44+CD25- library was also relatively enriched for sequences associated with cell adhesion and cytoskeletal components, whereas the CD44-CD25+ library was enriched for sequences associated with metabolic functions.
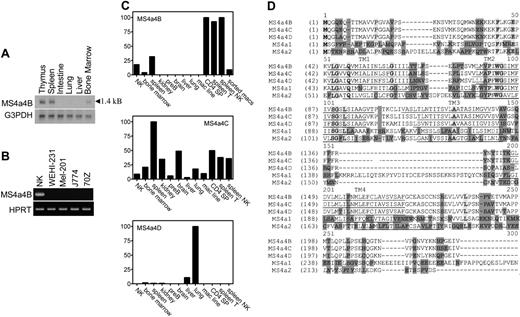
The gene selected for further analysis in this report matched a previously identified gene named Ms4a4b. Ms4a4b was expressed in uncommitted CD25-CD44+CD4-CD8- thymocytes and silenced in the CD25+CD44-CD4-CD8- thymocyte population. Northern blot analysis of tissue samples showed an MS4a4B mRNA transcript of 1.4 kB (kilobase) in thymus, spleen, and bone marrow but not in intestine, liver, and lung, suggesting that Ms4a4b expression is restricted to immune cells (Figure 1A). Expression of Ms4a4b mRNA in cultured cell lines was measured using RT-PCR with Ms4a4b-specific primers. A correctly sized PCR product was detected in T cells (not shown) and NK cells but not in WEHI 231 (a B-cell line), MEL-201 (erythroleukemia cell line), and 70Z/3 (pre-B-cell line) (Figure 1B). Using quantitative real-time RT-PCR, high levels of Ms4a4b RNA were found in mouse thymus, spleen, and NK cells but not in kidney, a pre-B-cell line, brain, liver, lung, and a macrophage cell line (Figure 1C). Analysis of FACS-sorted lymphocytes showed that Ms4a4b was highly transcribed in T cells and NK cells but not in B cells and only weakly in macrophages (Figure 1C).
Ms4a4b is a member of the Ms4a4 gene subfamily, which includes Ms4a4c and Ms4a4d.3 The sequence and homology of Ms4a4b to its closest mouse family members (Ms4a4c and Ms4a4d) as well as Ms4a1 (CD20) and Ms4a2 (FcϵRIβ) are displayed in Figure 1D. Because of the high sequence similarity among the proteins of this subfamily, the RNA expression profile of Ms4a4b with Ms4a4c and Ms4a4d was compared in both tissues and cells. Although Ms4a4b was highly expressed in T cells and NK cells, Ms4a4d was not detected in T cells and NK cells but was relatively restricted to lung and liver. Ms4a4c was more broadly expressed, with low expression in B cells, and its expression overlapped with that of Ms4a4b in T cells and NK cells (Figure 1C).
Characterization of anti-MS4a4B protein expression
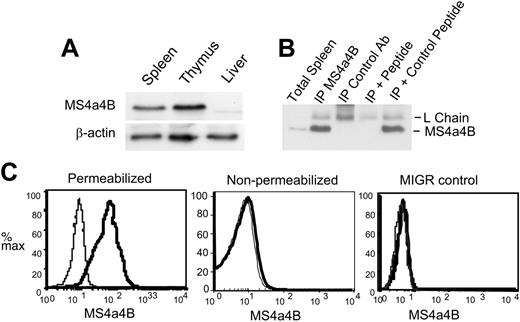
To detect MS4a4B protein, rabbit polyclonal antibodies were generated to a peptide from the C-terminal cytoplasmic domain of MS4a4B. The antibody reacted well with a band of about 24 kDa by Western blot of thymus or spleen but not liver (Figure 2A-B). Immunoprecipitation of MS4a4B was blocked by MS4a4B C-terminal peptide but not an irrelevant peptide (Figure 2B). The lack of reactivity in liver by Western blot and B cells (by flow cytometry; Figure 4A) suggest that the antibody does not strongly crossreact with MS4a4C or MS4a4D. Nevertheless, antibody recognition of MS4a4C or MS4a4D (which is not expressed in lymphoid cells or tissue) cannot be completely ruled out.
NIH 3T3 cells, which do not express Ms4a4b mRNA (not shown), were used to examine ectopically expressed MS4a4B protein by retroviral-mediated transduction. MS4a4B was detected in infected 3T3 cells but not control vector-infected cells and only after permeabilization (Figure 2C). Because the epitope chosen for antibody generation was C-terminal, the topology of the molecule is likely as predicted, based on its homology with CD20, with the N- and C-terminal regions within the cells, and 4 transmembrane domains.
Selective MS4a4B protein expression in hematopoietic cells
Because MS4a4B was cloned from uncommitted thymocytes and its mRNA was detected in both bone marrow and fetal thymus (data not shown), we further determined the expression of MS4a4B protein during T-cell development. In the thymus, T-cell precursors pass through several stages to develop into mature T lymphocytes. These are characterized by ordered changes in expression of cell-surface proteins; CD4-/CD8- (double negative, DN) → CD4+/CD8+ (double positive, DP) → CD4+ or CD8+ (single positive, SP).39 The DN stage can be further subdivided by the regulated expression of CD44 and CD25 in order of increasing maturity: CD44+/CD25- → CD44+/CD25+ → CD44-/CD25+ → CD44-/CD25-.40
MS4a4B mRNA is expressed in mouse tissues and cells. (A) Northern blot: 10 μg RNA was separated on 1% denatured agarose gel and transferred to membrane, probed using Ms4a4b or G3PDH probes, washed, and exposed overnight. The length of the transcript is shown. (B) RT-PCR: samples were normalized by HPRT PCR and then were amplified with the primers specific for Ms4a4b, separated by gel, stained with ethidium bromide, and photographed. Samples were IL-2-expanded NK cells from severe combined immunodeficient (SCID) bone marrow, WEHI-231 B cells, Mel-201 erythroid line, J774 macrophage line, and 70Z-13 pre-B-cell line. (C) Quantitative PCR for expression of Ms4a4b, Ms4a4c, and Ms4a4d was performed using the Light-Cycler system (see “Materials and methods”). Results are in arbitrary units normalized to HPRT, and ribosomal RNA standards were done in parallel. Results shown are representative of 2 experiments. Samples are (1) expanded primary NK cells, (2) bone marrow, (3) whole spleen, (4) kidney, (5) pre-B-cell line 70Z13, (6) brain, (7) liver, (8) lung, (9) macrophage line (J774) FACS-sorted cells, (10) CD4 SP (single-positive thymocytes), (11) spleen T cells (CD3+), (12) spleen NK cells (NK1.1+), and (13) macrophages (Mac1+). (D) Alignments of deduced amino acid sequences for the mouse Ms4a4 subfamily, Ms4a1 (CD20), and Ms4a2 (FcϵRIβ). The membrane-spanning domains (TM) are predicted by TMHMM 2.0 program and underlined. Identical sequences are indicated by bold letters in light shade; conservative sequences by unshaded letters; nonhomologous sequences by dark shade. ITAM motif in cytoplasmic domain of MS4a2 is indicated as italic.
MS4a4B mRNA is expressed in mouse tissues and cells. (A) Northern blot: 10 μg RNA was separated on 1% denatured agarose gel and transferred to membrane, probed using Ms4a4b or G3PDH probes, washed, and exposed overnight. The length of the transcript is shown. (B) RT-PCR: samples were normalized by HPRT PCR and then were amplified with the primers specific for Ms4a4b, separated by gel, stained with ethidium bromide, and photographed. Samples were IL-2-expanded NK cells from severe combined immunodeficient (SCID) bone marrow, WEHI-231 B cells, Mel-201 erythroid line, J774 macrophage line, and 70Z-13 pre-B-cell line. (C) Quantitative PCR for expression of Ms4a4b, Ms4a4c, and Ms4a4d was performed using the Light-Cycler system (see “Materials and methods”). Results are in arbitrary units normalized to HPRT, and ribosomal RNA standards were done in parallel. Results shown are representative of 2 experiments. Samples are (1) expanded primary NK cells, (2) bone marrow, (3) whole spleen, (4) kidney, (5) pre-B-cell line 70Z13, (6) brain, (7) liver, (8) lung, (9) macrophage line (J774) FACS-sorted cells, (10) CD4 SP (single-positive thymocytes), (11) spleen T cells (CD3+), (12) spleen NK cells (NK1.1+), and (13) macrophages (Mac1+). (D) Alignments of deduced amino acid sequences for the mouse Ms4a4 subfamily, Ms4a1 (CD20), and Ms4a2 (FcϵRIβ). The membrane-spanning domains (TM) are predicted by TMHMM 2.0 program and underlined. Identical sequences are indicated by bold letters in light shade; conservative sequences by unshaded letters; nonhomologous sequences by dark shade. ITAM motif in cytoplasmic domain of MS4a2 is indicated as italic.
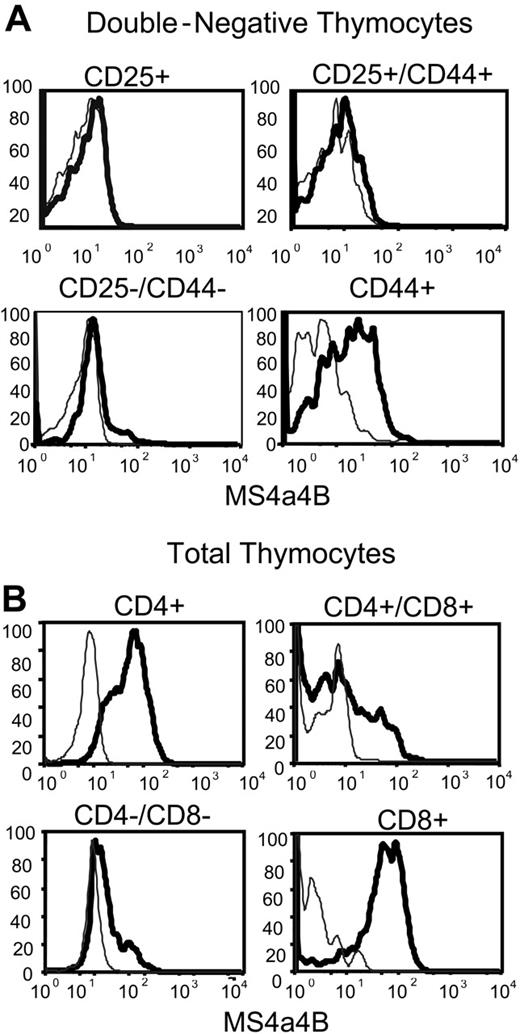
Analysis of MS4a4B expression in permeabilized DN thymocytes using flow cytometry supported the data from the cloning, in that the majority of CD44+/CD25- cells, but not CD44-/CD25+ cells, expressed MS4a4B protein (Figure 3A). The data suggest that MS4a4B is expressed in the uncommitted thymocyte progenitors and other resident cells of the hematopoietic lineage. However, once CD25 expression occurs coincident with commitment to the T lineage and the commencement of TCR gene rearrangement, expression of MS4a4B is down-regulated. The expression of MS4a4B protein resumed at a high level on all CD8+ and CD4+ single-positive (SP) thymocytes but was expressed in only a subset of CD4+CD8+ double-positive (DP) and double-negative (DN) thymocytes (Figure 3B). The results show that the expression of MS4a4B is highly regulated during T-cell development in thymus and are consistent with the original differential expression screen and the mRNA expression data.
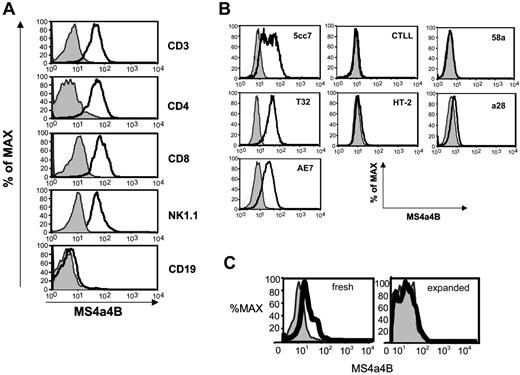
In peripheral hematopoietic cells, MS4a4B was highly expressed on T cells, including all primary mature CD4+ and CD8+ T cells and NK cells, but was not detected in B cells (Figure 4A). Consistent with previous results,31 MS4a4B protein expression was down-regulated in spleen T cells from a DO11.10 TCR transgenic mouse differentiated under Th2 conditions. MS4a4B expression was high prior to Th2 differentiation culture and absent after differentiation in Th2 conditions (data not shown).
MS4a4B protein was also detected in IL-2-dependent, clonal T-cell lines (5CC7, T32, and AE7) which remain responsive to TCR stimulation (Figure 4B). In IL-2-dependent T-cell lines that are no longer TCR responsive (CTLL and HT-2 cells) or in the T hybridoma cells, 58α and α28, MS4a4B expression was not observed (Figure 4B). Thus, MS4a4B was more likely to be expressed on IL-2-dependent, TCR-responsive cells than on cells which have lost their dependence on IL-2 (hybridomas) or TCR stimulation (CTLL and HT-2).
Low levels of MS4a4B protein were also detected in Mac1+ myeloid cells, including monocytes and macrophages, freshly isolated from bone marrow or spleen. Interestingly, when the macrophage cells were expanded in culture in the presence of MCSF (which also induces the maturation of immature myeloid cells), MS4a4B protein becomes undetectable (Figure 4C).
MS4a4B is present on the cell surface and is associated with membrane lipid rafts
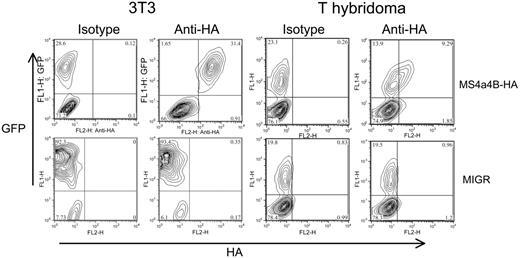
To confirm that MS4a4B was expressed on the cell surface, a cDNA chimera was engineered with the HA epitope tag in the region between the third and fourth transmembrane domains, predicted to be extracellular. The chimeric molecule was expressed by retroviral vector in 3T3 fibroblasts and a T-cell hybridoma. The cells were analyzed by flow cytometry using an anti-HA antibody, without fixation or permeabilization. The transduced cells, but not the vector controls, showed expression of the HA epitope at the cell surface (Figure 5). This finding is consistent with an intracellular orientation of MS4a4B's N- and C-termini.
Anti-MS4a4B antibody recognizes both native and retrovirus-expressed MS4a4B. (A) Western blot of MS4a4B in tissues. Tissue samples from mouse liver, spleen, and thymus were lysed, blotted, and probed with rabbit anti-MS4a4B antibody. Blots were stripped and reprobed with anti-β-actin as a loading control. The position of MS4a4B is shown with an apparent MW of 24 kDa. (B) Lysates from thymus were immunoprecipitated by anti-MS4a4B antibody (or rabbit Ig control)-coated Protein A-Sepharose 4B. The bound proteins were eluted and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and blotted by anti-MS4a4B antibody as described under “Materials and methods.” Expected positions of MS4a4B protein and Ig light chain are shown. The lanes are as follows: (1) total spleen protein (no immunoprecipitation), (2) IP with MS4a4B, (3) IP with control rabbit IgG, (4) MS4a4B IP blocked with MS4a4B immunizing peptide, (5) MS4a4B IP blocked with control MCC peptide. (C) NIH 3T3 cells were infected by MS4a4B retroviral vector or MIGR (empty vector) control. Staining and flow cytometry were performed as described in “Materials and methods.” The results are shown as fluorescence intensity of MS4a4B (solid heavy line) or rabbit IgG control (light line, shaded) in gated GFP+ populations. Results shown are representative of 2 experiments.
Anti-MS4a4B antibody recognizes both native and retrovirus-expressed MS4a4B. (A) Western blot of MS4a4B in tissues. Tissue samples from mouse liver, spleen, and thymus were lysed, blotted, and probed with rabbit anti-MS4a4B antibody. Blots were stripped and reprobed with anti-β-actin as a loading control. The position of MS4a4B is shown with an apparent MW of 24 kDa. (B) Lysates from thymus were immunoprecipitated by anti-MS4a4B antibody (or rabbit Ig control)-coated Protein A-Sepharose 4B. The bound proteins were eluted and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and blotted by anti-MS4a4B antibody as described under “Materials and methods.” Expected positions of MS4a4B protein and Ig light chain are shown. The lanes are as follows: (1) total spleen protein (no immunoprecipitation), (2) IP with MS4a4B, (3) IP with control rabbit IgG, (4) MS4a4B IP blocked with MS4a4B immunizing peptide, (5) MS4a4B IP blocked with control MCC peptide. (C) NIH 3T3 cells were infected by MS4a4B retroviral vector or MIGR (empty vector) control. Staining and flow cytometry were performed as described in “Materials and methods.” The results are shown as fluorescence intensity of MS4a4B (solid heavy line) or rabbit IgG control (light line, shaded) in gated GFP+ populations. Results shown are representative of 2 experiments.
Because MS4a4B is expressed on the cell surface and shows sequence similarity with CD20, which has been shown to localize to lipid rafts, we tested whether MS4a4B was localized to lipid rafts using a flow cytometric assay.37 T-hybridoma cells expressing HA-tagged MS4a4B were stained on ice with anti-HA, or anti-Thy-1 (a GPI-linked protein known to be raft associated), or the TCR-β chain (not associated with rafts unless cells are stimulated41-44 ). The relative resistance of staining to subsequent treatment with 1% Triton X-100 (TX) has been shown to be associated with raft localization.45 MS4a4B protein demonstrated resistance to TX solubilization, suggesting that MS4a4B is likely to be raft localized (Figure 6A). In contrast, TX treatment completely released the mostly nonraft-associated TCR from the cell surface (Figure 6B). The levels of Thy-1 protein, expected to be constitutively localized to lipid rafts, were almost the same after TX treatment as on untreated cells (Figure 6C). The TX resistance of both Thy-1 and MS4a4B was reduced by pretreatment of the cells with methyl-β-cyclodextrin (MbCD), a membrane cholesterol-extracting agent that disrupts lipid rafts (Figure 6D-E), and MbCD's effects were reversed in the presence of excess cholesterol (data not shown). In addition, treatment of cells with sphingomyelinase also reduced the detergent resistance of both MS4a4B (Figure 6F) and Thy-1 (not shown).
To extend these data to nontransfected primary T cells, the raft localization of MS4a4B was examined by sucrose density gradient ultracentrifugation. Mouse spleen cells were stimulated with ConA in culture, cell lysates were separated on a discontinuous sucrose density gradient, and the presence of MS4a4B protein in the fractions was detected by Western blot. Unlike in T-hybridoma cells, MS4a4B was found mostly in nonraft fractions in unstimulated spleen T cells (Figure 6G-H). However, a marked amount of MS4a4B translocated to the raft fractions after 24 to 48 hours of ConA stimulation (Figure 6G-H). This translocation was relatively slow, occurring between 12 and 48 hours after stimulation. Earlier time points (as little as 1 hour after stimulation) showed no accumulation of MS4a4B protein in the raft fractions (data not shown). Densitometric scanning indicated that in unstimulated cells, 20% of MS4a4B protein was found in raft fractions 4 to 6, whereas nearly 60% was in nonraft fractions 10 to 12. Conversely, after 48 hours of ConA stimulation, nearly 50% of MS4a4B was found in raft fractions, and 20% was in nonraft fractions.
MS4a4B overexpression enhances IL-2 and IFN-γ production by T cells
Because MS4a4B was highly and selectively expressed in primary T cells, but lost in T hybridomas, we tested the potential function of MS4a4B by expressing it in the α3A3 T hybridoma by retroviral transduction. Infected T hybridoma populations were FACS sorted based on the coexpressed GFP marker. MS4a4B expression in transduced cells was lower than that observed in mouse T cells or T-cell lines. Anti-MS4a4B staining showed only a 1.7-fold increase in mean fluorescence intensity (MFI) over isotype control. CD3+ splenic T cells showed a 2.5-fold increase in MS4a4B staining over isotype control, whereas for a T-cell line (T32) the ratio was 10.
MS4a4B protein is expressed in thymocytes. (A) Double-negative thymocytes were analyzed by flow cytometry using 4-color staining. The samples were gated first on the CD4/CD8 double-negative population and subsequently on the 4 subsets of CD44 and CD25 expression, and then analyzed for MS4a4B as shown. Fluorescence intensity of MS4a4B (solid heavy line) or rabbit IgG control (light line) in each selected subset. (B) Total thymocytes were analyzed by flow cytometry as described in “Materials and methods” using 3-color staining. CD4 and CD8 were used for gating thymocyte subpopulations, and anti-MS4a4B (bold line) or Ig as control (light line) was analyzed subsequently as shown. Results shown are representative of 2 separate experiments.
MS4a4B protein is expressed in thymocytes. (A) Double-negative thymocytes were analyzed by flow cytometry using 4-color staining. The samples were gated first on the CD4/CD8 double-negative population and subsequently on the 4 subsets of CD44 and CD25 expression, and then analyzed for MS4a4B as shown. Fluorescence intensity of MS4a4B (solid heavy line) or rabbit IgG control (light line) in each selected subset. (B) Total thymocytes were analyzed by flow cytometry as described in “Materials and methods” using 3-color staining. CD4 and CD8 were used for gating thymocyte subpopulations, and anti-MS4a4B (bold line) or Ig as control (light line) was analyzed subsequently as shown. Results shown are representative of 2 separate experiments.
After sorting, cells were stimulated using titrations of stimulating peptide (moth cytochrome-C peptide/I-Ek specificity) and irradiated spleen antigen-presenting cells. In some cases, cells were stimulated with plate-bound anti-TCR antibody, or PMA/ionomycin as controls. After 24 hours, supernatants were harvested, and IL-2 production was measured using CTLL bioassays or ELISA. In the α3A3 cells, MS4a4B expression induced a 3- to 4-fold increase (P < .05) in IL-2 production versus controls (Figure 7A). In other hybridoma lines, we found that MS4a4B expression had variable effects, including both decreases in antigen-induced IL-2 production, as well as no change (data not shown). Plate-bound TCR antibody-mediated stimulation mimicked the results of antigen stimulation, whereas treatment with PMA and ionomycin resulted in IL-2 production that was similar in all groups (not shown). These results suggested that MS4a4B could be playing a regulatory role for IL-2 production, but the biologic variability of T-hybridoma cell lines and the lower levels of MS4a4B expression indicated that experiments were needed in a more physiologic context such as primary T cells.
MS4a4B protein is detected by FACS analysis. Cells were isolated, stained for surface markers where indicated, fixed and permeabilized, and stained with anti-MS4a4B (solid line) or rabbit Ig (light line with shading) as control. (A) Spleen cells stained for the indicated lineage markers. (B) Staining results for T-cell clones (5cc7, T32, AE7, CTLL-2, HT-2) and hybridomas (58α and α28) are shown. (C) Staining results for Mac1+ cells, freshly isolated from spleen, analyzed immediately (fresh), or expanded in vitro with MCSF (expanded) for 7 days before analysis. Results shown are representative of 3 separate experiments.
MS4a4B protein is detected by FACS analysis. Cells were isolated, stained for surface markers where indicated, fixed and permeabilized, and stained with anti-MS4a4B (solid line) or rabbit Ig (light line with shading) as control. (A) Spleen cells stained for the indicated lineage markers. (B) Staining results for T-cell clones (5cc7, T32, AE7, CTLL-2, HT-2) and hybridomas (58α and α28) are shown. (C) Staining results for Mac1+ cells, freshly isolated from spleen, analyzed immediately (fresh), or expanded in vitro with MCSF (expanded) for 7 days before analysis. Results shown are representative of 3 separate experiments.
To test the effects of enhanced MS4a4B expression in a more physiologic context, CD4+ mouse T-cell blasts were infected with MS4a4B-expressing or control (MIGR) retroviral vector. The infected cell populations were restimulated with PMA and ionomycin in the presence of monensin, and cytokine production in GFP-positive cells was analyzed by intracellular staining and flow cytometry. Overexpression of MS4a4B significantly increased the percentage of cells producing IL-2 and IFN-γ (P < .05). There was no significant difference between MS4a4B and MIGR control vector-infected cells in the production of other intracellular cytokines (IL-4, IL-10, TNFα) (Figure 7B).
MS4a4B is expressed on the cell surface. NIH 3T3 cells or 58α T-hybridoma cells were infected with retrovirus encoding MS4a4B with an extracellular HA epitope tag or with an MIGR vector control. Viral transduction was detected by expression of green fluorescent protein (GFP). Cells were stained with anti-HA mAb and analyzed by flow cytometry. The inset numbers represent the percent of viable (propidium iodide-negative) cells in each quadrant. Results shown are representative of at least 5 separate experiments.
MS4a4B is expressed on the cell surface. NIH 3T3 cells or 58α T-hybridoma cells were infected with retrovirus encoding MS4a4B with an extracellular HA epitope tag or with an MIGR vector control. Viral transduction was detected by expression of green fluorescent protein (GFP). Cells were stained with anti-HA mAb and analyzed by flow cytometry. The inset numbers represent the percent of viable (propidium iodide-negative) cells in each quadrant. Results shown are representative of at least 5 separate experiments.
Discussion
An increasing body of evidence has demonstrated a role for the MS4A gene family, particularly CD20 and FcϵRIβ, in immune system function. Herein, we show that the family member Ms4a4b/Chandra is differentially expressed during thymic development. MS4a4B protein is not present during the DP stage at which thymic selection occurs, but it is reexpressed in mature thymocytes and in peripheral naive T cells. MS4a4B is expressed on NK cells but is not found in B cells. Our results confirm those of others that show that Ms4a4b mRNA is up-regulated in Th1-differentiated cells and down-regulated in Th2 cells. Our data further suggest that the overexpression of MS4a4B modifies cytokine production during T-cell activation and may enhance differentiation of naive T cells to the Th1 lineage.
MS4a4B is located in lipid rafts. (A-F) T-hybridoma cells expressing the MS4a4B-HA tag were stained with antibodies before FACS analysis as indicated. (A) HA staining of cells untreated (bold line) or treated with Triton X-100 (TX; light line) compared with control vector-infected cells (dashed line). (B) TCR staining of cells untreated (bold line) or TX treated (light line). (C) Thy-1-stained cells untreated (bold line) or TX treated (light line). (D) HA staining for cells untreated (dash line), treated with TX (light line), treated with 10 mM MbCD (bold line), treated with 10 mM MbCD and subsequently TX (shaded line). (E) Thy-1 staining of cells treated as in panel G (MbCD alone not shown). (F) HA staining of cells untreated (dashed line) or treated with sphingomyelinase, 5 U/mL (light line), 10 U/mL (shaded line). (G-H) MS4a4B translocates to lipid rafts in activated spleen cells. Spleen cells were cultured for the indicated times in the presence or absence of concanavalin A (ConA). Cells lysates were separated on discontinuous sucrose gradients, fractionated and selected raft (R) and non-raft (N) fractions were pooled (G), or all fractions were analyzed separately (H). A corresponding amount of the total lysate was reserved before fractionation for comparative purposes (T). Fractions were separated by PAGE and immunoblotted with antibodies to MS4a4B, LAT, or transferrin receptor (CD71) or blotted with labeled cholera toxin B (to detect raft glycoprotein GM1). Results shown are representative of 3 separate experiments.
MS4a4B is located in lipid rafts. (A-F) T-hybridoma cells expressing the MS4a4B-HA tag were stained with antibodies before FACS analysis as indicated. (A) HA staining of cells untreated (bold line) or treated with Triton X-100 (TX; light line) compared with control vector-infected cells (dashed line). (B) TCR staining of cells untreated (bold line) or TX treated (light line). (C) Thy-1-stained cells untreated (bold line) or TX treated (light line). (D) HA staining for cells untreated (dash line), treated with TX (light line), treated with 10 mM MbCD (bold line), treated with 10 mM MbCD and subsequently TX (shaded line). (E) Thy-1 staining of cells treated as in panel G (MbCD alone not shown). (F) HA staining of cells untreated (dashed line) or treated with sphingomyelinase, 5 U/mL (light line), 10 U/mL (shaded line). (G-H) MS4a4B translocates to lipid rafts in activated spleen cells. Spleen cells were cultured for the indicated times in the presence or absence of concanavalin A (ConA). Cells lysates were separated on discontinuous sucrose gradients, fractionated and selected raft (R) and non-raft (N) fractions were pooled (G), or all fractions were analyzed separately (H). A corresponding amount of the total lysate was reserved before fractionation for comparative purposes (T). Fractions were separated by PAGE and immunoblotted with antibodies to MS4a4B, LAT, or transferrin receptor (CD71) or blotted with labeled cholera toxin B (to detect raft glycoprotein GM1). Results shown are representative of 3 separate experiments.
MS4a4B was isolated based on its regulated expression during thymocyte development. MS4a4B expression in CD44+CD25- CD4-CD8- thymocytes may be due to NK cells and precursors in this population, or MS4a4B may be expressed on true uncommitted T-cell progenitors. MS4a4B was not highly expressed in CD4+CD8+ thymocytes undergoing selection but was induced to its highest levels when thymocytes become mature single-positive. A small subpopulation of DP thymocytes did express MS4a4B, but it is unclear whether these cells are postselection intermediates “on their way” to SP development.
Ms4a4b mRNA was previously detected in hematopoietic tissues2 ; however, we found MS4a4B expression was relatively restricted to T and NK lineages but not in B cells. A low level of MA4a4B protein was found on freshly isolated spleen myeloid cells but disappeared on brief in vitro culture. Because mRNA expression in myeloid cells is barely detectable, myeloid cells may pick up MS4a4B protein from the cellular environment. Other MS4A proteins, such as CD20, have been previously shown to be concentrated in exosomes, small membrane bodies containing lipid raft-associated proteins that are extruded from cells and may fuse to neighboring cells.46-48
The presence of MS4a4B on the cell surface was shown by staining of cells expressing the HA-tagged protein without permeabilization. On the basis of the membrane-spanning orientation of CD20, the HA tag should be located in a short extracellular loop between the third and fourth transmembrane domains. Detection of MS4a4B with an antibody raised to a peptide corresponding to the C-terminus of the protein, however, did require prior permeabilization. Previous data suggested that the C-terminus of MS4a4B was extracellular,31 but the current results indicate that both endogenous and ectopically expressed MS4a4B spans the membrane with its N- and C-termini within the cell. This topology is consistent with that of the other MS4A family members.49,50
Because MS4a4B does not have ITAM domains like FcϵRIβ, it may function more similarly to CD20. CD20 is known to associate with other proteins in rafts, such as kinases51,52 and Cbp/PAG,53 and may contribute to CD20's potential signaling function. However, more recent studies, using milder detergents such as Brij58, show that CD20 is constitutively associated with lipid rafts.29 The current results suggest MS4a4B also localizes in detergent-resistant lipid microdomains.
When expressed in a T-cell hybridoma, MS4a4B is constitutively associated with detergent-resistant membranes, similar to the GPI-linked Thy-1 protein. Our experiments do not rule out the possibility that antibody binding induces or enhances raft association as was shown for CD20.30 Interestingly, little MS4a4B was raft associated in unstimulated primary spleen T cells. However, within 24 to 48 hours after activation, a high percentage of MS4a4B protein was localized to lipid rafts. MS4a4B was not increased in raft fractions in early time points, which is inconsistent with TCR localization to lipid rafts that occurs within minutes of receptor stimulation. We speculate that the raft localization of MS4a4B may be related to the proliferative state of the cell. Activated cells strongly up-regulate MS4a4B protein expression, and this may also promote its association with lipid microdomains. Taken together, these results show that MS4a4B can be raft localized and that this localization is subject to regulation.
MS4a4B augments IL-2 production from stimulated T cells. (A) T-cell hybridoma cells (α3A3) were infected with retrovirus encoding MS4a4B or with an MIGR vector control. GFP+ infected cells were FACS sorted and stimulated with graded amounts of antigen (moth cytochrome C [MCC] peptide) presented by antigen-presenting cells (APCs). Supernatants were harvested after 24 hours, and IL-2 production was measured by CTLL-2 bioassay. The data are representative of 4 independent experiments, and points are averages and SDs of quadruplicate wells. (B) Primary CD4+ mouse T-cell blasts were infected with retrovirus-encoding MS4a4B or with a MIGR vector control. Cells were restimulated with PMA and ionomycin in the presence of monensin for 4 hours, and cytokine production was determined by intracellular cytokine staining in GFP+ cells using flow cytometry. The data represent the percentage of GFP+ cells that were cytokine positive. The results shown are the averages plus SE of 4 separate experiments. *Significantly different from vector control (P < .05).
MS4a4B augments IL-2 production from stimulated T cells. (A) T-cell hybridoma cells (α3A3) were infected with retrovirus encoding MS4a4B or with an MIGR vector control. GFP+ infected cells were FACS sorted and stimulated with graded amounts of antigen (moth cytochrome C [MCC] peptide) presented by antigen-presenting cells (APCs). Supernatants were harvested after 24 hours, and IL-2 production was measured by CTLL-2 bioassay. The data are representative of 4 independent experiments, and points are averages and SDs of quadruplicate wells. (B) Primary CD4+ mouse T-cell blasts were infected with retrovirus-encoding MS4a4B or with a MIGR vector control. Cells were restimulated with PMA and ionomycin in the presence of monensin for 4 hours, and cytokine production was determined by intracellular cytokine staining in GFP+ cells using flow cytometry. The data represent the percentage of GFP+ cells that were cytokine positive. The results shown are the averages plus SE of 4 separate experiments. *Significantly different from vector control (P < .05).
Expression of MS4a4B was absent on T-cell hybridomas and is apparently not required for T-hybridoma growth or signaling. However, other Ms4a4 genes (most likely the coexpressed Ms4a4c) may compensate for the loss of Ms4a4b in hybridomas. It is also possible that Ms4a4b expression is turned off in hybridomas because its expression is incompatible with continuous transformed growth. Although we did not detect reductions in cell growth of Ms4a4b-expressing hybridomas relative to controls, the amount of MS4a4B protein in transfected T hybridomas was low relative to that in spleen T cells or T-cell clones.
Nevertheless, expression of MS4a4B in the α3A3 T-cell hybridoma cell line enhanced stimulated IL-2 production. These data are consistent with selective expression of Ms4a4b in Th1 cells and were supported by results in primary CD4+ T-cell blasts. MS4a4B overexpression increased the frequency of cells that produce IL-2 and IFN-γ without changing the percentage of cells that produce Th2 cytokines (IL-4, IL-5). MS4a4B expression did not enhance Th1 cytokine production in cells that differentiated to the Th2 lineage, because IFN-γ+ populations did not coexpress IL-4 (not shown). In addition, MS4a4B expression did not interfere with Th2-type cytokine production, although MS4a4B is down-regulated in Th2 cells.31 Thus, enforced expression of MS4a4B may promote Th1 development. For example, MS4a4B protein expression may increase the survival of cells destined to become Th1 effectors, or it could enhance T-cell activation and cytokine production upstream of only IL-2 and IFN-γ expression.
In conclusion, our data show the highly regulated expression of a newer MS4 family member in the T-cell lineage during thymic development and later during effector cell differentiation. Our data further suggest that MS4a4B plays a functional role in regulating either TCR-stimulated T-lineage commitment or Th1-type cytokine production. MS4a4B shows features of a regulated lipid raft localization which underscores its potential functional similarity to CD20. Finally, as a functional protein in the MS4A family, showing regulated expression in immune cells, and whose gene is linked with susceptibility to allergy and atopic asthma, our data suggest that human homologs of Ms4a4b could be considered candidates for conferring disease susceptibility directly.
Prepublished online as Blood First Edition Paper, November 17, 2005; DOI 10.1182/blood-2005-08-3340.
Supported by grants from the American Cancer Society (RPG-00-094-01-LBC) (L.M.S.), the National Institutes of Health (grant AI49807) (L.M.S. and M.S.W.), and the Lymphoma Research Foundation (H.X.).
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
H.X. was a Lymphoma Research Foundation Fellow.


![Figure 7. MS4a4B augments IL-2 production from stimulated T cells. (A) T-cell hybridoma cells (α3A3) were infected with retrovirus encoding MS4a4B or with an MIGR vector control. GFP+ infected cells were FACS sorted and stimulated with graded amounts of antigen (moth cytochrome C [MCC] peptide) presented by antigen-presenting cells (APCs). Supernatants were harvested after 24 hours, and IL-2 production was measured by CTLL-2 bioassay. The data are representative of 4 independent experiments, and points are averages and SDs of quadruplicate wells. (B) Primary CD4+ mouse T-cell blasts were infected with retrovirus-encoding MS4a4B or with a MIGR vector control. Cells were restimulated with PMA and ionomycin in the presence of monensin for 4 hours, and cytokine production was determined by intracellular cytokine staining in GFP+ cells using flow cytometry. The data represent the percentage of GFP+ cells that were cytokine positive. The results shown are the averages plus SE of 4 separate experiments. *Significantly different from vector control (P < .05).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/107/6/10.1182_blood-2005-08-3340/4/m_zh80060692970007.jpeg?Expires=1769806041&Signature=Y22bMqYVZ-mnuv1PZM8MHMFZVs~PxM-4pp~xIPn3cwrWndsitSmcl-3YzVKgrrCcxRqcNajlCJAm6rYabs7V8M1t6rv4OFJbf~qxGFKbR2ByyUplJua2409WJvJFYy9qWUuieGAhEsPeoZkckFgIZ-np3URknOxtRbc-zyaUFMkBt8h1PWsIGDd5VUzK0m4SZsGXwn3FxT3G1RBDoEEFH~WqZ9-u7YeY1U7m8n0ytqCtlInKrTAe~Y45xakddpNvaN1SpLKzE-kRjhSH6ni8WZ9VS5yCJ6jlkfsoCCs4cDHl1NBiSkoWozL3u1f3t6az8Mk1ysBGWVKhCDcJuxocNQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal