Binding of the stage selector protein (SSP) to the stage selector element (SSE) in the human γ-globin promoter contributes to the preferential expression of the γ-gene in fetal erythroid cells. The SSP contains the transcription factor CP2 and an erythroid-specific partner, NF-E4. The NF-E4 gene encodes a 22-kDa polypeptide employing a non-AUG initiation codon. Antisera specific to NF-E4 detects this species and an additional 14 kDa protein, which initiates from an internal methionine. Enforced expression of p14 NF-E4 in the K562 fetal/erythroid cell line, and in primary erythroid cord blood progenitors, results in repression of γ-gene expression. Biochemical studies reveal that p14 NF-E4 interacts with CP2, resulting in diminished association of CP2 with the SSE in chromatin immunoprecipitation assays. p45 NF-E2 recruitment to the γ-promoter is also lost, resulting in a reduction in RNA polymerase II and TBP binding and a fall in promoter transcriptional activity. This effect is specific, as enforced expression of a mutant form of p14 NF-E4, which fails to interact with CP2, also fails to repress γ-gene expression in K562 cells. These findings provide one potential mechanism that could contribute to the autonomous silencing of the human γ-genes in adult erythroid cells.
Introduction
The human β-like globin genes (ϵ [HBE1], Gγ [HBG2], Aγ [HBG1], δ [HBD], β [HBB]) are expressed in a tissue-specific, developmentally regulated pattern.1-3 The high level of gene activation observed at the embryonic, fetal, and adult developmental stages is dependent on the presence of key regulatory sequences upstream of the ϵ-gene, the locus control region (LCR),4-8 characterized by the presence of multiple DNaseI hypersensitivity sites (HS).9-11 However, the LCR does not provide the temporal specificity of globin gene expression, because transgenic mice lacking this region still exhibit appropriate developmental switching.12 Similarly, mice lacking the endogenous LCR retain the normal developmental profile of globin gene expression, albeit at markedly reduced levels.13 From these studies it is evident that the sequences conferring stage-specific expression reside in the regions immediately flanking the globin genes.
Several diverse regulatory mechanisms govern the highly restricted pattern of globin gene expression. These include the gene order within the locus,14,15 competition between globin gene promoters for the enhancer sequences of the LCR,16-18 the transcription factor milieu at different developmental stages;19-24 tissue- and stage-specific chromatin remodeling,25-29 and nuclear compartmentalization of the active genes.30,31 Studies in transgenic mice have emphasized the importance of gene order and promoter competition. Transgenic lines carrying a β-gene in a more proximal position or lines in which an isolated β-gene is linked to the LCR show promiscuous adult globin expression in yolk sac and fetal liver.17,18,32 Correct temporal regulation of β-gene expression is restored in lines in which the ϵ- and/or γ-genes are linked more proximally in cis. In contrast, the ϵ- and γ-globin genes are silenced autonomously in transgenic mice, requiring no linked globin gene for correct stage-specific expression.32.33
Additional insight into γ-gene repression comes from the study of mutations that result in continued fetal hemoglobin production into adult life. This clinical syndrome, known as hereditary persistence of fetal hemoglobin (HPFH), is seen with specific deletions in the 3′ half of the locus (deletional form) and also with some point mutations in the γ-promoters (the nondeletional form).34 Studies of the molecular mechanisms of nondeletional forms of HPFH have largely focused on potential alterations in transcription factor binding sites with loss of binding of repressors or enhanced binding of activators being pursued. No specific factor with activator or repressor function has been conclusively linked to these mutations, and several studies suggest that silencing of the γ-promoter in this setting may be multifactorial, involving a combination of specific factors and promoter structure.35,36 Recently, a direct repeat element in the Aγ-globin proximal promoter that is altered in the -117 G→A HPFH mutation has been shown to mediate adult stage silencing of the γ-gene.37 This effect is presumably due to COUP-TFII,38 or DRED,39 the 2 protein complexes shown to bind to this element, although this is yet to be established.
Another factor complex that is a candidate for stage-specific silencing of the γ-genes is the stage selector protein (SSP).23,24,40 Although loss of SSP expression in adult erythroid cells would provide one potential mechanism for autonomous silencing of the γ-genes, this does not occur, because abundant levels of CP2 and p22 NF-E4 protein are detectable in bone marrow.24 An alternate hypothesis is that a component of the SSP is modified in the adult erythroid environment, allowing it to act as an inhibitor of the native complex. In our previous studies, we observed a short isoform of the NF-E4 protein (p14 NF-E4) present at higher levels in bone marrow than cord blood.24 We now report that this isoform acts in direct contrast to p22 NF-E4, suppressing γ-gene expression in fetal/erythroid cells.
Materials and methods
Extract preparation, Western analysis, immunoprecipitation, and N-terminal sequencing
Nuclear extracts were prepared by the method of Dignam as previously described.41 For Western analysis, CD34+ cells were isolated from fresh bone marrow or cord blood and cultured for 7 days in an erythroid burst-forming unit (BFU-E) mix. Cell surface marker analysis indicated that these cells were more than 90% erythroid lineage.28 Nuclear extracts were prepared from both samples and from K562 cells and resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) using a 12% gel. After transfer to PVDF, the samples were immunoblotted with specific antiserum and developed with ECL according to the manufacturer's instructions (Amersham Pharmacia Biotech, Piscataway, NJ).
For immunoprecipitation studies, nuclear extracts were initially precleared with normal rabbit serum (10 μg/mL) and then incubated with preimmune serum or antisera to CP2, NF-E4, or HA overnight at 4°C. A 50% slurry of protein G–Sepharose was added and incubated at 4°C for 1 hour. The mixture was then centrifuged at 3000g for 1 minute and the pellet washed in 50 mM Tris-HCl, pH 7.9, containing 150 mM NaCl prior to being resuspended in SDS loading buffer. Samples were subjected to SDS-PAGE, transferred to PVDF membranes, and blotted with the relevant antisera. Signal detection was achieved with the ECL system as per the manufacturer's instructions (Amersham Pharmacia Biotech).
For N-terminal sequencing, immunoprecipitated p14 NF-E4-FLAG was subjected to SDS-PAGE, transferred to PVDF membranes, and stained with 0.1% Coomassie brilliant blue. After destaining with 50% methanol, the band of interest was excised and subjected to automated Edman degradation and sequencing using an Applied Biosystems 494 Precise Protein Sequencing System through the Australian Proteome Analaysis Facility (Sydney, Australia).
5′ RACE and Northern analysis
A marathon 5′ rapid amplification of complementary DNA ends (RACE) cDNA library was constructed from mRNA from K562 cells according to the manufacturer's instructions (Clontech, Mountain View, CA). Nested PCR was performed using vector- and gene-specific primers. Primer sequences were as follows: gene-specific 1, 5′-CCCTTGGCTCAGATGAAGCGATGGTAGT-3′; gene-specific 2, 5′-GGTGTCTGTTCCAAGGCACTATCAGTC-3′; vector-specific 1, 5′-CCATCCTAATACGACTCACTATAGGGC-3′; and vector-specific 2, 5′-ACTCACTATAGGGCTCGAGCGGC-3′.
Polymerase chain reaction (PCR) conditions used were 95°C for 1 minute, 1 cycle; 94°C for 10 seconds and 68°C for 2 minutes, 30 cycles; and 68°C for 5 minutes, 1 cycle. PCR products were electrophoresed on 1% agarose, blotted onto nitrocellulose, and probed with internal genespecific oligonucleotides. Final PCR products were cloned into the TOPO 2.1 vector according to the manufacturer's instructions (Invitrogen, Carlsbad, CA) and sequenced.
Northern analysis of K562 pools was performed as described previously.42
Generation of MSCV-based amphotropic retroviral supernatant and transduction of mammalian cell lines
The p22 NF-E4 or p14 NF-E4 coding regions were cloned into the retroviral vector plasmid MSCV-HA at unique XhoI or EcoRI sites and amphotropic viral supernatant obtained as described previously.28 Filtered supernatant was added to K562 cells every 12 hours for 3 days. The cells were allowed to recover for 72 hours and then analyzed for GFP expression by flow cytometry. The highest-expressing 10% of cells were sterilely sorted, expanded, and resorted and subsequently expanded in oligoclonal pools. The p14 NF-E4–expressing retrovirus was also used to transduce CD34+ cells from human cord blood as detailed previously.24 Human cord blood was generously provided by the Bone Marrow Donor Institute Cord Blood Bank.
Chromatin immunoprecipitation (ChIP) assay
Chromatin immunoprecipitation (ChIP) assays were performed as described previously.43,44 Isolated DNA fragments were purified with a QIAquick spin kit (Qiagen, Valencia, CA), and 2 μL from a 40 μL DNA extraction was amplified quantitatively by real-time PCR with the γ-globin gene promoter–specific primers or MYOD primers as a negative control.45
Primers were as follows: γ-globin forward, 5′-AATTAAGCAGCAGTATCCTCTTGGG-3′; γ-globin reverse, 5′-GTTCCAGAAGCGAGTGTGTGG-3′; γ-globin probe, 5′-AGTCTTAGAGTATCCAGTGAGG-3′; MyoD forward, 5′-TGCAAGGCGTGCAAGCGCAAGAC-3′; MYOD reverse, 5′-CTCGATATAGCGGATGGCGTTG-3′; and MYOD probe, 5′-GGCCTTTGAGACACTCAAGC-3′.
Reverse 2-hybrid screen and site-directed mutagenesis
The reverse 2-hybrid screen was performed as described previously.46 The p22 NF-E4 cDNA in pACT2-NF-E4 was randomly mutated by PCR using the GeneMorph PCR mutagenesis system (Stratagene, La Jolla, CA).
Primers were as follows: Up1, 5′-AACGGTCCGAACCTCATAAC-3′; and Dn1, 5′-GTCAACAACGTATCTACCAAC-3′.
For in vivo homologous recombination, the purified PCR products were cotransformed with the pACT2 plasmid digested with EcoRI and XhoI into the yeast strain MAV103 carrying a second plasmid, pGBT9-TFCP2, which encodes a GAL4-DNA binding domain/CP2 fusion protein.28 Yeast transformants were selected on LT– plates containing 0.2% FOA (LT-+FOA). Colonies were then plucked from these plates and replica plated on LT- (for X-gal assays), LTH- containing 3-AT, and LT-+FOA plates. Mutants of p22 NF-E4 (p22 NF-E4m) that failed to interact with CP2 grew on LT-+FOA plates but not LTH- plates. The loss of interaction was confirmed by X-gal assays of the LT– plates in which colonies containing the noninteracting mutants did not turn blue. These colonies were expanded and analyzed by Western blot with p22 NF-E4 antisera to ensure that the mutants encode full-length proteins. Mutants encoding truncated p22 NF-E4 were discarded. Plasmid DNA was isolated from the remaining yeast and sequenced. The deduced amino acid sequence was compared with that of the wild-type protein to determine the position of the amino acid substitutions.
For site-directed mutagenesis, nucleotide changes were introduced into the MSCV-NF-E4-FLAG plasmid using Quik Change (Stratagene) according to the manufacturer's instructions.
Primers were as follows: NF-E4 forward, 5′-TCAAGTGCGGCATTGAAGGCCACTGGC-3′; and NF-E4 reverse, 5′-GCCAGTGGCCTTCAATGCCGCACTTGA-3′.
Results
Identification of 2 immunoreactive NF-E4 species of 22 and 14 kDa
The NF-E4 cDNA was originally isolated from a yeast 2-hybrid screen of a library derived from the fetal/erythroid cell line K562 using CP2 as the “bait.”24 The cDNA encodes a 179–amino acid protein initiated from a non-AUG codon with a predicted molecular weight of 22 kDa. Western analysis of nuclear extract from K562 cells and erythroid progenitors from cord blood and bone marrow using an antibody raised against peptides from the COOH-terminus of NF-E4 demonstrated 2 immunoreactive species, the predicted 22-kDa protein and a smaller form at 14 kDa.24 The ratio of p22 NF-E4 to p14 NF-E4 was approximately 2:1 in bone marrow and 4:1 in cord blood, and the 2 isoforms were expressed predominantly in the nucleus (Zhou et al24 and data not shown).
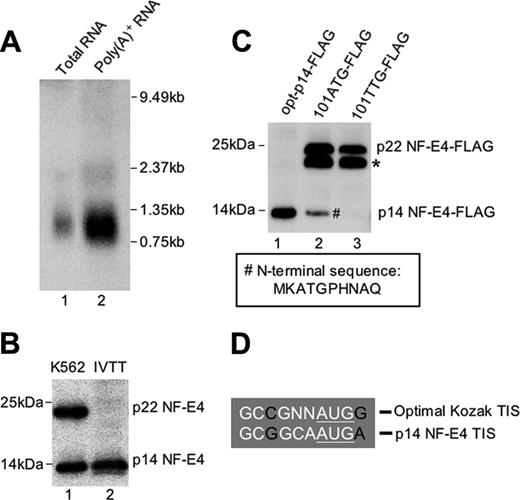
To determine whether the p14 NF-E4 isoform arose from a unique transcript, we performed Northern analysis on RNA from K562 cells. As we had reported previously,24 the NF-E4 transcript is rare, and hence we utilized polyA-positive RNA derived from 1.4 mg total RNA for the blot (Figure 1A). Despite a prolonged exposure time, only a single transcript corresponding to full-length NF-E4 was observed. To ensure that we had not missed a very low abundance alternate transcript, we performed 5′ RACE using different primers that annealed to the NF-E4 cDNA sequence at its 3′ end. Despite multiple attempts from cDNA derived from K562 cells, bone marrow, and cord blood, we only amplified a product that corresponded to our full-length sequence (data not shown). We therefore postulated that p14 NF-E4 could only arise from the full-length NF-E4 transcript via alternate translation initiation. Examination of the predicted amino acid sequence of p22 NF-E4 revealed the presence of a single methionine at codon 101. Translation initiation at this site would generate a protein of approximately 14 kDa. To confirm codon 101 as a potential initiation site of p14 NF-E4, we performed in vitro transcription/translation from the NF-E4 cDNA truncated at the N-terminal to codon 101. The product from this reaction was then electrophoresed and blotted with anti–NF-E4 antisera. As shown in Figure 1B, the protein species generated from the truncated template (lane 2) comigrated with the 14-kDa species observed in crude K562 cell nuclear extract (lane 1). To further confirm the methionine at codon 101 as the initiation site of p14 NF-E4, we generated mammalian expression vectors containing either the NF-E4 cDNA truncated to codon 101 and linked to an optimal Kozak sequence (opt-p14-FLAG), or the full-length NF-E4 cDNA with codon 101 unchanged (101ATG-FLAG), or the full-length NF-E4 cDNA with codon 101 altered to a TTG (101TTG-FLAG). All 3 cDNAs were tagged at the 3′ end with the FLAG epitope. The 293T cell line was transfected with these vectors, and Western analysis of extract from these cells was performed with anti-FLAG antisera (Figure 1C). The truncated cDNA generated a protein species of 14 kDa (lane 1). The native cDNA generated both p22 and p14 isoforms (lane 2) as well as an additional 20 kDa band (Figure 1C, *) that we observed with all tagged constructs and presumably reflects a degradation product or an alternatively modified NF-E4 species.43 The NF-E4 cDNA in which the internal ATG had been mutated to TTG failed to generate the p14 isoform of NF-E4 (lane 3), again consistent with the internal ATG being an alternative initiation codon for p14 NF-E4. To further establish this, we purified the FLAG-tagged p14 NF-E4 from the 101ATG-FLAG cells (Figure 1C, #) by immunoprecipitation with anti-FLAG antisera and sent the protein for N-terminal sequencing. The resulting sequence, MKATGPHNAQTQ, was identical to the predicted amino acid sequence of the p14 isoform initiated at codon 101 (Figure 1C, box). Further examination of the native DNA sequence immediately 5′ to codon 101 revealed the presence of a Kozak sequence that displayed only minor nucleotide changes from an optimal Kozak sequence (Figure 1D).47 Taken together, these findings indicate that the p14 isoform of NF-E4 arises from the full-length NF-E4 transcript as an alternately translated product from the internal AUG codon at position 101.
Characterization of the p14 isoform of NF-E4. (A) Northern analysis of NF-E4. The full-length NF-E4 cDNA was used to probe 20 μg total RNA (lane 1) and 2.6 μg polyA-positive RNA (lane 2) derived from K562 cells. The size standards are indicated. (B) In vitro transcription/translation of p14 NF-E4. The NF-E4 cDNA truncated to codon 101 was cloned into pSP72, and 35S methionine–labeled protein was produced. This protein (lane 2) and K562 cell nuclear extract (lane 1) were resolved by SDS-PAGE on a 12% gel, transferred to PVDF, and immunoblotted with anti–NF-E4 antiserum. The membrane was developed with ECL. (C) Mutation of codon 101 prevents formation of p14 NF-E4. pCAGGS vectors containing FLAG-tagged NF-E4 cDNA truncated to codon 101 and linked to an optimal Kozak sequence (opt-p14-FLAG) (lane 1), or the full-length NF-E4 cDNA with codon 101 unchanged (101ATG-FLAG) (lane 2), or the full-length NF-E4 cDNA with codon 101 mutated to a TTG (101TTG-FLAG) (lane 3) were transfected into 293T cells, and Western analysis of extract from these cells was performed with anti-FLAG antisera. The p22 NF-E4 and p14 NF-E4 isoforms are marked. The band marked with an asterisk was observed with all tagged constructs and presumably reflects a degradation product or an alternatively modified NF-E4 species. The p14 NF-E4-FLAG band (#) was used as the source of protein for sequence analysis. The boxed amino acid sequence was obtained from Edman sequencing of this band. The size markers are indicated. (D) Kozak sequence 5′ to codon 101 of NF-E4. The sequence 5′ to codon 101 of NF-E4 is compared with an optimal Kozak translation initiation site (TIS).53
Characterization of the p14 isoform of NF-E4. (A) Northern analysis of NF-E4. The full-length NF-E4 cDNA was used to probe 20 μg total RNA (lane 1) and 2.6 μg polyA-positive RNA (lane 2) derived from K562 cells. The size standards are indicated. (B) In vitro transcription/translation of p14 NF-E4. The NF-E4 cDNA truncated to codon 101 was cloned into pSP72, and 35S methionine–labeled protein was produced. This protein (lane 2) and K562 cell nuclear extract (lane 1) were resolved by SDS-PAGE on a 12% gel, transferred to PVDF, and immunoblotted with anti–NF-E4 antiserum. The membrane was developed with ECL. (C) Mutation of codon 101 prevents formation of p14 NF-E4. pCAGGS vectors containing FLAG-tagged NF-E4 cDNA truncated to codon 101 and linked to an optimal Kozak sequence (opt-p14-FLAG) (lane 1), or the full-length NF-E4 cDNA with codon 101 unchanged (101ATG-FLAG) (lane 2), or the full-length NF-E4 cDNA with codon 101 mutated to a TTG (101TTG-FLAG) (lane 3) were transfected into 293T cells, and Western analysis of extract from these cells was performed with anti-FLAG antisera. The p22 NF-E4 and p14 NF-E4 isoforms are marked. The band marked with an asterisk was observed with all tagged constructs and presumably reflects a degradation product or an alternatively modified NF-E4 species. The p14 NF-E4-FLAG band (#) was used as the source of protein for sequence analysis. The boxed amino acid sequence was obtained from Edman sequencing of this band. The size markers are indicated. (D) Kozak sequence 5′ to codon 101 of NF-E4. The sequence 5′ to codon 101 of NF-E4 is compared with an optimal Kozak translation initiation site (TIS).53
Enforced expression of the truncated form of NF-E4 represses γ- and ϵ-globin gene expression
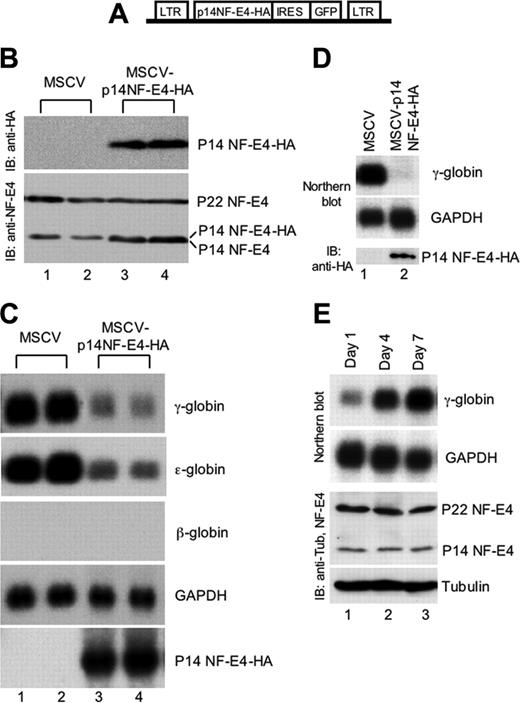
To determine the potential functional role of p14 NF-E4, we generated an MSCV-IRES-GFP–based retroviral construct containing the NF-E4 cDNA truncated to codon 101 tagged at the 3′ end with 3 hemagglutinin (HA) epitopes (MSCVp14 NF-E4-HA) (Figure 2A). The vector lacking the NF-E4 cDNA (MSCV) was used as a control. The K562 cell line was transfected with viral supernatant derived from both vectors and GFP-positive oligoclonal pools isolated by fluorescence-activated cell sorting (FACS). As shown in the Western analysis in Figure 2B, MSCV-transduced cell lysate contained no immunoreactive species when blotted with anti-HA antisera (upper panel, lanes 1-2). In contrast, the pools generated with the MSCVp14 NF-E4-HA virus expressed the truncated form of the NF-E4 protein (upper panel, lanes 3-4). Immunoblotting of the respective lysates with anti–NF-E4 antisera revealed that K562 cells transduced with the p14 NF-E4-HA virus now displayed total levels of p14 NF-E4 that were equivalent to or slightly higher than the levels of p22 NF-E4 (Figure 2A, bottom panel, lanes 3-4), whereas the MSCV-transduced cells retained the original p22 NF-E4/p14 NF-E4 ratio (bottom panel, lanes 1-2). RNA from these pools was then analyzed by Northern blot (Figure 2C). The pools containing the overexpressed truncated protein exhibited a dramatic reduction in both γ- and ϵ-globin gene expression when compared with the MSCV-transduced cells (compare lanes 3-4 with lanes 1-2). No reciprocal change in β-globin gene expression was observed, and the levels of expression of this gene were undetectable in both control and experimental cells. Expression of the housekeeping gene, GAPDH, and several other unrelated genes, including ding, LMO2, 28, and 18S rRNA, were also unaltered (data not shown). This finding was consistent with specific p14-dependent repression of γ- and ϵ-globin gene expression, in keeping with the SSP binding sites that have been demonstrated in the promoters of both these genes.48,49
Enforced expression of p14 NF-E4 represses γ- and ϵ-globin gene expression. (A) Diagrammatic representation of the MSCV-based p14 NF-E4 retroviral vector. (B) Western analysis of K562 cell pools transduced with MSCV vectors. K562 cells were transduced with MSCV p14 NF-E4-HA or MSCV alone and GFP-positive cells obtained by FACS. Cells were expanded, resorted, and cultured in 2 pools. Nuclear extract was prepared from MSCV (lanes 1-2) and MSCV p14 NF-E4 (lanes 3-4) and resolved by SDS-PAGE on a 12% gel. The proteins were then transferred to a PVDF membrane that was immunoblotted with anti-HA antisera (top panel) and anti–NF-E4 antisera (bottom panel) and developed by ECL. The p14 NF-E4-HA, p22 and p14 NF-E4 bands, and the molecular weight standards are indicated. (C) Northern analysis of K562 cell pools overexpressing p14 NF-E4. Total RNA was prepared from the pools derived in panel B. Ten micrograms of total RNA from the 2 MSCV pools (lanes 1-2) and 2 MSCV p14 NF-E4-HA pools (lanes 3-4) was analyzed by Northern blot with γ-, ϵ-, and β-globin and NF-E4 probes. GAPDH served as the control. (D) Northern and Western analysis of CD34+ cord blood progenitors transduced with either the MSCV or MSCV-p14 NF-E4-HA retrovirus and expanded in vitro in BFU-E mix for 7 days (see “Materials and methods”). For the Northern blot, 3 μg total RNA was used in each lane, and the probes used are shown on the right. For the Western blot, nuclear extracts were prepared from both samples and resolved by SDS-PAGE using a 12% gel. After transfer to PVDF, the samples were immunoblotted with anti-HA antiserum and developed with ECL. Positions of migration of the p14 NF-E4-HA isoform are indicated. (E) Northern and Western analysis of CD34+ cord blood progenitors expanded in vitro in BFU-E mix for 1, 3, and 7 days (see “Materials and methods”). For the Northern blot, 3 μg total RNA was used in each lane, and the probes used are shown on the right. For the Western blot, cell extracts were prepared from all samples and resolved by SDS-PAGE using a 12% gel. After transfer to PVDF, the samples were immunoblotted with anti–NF-E4 or antitubulin antiserum and developed with ECL. Positions of migration of the NF-E4 isoforms and tubulin are indicated.
Enforced expression of p14 NF-E4 represses γ- and ϵ-globin gene expression. (A) Diagrammatic representation of the MSCV-based p14 NF-E4 retroviral vector. (B) Western analysis of K562 cell pools transduced with MSCV vectors. K562 cells were transduced with MSCV p14 NF-E4-HA or MSCV alone and GFP-positive cells obtained by FACS. Cells were expanded, resorted, and cultured in 2 pools. Nuclear extract was prepared from MSCV (lanes 1-2) and MSCV p14 NF-E4 (lanes 3-4) and resolved by SDS-PAGE on a 12% gel. The proteins were then transferred to a PVDF membrane that was immunoblotted with anti-HA antisera (top panel) and anti–NF-E4 antisera (bottom panel) and developed by ECL. The p14 NF-E4-HA, p22 and p14 NF-E4 bands, and the molecular weight standards are indicated. (C) Northern analysis of K562 cell pools overexpressing p14 NF-E4. Total RNA was prepared from the pools derived in panel B. Ten micrograms of total RNA from the 2 MSCV pools (lanes 1-2) and 2 MSCV p14 NF-E4-HA pools (lanes 3-4) was analyzed by Northern blot with γ-, ϵ-, and β-globin and NF-E4 probes. GAPDH served as the control. (D) Northern and Western analysis of CD34+ cord blood progenitors transduced with either the MSCV or MSCV-p14 NF-E4-HA retrovirus and expanded in vitro in BFU-E mix for 7 days (see “Materials and methods”). For the Northern blot, 3 μg total RNA was used in each lane, and the probes used are shown on the right. For the Western blot, nuclear extracts were prepared from both samples and resolved by SDS-PAGE using a 12% gel. After transfer to PVDF, the samples were immunoblotted with anti-HA antiserum and developed with ECL. Positions of migration of the p14 NF-E4-HA isoform are indicated. (E) Northern and Western analysis of CD34+ cord blood progenitors expanded in vitro in BFU-E mix for 1, 3, and 7 days (see “Materials and methods”). For the Northern blot, 3 μg total RNA was used in each lane, and the probes used are shown on the right. For the Western blot, cell extracts were prepared from all samples and resolved by SDS-PAGE using a 12% gel. After transfer to PVDF, the samples were immunoblotted with anti–NF-E4 or antitubulin antiserum and developed with ECL. Positions of migration of the NF-E4 isoforms and tubulin are indicated.
To extend the functional studies of p14 NF-E4, we utilized the MSCV and MSCVp14 NF-E4-HA retroviruses to transduce CD34+ progenitors derived from human cord blood.24 The cells were then cultured in erythroid differentiation medium (IL-3, SCF, and erythropoietin) for 7 days and RNA prepared and analyzed by Northern blot (Figure 2D). Enforced expression of p14 NF-E4 (as demonstrated by the Western blot with anti-HA antisera in the lower panel) led to a marked reduction in γ-gene expression (upper panel, lane 2) compared with control cells (lane 1). No difference was observed in the expression of the GAPDH housekeeping gene (middle panel). To establish whether changes in the relative levels of p22 and p14 NF-E4 correlated with increased γ-gene expression during CD34+ cord blood progenitor erythroid differentiation, we cultured the cells for varying time intervals (1, 3, and 7 days) in IL-3, SCF, and erythropoietin. As shown in Figure 2E, the levels of γ-gene expression (assessed by Northern blot) increased over the period in culture (upper panel). No change was observed in the relative levels of p22 and p14 NF-E4 during this period or in the RNA (GAPDH) or protein (tubulin) controls, suggesting that NF-E4 plays a key role in the developmental regulation of γ-gene expression but is not essential during erythroid differentiation.
One explanation for the repression of γ- (and ϵ-globin) gene expression we observed with p14 NF-E4 was that the truncated protein participated in the SSP complex but lacked a key transactivation domain and thereby functioned as a dominant negative. We excluded this possibility using a GAL4-dependent/reporter gene transactivation assay (data not shown). We also demonstrated that enforced expression of p14 NF-E4 did not alter the chromatin structure of the γ-promoter in DNAse I hypersensitivity assays (data not shown). Taken together, these findings suggested that the effects of p14 NF-E4 may be on transcription and mediated through an interaction with a component of the SSP complex.
The truncated form of NF-E4 binds CP2 but not p22 NF-E4 in vivo
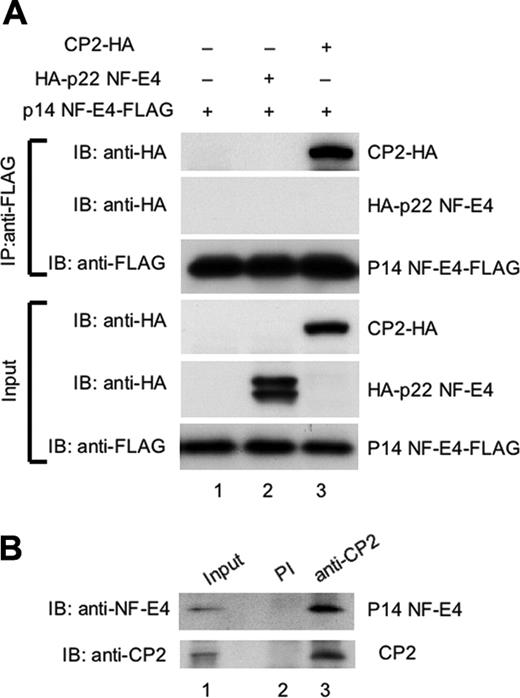
To examine whether p14 NF-E4 could bind CP2 or p22 NF-E4 in vivo, we performed coimmunoprecipitation experiments. Extract from 293T cells transduced with either an MSCV p14 NF-E4-FLAG retrovirus alone (p14 NF-E4-FLAG) or in combination with expression vectors containing the CP2 cDNA tagged at the 3′ end with an HA epitope (CP2-HA) or the p22 NF-E4 cDNAs tagged at the 5′ end with an HA epitope (HA-p22 NF-E4) was immunoprecipitated with antisera to the FLAG epitope. The precipitates were then electrophoresed and transferred to a membrane and blotted with either anti-FLAG antisera as a loading control or anti-HA to detect CP2 or p22 NF-E4. As shown in Figure 3, CP2 was detected in immunoprecipitates derived from the p14 NF-E4-FLAG/CP2-HA–transduced cells using anti-HA antisera (upper panels, lane 3). In contrast, p22 NF-E4 was not detected in immunoprecipitates derived from the p14 NF-E4-FLAG/p22 NF-E4-HA–transduced cells using anti-HA antisera (upper panels, lane 2). Neither protein was detected in immunoprecipitate-derived cells transduced with only the p14 NF-E4-FLAG vector (upper panel, lane 1). Abundant quantities of p14 NF-E4-FLAG, p22 NF-E4-HA, and CP2-HA were present in the input extracts (lanes 1-3, respectively). These results indicate that p14 NF-E4 forms a complex with CP2 but not p22 NF-E4 in vivo.
We had previously shown that endogenous CP2 and p22 NF-E4 formed a complex in a cellular milieu in K562 cells.24 However, the low levels of p14 NF-E4 in these cells precluded a clear visualization of this isoform in these experiments. We therefore repeated the coimmunoprecipitations using significantly greater quantities of starting extract and examined the precipitates for the presence of p14 NF-E4. As shown in Figure 3B, immunoprecipitation with anti-CP2 antisera and subsequent blotting with anti–NF-E4 antisera revealed the presence of endogenous p14 NF-E4 (lane 3). No p14 NF-E4 or CP2 was detected with immunoprecipitation with preimmune sera (lane 2).
p14 NF-E4 binds CP2 but not p22 NF-E4 in vivo. (A) Coimmunoprecipitation studies of CP2 and the NF-E4 isoforms. 293T cells were transduced with the MSCV p14 NF-E4-FLAG retrovirus alone (lane 1) or in combination with a pCAGGS vector containing the p22 NF-E4 cDNA tagged at the 5′ end with an HA epitope (HA-p22 NF-E4) (lane 2) or the CP2 cDNA tagged at the 3′ end with an HA epitope (CP2-HA) (lane 3). Cell extract from the 3 lines was immunoprecipitated with anti-FLAG antisera, and the precipitates were fractionated by SDS-PAGE and blotted with antisera to HA or FLAG as indicated (top panels). The input extracts for these experiments were also immunoblotted with antisera to HA and FLAG as loading controls (bottom panels). The migration of CP2-HA, HA-p22 NF-E4, and p14 NF-E4-FLAG are indicated. (B) Coimmunoprecipitation of endogenous CP2 and p14 NF-E4 from K562 cells. Nuclear extract from K562 cells was immunoprecipitated with polyclonal antiserum to CP2 (lane 3) or preimmune serum (lane 2) and the precipitates fractionated by SDS-PAGE prior to blotting with antiserum to NF-E4 (top panel) or CP2 (bottom panel). The first lane shows the levels of p14 and CP2 in the input extract.
p14 NF-E4 binds CP2 but not p22 NF-E4 in vivo. (A) Coimmunoprecipitation studies of CP2 and the NF-E4 isoforms. 293T cells were transduced with the MSCV p14 NF-E4-FLAG retrovirus alone (lane 1) or in combination with a pCAGGS vector containing the p22 NF-E4 cDNA tagged at the 5′ end with an HA epitope (HA-p22 NF-E4) (lane 2) or the CP2 cDNA tagged at the 3′ end with an HA epitope (CP2-HA) (lane 3). Cell extract from the 3 lines was immunoprecipitated with anti-FLAG antisera, and the precipitates were fractionated by SDS-PAGE and blotted with antisera to HA or FLAG as indicated (top panels). The input extracts for these experiments were also immunoblotted with antisera to HA and FLAG as loading controls (bottom panels). The migration of CP2-HA, HA-p22 NF-E4, and p14 NF-E4-FLAG are indicated. (B) Coimmunoprecipitation of endogenous CP2 and p14 NF-E4 from K562 cells. Nuclear extract from K562 cells was immunoprecipitated with polyclonal antiserum to CP2 (lane 3) or preimmune serum (lane 2) and the precipitates fractionated by SDS-PAGE prior to blotting with antiserum to NF-E4 (top panel) or CP2 (bottom panel). The first lane shows the levels of p14 and CP2 in the input extract.
CP2 fails to bind to the SSE in p14 NF-E4–overexpressing K562 cells
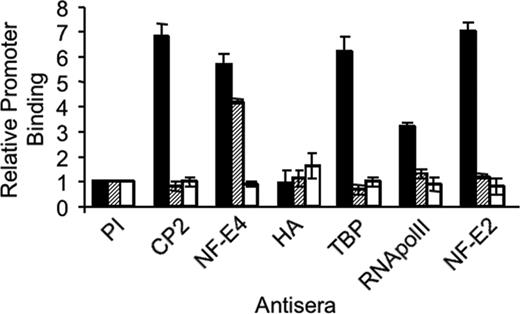
To examine the consequences of the p14 NF-E4/CP2 interaction on the formation of the SSP on the γ-promoter, we performed quantitative ChIP experiments using real-time PCR with K562 cells transduced with the MSCV (K562 [hatched bars]) or an MSCV p14 NF-E4-HA retrovirus (K562 p14-HA [striped bars]) (Figure 4). Utilizing anti–NF-E4 antisera we demonstrated similar binding of NF-E4 to the SSE in both cell lines (NF-E4 lane). This binding was enriched more than 6-fold compared with that observed with the negative control, the MYOD gene (open bars).45 This binding activity can be attributed to p22 NF-E4, because no significant binding of p14 NF-E4 was observed when anti-HA antiserum was employed (HA lane). CP2 binding to the SSE was demonstrable in the MSCV-transduced cells (more than 5-fold greater than MyoD [CP2 lane]) but was absent in the K562 line in which p14 NF-E4 was overexpressed with levels equivalent to the control MYOD gene. Because CP2 is known to interact with the TATA-binding protein (TBP) associated factor (TAFII130) subunit of TFIID,50 we examined whether the recruitment of the basal transcriptional machinery to the γ-promoter was altered in the p14 NF-E4–overexpressing cells. Binding of RNA polymerase II (RNA polII) was selectively reduced to almost background levels in the p14 NF-E4-HA–transduced K562 cells compared with those transduced with MSCV. Similarly, the binding of TBP to the γ-promoter was also lost in these cells compared with the MSCV-transduced cells. Previous studies have demonstrated the importance of p45 NF-E2 in the recruitment of RNA polII to the globin gene promoters.43-45,51 Consistent with this, we demonstrated abundant p45 NF-E2 at the γ-promoter in the MSCV-transduced K562 cells. In contrast, only background levels of this factor were observed at the promoter in p14 NF-E4–expressing cells. No difference in p45 NF-E2 binding to the LCR was observed in the MSCV- or p14 NF-E4–transduced cells (data not shown). No significant signal was detected from any line with preimmune sera (PI).
Repression of γ-gene expression by p14 NF-E4 is dependent on the interaction with CP2
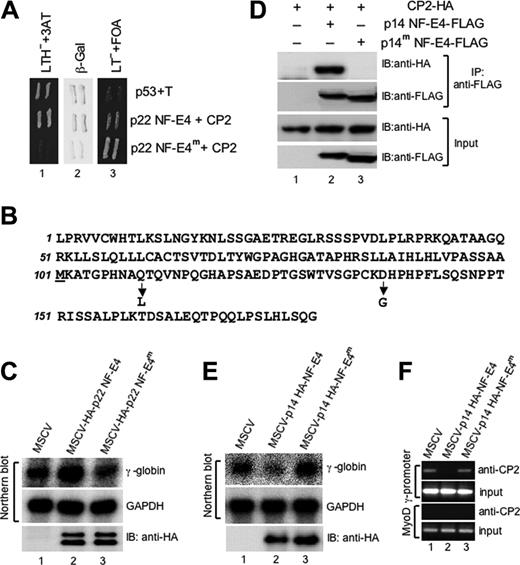
To confirm the specificity of the effects of p14 NF-E4 we initially generated a mutant form of the p22 NF-E4 protein that failed to interact with CP2 using a reverse 2-hybrid screen (see “Materials and methods” and Kozak47 ) (Figure 5A). The mutant we identified (p22 NF-E4m) contained 2 amino acid substitutions, a glutamine to leucine at codon 110 and an aspartic acid to glycine at codon 137, both of which are contained within p14 NF-E4 (Figure 5B). Coimmunopreciptation experiments confirmed that p22 NF-E4m was unable to interact with CP2 in a cellular milieu (data not shown). To examine whether this mutant form of p22 NF-E4 could induce γ-gene expression, as we had shown previously for wild-type p22 NF-E4,24 we transduced K562 cells with an MSCV retrovirus containing HA-tagged wild-type or mutant p22 NF-E4 cDNA. Cells transduced with MSCV alone served as the control. As shown in Figure 5C, wild-type p22 NF-E4 induced γ-gene expression compared with MSCV-transduced cells (compare lanes 1 and 2). Expression of the p22 NF-E4m failed to induce γ-gene expression (lane 3). Both wild-type and mutant forms of p22 NF-E4 were expressed as detected by immunoblot (lower panel). To confirm that a mutant form of p14 NF-E4 was also unable to bind CP2 in a cellular context, we performed coimmunoprecipitation experiments (Figure 5D). Extract from 293T cells transfected with an expression vector containing CP2 tagged at the 3′ end with an HA-epitope alone (lane 1) or in combination with either an MSCV p14 NF-E4-FLAG retrovirus (p14 NF-E4-FLAG) (lane 2) or an MSCV p14 NF-E4 mutant (p14 NF-E4m-FLAG) (lane 3) was immunoprecipitated with antisera to the FLAG epitope. The precipitates were then electrophoresed and transferred to a membrane and blotted with either anti-FLAG antisera to detect wild-type and mutant p14 NF-E4 or anti-HA to detect CP2. CP2 was detected in immunoprecipitates derived from the wild-type p14 NF-E4-FLAG/CP2-HA–transduced cells (upper panel, lane 2), as we had observed in Figure 3. In contrast, no CP2-HA was detected in immunoprecipitates from the p14 NF-E4m-FLAG/CP2-HA–transduced cells (upper panel lane 3) despite the presence of abundant CP2-HA in the input (lower panel, lane 3). As expected, no CP2-HA was detected in the immunoprecipitates from cells lacking either form of FLAG-tagged NF-E4 (upper panel, lane 1), despite the presence of abundant CP2-HA in the input extract (lower panel, lane 1). The levels of FLAG-tagged wild-type and mutant p14 NF-E4 were equivalent in the cellular extract inputs (lower panel, lanes 2-3). These results indicate that mutation of 2 amino acid residues in p14 NF-E4 can prevent binding of the protein to CP2 in vivo.
Reduced binding of CP2 and RNA polymerase II to the SSE in K562 cells overexpressing p14 NF-E4 demonstrated by ChIP. Chromatin from K562 cells transduced with either the MSCV (▪) or MSCV p14 NF-E4-HA ( , □) retrovirus was immunoprecipitated using antisera to CP2, NF-E4, HA, TBP, RNA polII, and p45 NF-E2 as indicated. Preimmune sera served as the control (PI). Quantitative PCR was performed with primer pairs to amplify the SSE in the γ-promoter SSE (▪,
, □) retrovirus was immunoprecipitated using antisera to CP2, NF-E4, HA, TBP, RNA polII, and p45 NF-E2 as indicated. Preimmune sera served as the control (PI). Quantitative PCR was performed with primer pairs to amplify the SSE in the γ-promoter SSE (▪,  ) or the MYOD gene (□).
) or the MYOD gene (□).
Reduced binding of CP2 and RNA polymerase II to the SSE in K562 cells overexpressing p14 NF-E4 demonstrated by ChIP. Chromatin from K562 cells transduced with either the MSCV (▪) or MSCV p14 NF-E4-HA ( , □) retrovirus was immunoprecipitated using antisera to CP2, NF-E4, HA, TBP, RNA polII, and p45 NF-E2 as indicated. Preimmune sera served as the control (PI). Quantitative PCR was performed with primer pairs to amplify the SSE in the γ-promoter SSE (▪,
, □) retrovirus was immunoprecipitated using antisera to CP2, NF-E4, HA, TBP, RNA polII, and p45 NF-E2 as indicated. Preimmune sera served as the control (PI). Quantitative PCR was performed with primer pairs to amplify the SSE in the γ-promoter SSE (▪,  ) or the MYOD gene (□).
) or the MYOD gene (□).
Repression of γ-gene expression by p14 NF-E4 is dependent on the interaction with CP2. (A) Reverse 2-hybrid assay of wild-type and mutant p22 NF-E4. A p22 NF-E4 mutant that fails to interact with CP2 was identified using the reverse 2-hybrid assay (see “Materials and methods”). A GAL4-AD–based yeast expression vector containing this mutant (p22 NF-E4m) or the pACT vector containing the wild-type p22 NF-E4 (p22 NF-E4) was cotransformed into the yeast strain MAV203 with the GAL4-DBD–based plasmid pGBT9-CP2. Cotransformation of pGBp53 and pACT SV40 large T antigen served as the positive control (P53+T). The resultant transformants were initially plated on selective media plates lacking leucine/tryptophan but containing FOA. Colonies were then replica plated on selective media plates lacking leucine/tryptophan/histidine but containing 3AT (LTH-+3AT) or plates lacking leucine/tryptophan but containing FOA (LT-+FOA) or plates lacking leucine/tryptophan for X-gal assays (β-Gal). (B) Amino acid substitutions in p22 NF-E4m The predicted amino acid sequence of p22 NF-E4 is shown with the position and identity of the 2 substitutions in p22 NF-E4m indicated by an arrow. The initiating methionine of p14 NF-E4 is underlined. (C) Enforced expression of p22 NF-E4m in K562 cells fails to induce γ-gene expression. Total RNA from K562 cells transduced with either the MSCV (lane 1), MSCV-HA-p22 NF-E4 (lane 2), or MSCV-HA-p22 NF-E4m (lane 3) was analyzed by Northern blot (top panels) using a γ-globin gene probe or a GAPDH control probe as indicated. The expression of the wild-type and mutant NF-E4 proteins in the respective cell line extracts is demonstrated by Western analysis using anti-HA antisera (bottom panel). (D) Coimmunoprecipitation of p14 NF-E4 and p14 NF-E4m with CP2. Cell extract from 293T cells transduced with an expression vector containing CP2 tagged at the 3′ end with an HA-epitope alone (lane 1) or in combination with either an MSCV p14 NF-E4-FLAG retrovirus (p14 NF-E4-FLAG) (lane 2) or an MSCV p14 NF-E4 mutant (p14 NF-E4m-FLAG) (lane 3) was immunoprecipitated with antisera to the FLAG epitope. The precipitates were then electrophoresed and transferred to a membrane and blotted with either anti-FLAG antisera to detect wild-type and mutant p14 NF-E4 or anti-HA to detect CP2 (top panels). The input extracts for these experiments were also immunoblotted with antisera to HA and FLAG as loading controls (bottom panels). (E) Enforced expression of p14 NF-E4m in K562 cells fails to repress γ-globin gene expression. Total RNA from K562 cells transduced with either the MSCV (lane 1), MSCV-HA-p14 NF-E4 (lane 2), or MSCV-HA-p14 NF-E4m (lane 3) was analyzed by Northern blot (top panels) using a γ-globin gene probe or a GAPDH control probe as indicated. The expression of the wild-type and mutant NF-E4 proteins in the respective cell line extracts is demonstrated by Western analysis using anti-HA antisera (bottom panel). (F) ChIP analysis of CP2 at the γ-promoter in K562 cell lines. Chromatin from K562 cells transduced with either the MSCV (lane 1), MSCV p14 HA-NF-E4 (lane 2), or MSCV p14 HA-NF-E4m retrovirus was immunoprecipitated using antisera to CP2. Quantitative PCR was performed with primer pairs to amplify the SSE in the γ-promoter (top panels) or the MYOD gene (bottom panels). The PCR of the input for each experiment is shown.
Repression of γ-gene expression by p14 NF-E4 is dependent on the interaction with CP2. (A) Reverse 2-hybrid assay of wild-type and mutant p22 NF-E4. A p22 NF-E4 mutant that fails to interact with CP2 was identified using the reverse 2-hybrid assay (see “Materials and methods”). A GAL4-AD–based yeast expression vector containing this mutant (p22 NF-E4m) or the pACT vector containing the wild-type p22 NF-E4 (p22 NF-E4) was cotransformed into the yeast strain MAV203 with the GAL4-DBD–based plasmid pGBT9-CP2. Cotransformation of pGBp53 and pACT SV40 large T antigen served as the positive control (P53+T). The resultant transformants were initially plated on selective media plates lacking leucine/tryptophan but containing FOA. Colonies were then replica plated on selective media plates lacking leucine/tryptophan/histidine but containing 3AT (LTH-+3AT) or plates lacking leucine/tryptophan but containing FOA (LT-+FOA) or plates lacking leucine/tryptophan for X-gal assays (β-Gal). (B) Amino acid substitutions in p22 NF-E4m The predicted amino acid sequence of p22 NF-E4 is shown with the position and identity of the 2 substitutions in p22 NF-E4m indicated by an arrow. The initiating methionine of p14 NF-E4 is underlined. (C) Enforced expression of p22 NF-E4m in K562 cells fails to induce γ-gene expression. Total RNA from K562 cells transduced with either the MSCV (lane 1), MSCV-HA-p22 NF-E4 (lane 2), or MSCV-HA-p22 NF-E4m (lane 3) was analyzed by Northern blot (top panels) using a γ-globin gene probe or a GAPDH control probe as indicated. The expression of the wild-type and mutant NF-E4 proteins in the respective cell line extracts is demonstrated by Western analysis using anti-HA antisera (bottom panel). (D) Coimmunoprecipitation of p14 NF-E4 and p14 NF-E4m with CP2. Cell extract from 293T cells transduced with an expression vector containing CP2 tagged at the 3′ end with an HA-epitope alone (lane 1) or in combination with either an MSCV p14 NF-E4-FLAG retrovirus (p14 NF-E4-FLAG) (lane 2) or an MSCV p14 NF-E4 mutant (p14 NF-E4m-FLAG) (lane 3) was immunoprecipitated with antisera to the FLAG epitope. The precipitates were then electrophoresed and transferred to a membrane and blotted with either anti-FLAG antisera to detect wild-type and mutant p14 NF-E4 or anti-HA to detect CP2 (top panels). The input extracts for these experiments were also immunoblotted with antisera to HA and FLAG as loading controls (bottom panels). (E) Enforced expression of p14 NF-E4m in K562 cells fails to repress γ-globin gene expression. Total RNA from K562 cells transduced with either the MSCV (lane 1), MSCV-HA-p14 NF-E4 (lane 2), or MSCV-HA-p14 NF-E4m (lane 3) was analyzed by Northern blot (top panels) using a γ-globin gene probe or a GAPDH control probe as indicated. The expression of the wild-type and mutant NF-E4 proteins in the respective cell line extracts is demonstrated by Western analysis using anti-HA antisera (bottom panel). (F) ChIP analysis of CP2 at the γ-promoter in K562 cell lines. Chromatin from K562 cells transduced with either the MSCV (lane 1), MSCV p14 HA-NF-E4 (lane 2), or MSCV p14 HA-NF-E4m retrovirus was immunoprecipitated using antisera to CP2. Quantitative PCR was performed with primer pairs to amplify the SSE in the γ-promoter (top panels) or the MYOD gene (bottom panels). The PCR of the input for each experiment is shown.
To determine the functional consequences of specifically abolishing the p14 NF-E4/CP2 interaction, we transfected K562 cells with MSCV wild-type p14 NF-E4 or MSCV p14 NF-E4m Cells transduced with MSCV alone served as the control. Cells sorted by FACS for GFP were shown by Western analysis with anti-HA antisera to have equivalent levels of expression of p14 NF-E4 and p14 NF-E4m (Figure 5E, lower panel). Despite this, the effects on γ-globin gene expression were markedly different between the 2 proteins. Cells transduced with wild-type NF-E4 showed the previously demonstrated (Figure 2C) reduction in γ-globin gene transcription compared with the vector alone control (compare lanes 1 and 2, top panel). In contrast, no effect was observed with p14 NF-E4m (lane 3, top panel), indicating that specific amino acid substitutions, which prevent the interaction between p14 NF-E4 and CP2, block the ability of p14 NF-E4 to repress γ-globin gene expression. No effect was observed on GAPDH expression (lanes 1-3, middle panel) or several other housekeeping genes (data not shown). To confirm that p14 NF-E4m failed to repress γ-gene expression because it no longer sequestered CP2 away from the γ-promoter, we performed ChIP analysis on K562 cells transduced with wild-type and mutant p14 NF-E4 (Figure 5F). As shown in Figure 4, no CP2 was detectable on the γ-promoter in the MSCV p14 NF-E4–transduced K562 cells (lane 2). In contrast, anti-CP2 antisera was able to ChIP down the γ-promoter in the cells expressing the p14 NF-E4m (lane 3). No CP2 was detected on the control MyoD promoter in either cell line.
Discussion
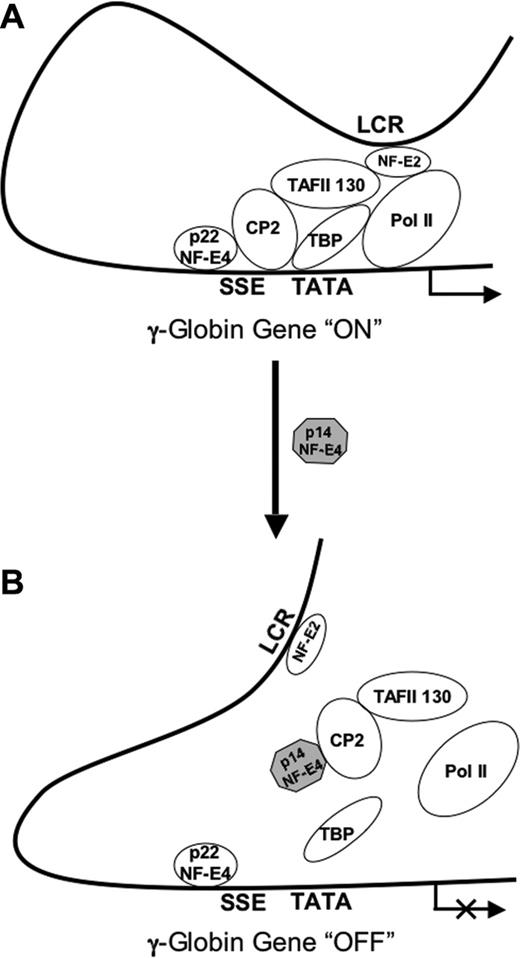
This report details the characterization of a short isoform of the fetal globin regulatory protein NF-E4. Unlike the full-length protein, p14 NF-E4 functions as repressor of ϵ- and γ-gene expression when transfected into a fetal/erythroid K562 cell line. The effects observed on both ϵ- and γ-genes are consistent with the presence of binding sites for the SSP in both promoters.48,49 The repressor effects of p14 NF-E4 on γ-gene expression are also observed in primary erythroid progenitors derived from cord blood. The dominant negative role of p14 NF-E4 is achieved through sequestration of CP2, with subsequent loss of that protein's ability to interact with the γ-promoter. Abrogation of p45 NF-E2 binding to the γ-promoter also occurs in the p14 NF-E4–expressing cells. p45 NF-E2 has previously been shown to play a key role in recruitment of the RNA polII complex to the globin promoters, even though the promoters lack binding sites for this factor.43-45,51 Consistent with this, we observed a marked reduction in RNA polII binding to the γ-promoter in p14 NF-E4–expressing cells. The mechanism by which p45 NF-E2 is recruited to the globin promoters has not been defined, but Sawado et al45 postulated this may occur through a protein-protein interaction between p45 NF-E2 and TAFII130, which has been shown to be critical for the activating function of NF-E2.52 Our results suggest that at the γ-promoter, loss of CP2 binding induced by p14 NF-E4 leads to disruption of the TFIID complex association with the promoter (as evidenced by loss of TBP binding), thereby preventing recruitment of p45 NF-E2 (and subsequently RNA polII) to the promoter (Figure 6B). The interaction between p14 NF-E4 and CP2 is essential for this effect, because transduction of K562 cells with a mutant form of p14 NF-E4 that fails to bind CP2 does not alter γ-gene expression. Consistent with this is our observation that the interaction between wild-type p22 NF-E4 and CP2 is essential for γ-globin gene activation. We propose that the previously reported interactions between CP2 and NF-E2 and TAFII130 are integral to the recruitment of p45 NF-E2 to the γ-promoter and the subsequent recruitment of RNA polII (Figure 6A). The location of the SSE immediately adjacent to the TATA box is consistent with this role for the SSP. Despite the sequestration of CP2 away from the SSPcomplex, p22 NF-E4 continues to bind to the SSE (as evidenced by the ChIP analysis with anti–NF-E4 antisera) and the promoter HS still forms. This suggests that additional steps, which may be p22 NF-E4 dependent, are required for the complete silencing, chromatin condensation, and methylation of the γ-promoter. The presence of 2 of the 3 CpG dinucleotides in the first 400 bp of the γ-promoter within the SSE, and the correlation between methylation of these cytosines and permanent silencing of γ-gene expression, suggests that after disruption of the activator complex by p14 NF-E4, a methyltransferase-containing repressor complex could assemble on the SSE.
Model of p14 NF-E4 repression of γ-globin gene expression. (A) At the actively transcribing γ-globin gene promoter p45 NF-E2 is recruited through its interaction with TAFII130. This interaction is stabilized by the assembly of the SSP complex on the SSE adjacent to the TATA box and, in particular, the CP2/TAFII130 interaction. Subsequent to this, RNA polII is recruited to the promoter to mediate transcription. (B) In the context of increased levels of p14 NF-E4, CP2 and TAFII130 are sequestered away from the SSE and the recruitment of p45 NF-E2 is lost. As a result, recruitment of RNA polII is also lost and γ-globin gene transcription is repressed.
Model of p14 NF-E4 repression of γ-globin gene expression. (A) At the actively transcribing γ-globin gene promoter p45 NF-E2 is recruited through its interaction with TAFII130. This interaction is stabilized by the assembly of the SSP complex on the SSE adjacent to the TATA box and, in particular, the CP2/TAFII130 interaction. Subsequent to this, RNA polII is recruited to the promoter to mediate transcription. (B) In the context of increased levels of p14 NF-E4, CP2 and TAFII130 are sequestered away from the SSE and the recruitment of p45 NF-E2 is lost. As a result, recruitment of RNA polII is also lost and γ-globin gene transcription is repressed.
Our previous studies had demonstrated an activator role for p22 NF-E4.24 Transcription factor isoforms playing contrasting roles is a common mechanism of developmental gene regulation. This may be achieved through changes in cellular localization, transcriptional activation potential, DNA binding properties, or protein dimerization.53 Our studies suggest that although p14 NF-E4 retains its nuclear localization (data not shown), it is unable to bind to the SSE when complexed with CP2. As a result, the p14NF-E4/CP2 complex remains untethered, and this impedes the recruitment of p45 NF-E2 and the basal transcriptional machinery to the promoter.
The p14 isoform of NF-E4 appeared to be generated by alternate translation initiation from the single internal AUG codon at position 101. This was evidenced by the comigration of a protein product from in vitro transcription/translation from a cDNA construct truncated to codon 101, loss of the p14 NF-E4 isoform in the setting of a mutation of the internal AUG codon, and N-terminal sequencing of a tagged p14 NF-E4 generated from a full-length NF-E4 cDNA. Although sequencing of the endogenous p14 NF-E4 would have provided further evidence of its origins, we were unable to achieve this due to low expression levels in K562 cells. Alternate translation initiation from internal AUG codons has been described as a mechanism for generating short isoforms with distinct functional properties for a number of proteins.54,55 This is particularly evident in factors in which translation initiation of the larger isoform is from a non-AUG codon.54,56 For example, the CUG- and AUG-initiated isoforms of the steroid receptor binding protein Bag-1, and the proto-oncogenes Int2 and Hck-1, differ in their subcellular localization.56-58 In addition, the CUG isoform of Bag-1 interacts with different protein partners and consequently has a unique functional role.59
Our enforced expression studies of p14 NF-E4 in K562 cells suggest that modest changes in the p22/p14 NF-E4 ratio are sufficient to perturb γ-gene expression. The concept of a dynamic balance of transcription factors regulating developmental expression of the globin genes is not novel. Studies of the γ- to β-globin switch in the fetal livers of mice transgenic for the β-globin locus YAC by primary transcript in situ hybridization reveal that the LCR flip-flops back and forward between the 2 genes as the transcription factor milieu changes to favor the adult gene.60 In the EKLF nullizygous mice carrying the β-YAC an increase is observed in the percentage of loci actively transcribing the γ-genes in day 13.5 fetal liver cells. However, most day 13.5 fetal liver cells in these mice still have only one active γ-gene, suggesting that the other locus in these cells has already been silenced.19 From these results it appears that the presence of a γ-gene repressor in the transcription factor environment coupled with promoter competition are the key mechanisms involved in γ-gene silencing. Our studies of enforced p14 NF-E4 expression in the β-YAC mice will further address this issue.
Prepublished online as Blood First Edition Paper, November 1, 2005; DOI 10.1182/blood-2005-06-2497.
Supported by the National Health and Medical Research Council of Australia, National Institutes of Health grants PO1 HL53749-03 and RO1 HL69232-01, Cancer Centre Support CORE grant P30 CA 21765, the American Lebanese Syrian Associated Charities (ALSAC), the Assisi Foundation of Memphis, and the National Natural Science Foundation of China (no. 30570392).
Q.Z., W.Z., G.R., R.S., X.W., H.C., and L.C. performed the research; Q.Z., W.Z., G.R., J.M.C., and S.M.J. designed the research and analyzed the data; Q.Z. and S.M.J. wrote the paper; and all authors checked the final version of the manuscript.
Q.Z. and W.Z. contributed equally to this study.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank members of the Jane and Cunningham Laboratories for providing many useful suggestions.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal