It is often predicted that stem cells divide asymmetrically, creating a daughter cell that maintains the stem-cell capacity, and 1 daughter cell committed to differentiation. While asymmetric stem-cell divisions have been proven to occur in model organisms (eg, in Drosophila), it remains illusive whether primitive hematopoietic cells in mammals actually can divide asymmetrically. In our experiments we have challenged this question and analyzed the developmental capacity of separated offspring of primitive human hematopoietic cells at a single-cell level. We show for the first time that the vast majority of the most primitive, in vitro–detectable human hematopoietic cells give rise to daughter cells adopting different cell fates; 1 inheriting the developmental capacity of the mother cell, and 1 becoming more specified. In contrast, approximately half of the committed progenitor cells studied gave rise to daughter cells, both of which adopted the cell fate of their mother. Although our data are compatible with the model of asymmetric cell division, other mechanisms of cell fate specification are discussed. In addition, we describe a novel human hematopoietic progenitor cell that has the capacity to form natural killer (NK) cells as well as macrophages, but not cells of other myeloid lineages.
Introduction
Somatic stem cells are defined as undifferentiated cells, which can self-renew over a long period of time and give rise to progenitor cells that are committed to differentiation upon their further development. Since both an uncontrolled expansion as well as loss of stem cells would be fatal for multicellular organisms, the decision of self-renewal versus differentiation needs to be tightly controlled. Therefore, a key question in stem-cell biology is how and which mechanisms govern this decision.
Hematopoietic stem cells (HSCs) are the most investigated mammalian stem cells. More than 30 years of clinical experience as well as experiments with animals demonstrated that neonatal and adult HSCs retain the ability to reconstitute the hematopoietic systems of patients after myeloablative treatment.1 Therefore, an important feature of HSCs is the capacity to replenish all lineages of mature blood cells. Beside this, they also have the potential to expand in vivo as revealed by sequential transplantation experiments using limiting numbers of mouse HSCs to reconstitute the hematopoietic systems of primary and secondary lethally irradiated hosts.2,3 However, although HSCs can be maintained in vitro in close contact to adequate stroma cells,4-8 no evaluated in vitro condition has been reported so far, which supports the expansion of these cells over a period of several weeks. These findings demonstrate that the surrounding environment has a major influence on the cell fate of HSCs and their daughter cells. In this context, it was recently shown that by participating in the formation of special HSC-supporting niches, osteoblasts regulate the size of the HSC pool in vivo.9-11
In addition to data supporting a hematopoietic stem-cell niche model, results of further studies suggest that cells of the hematopoietic stem- and progenitor-cell compartments are able to divide asymmetrically. In an initial set of experiments Ogawa's group analyzed the differentiation of murine and human hematopoietic progenitor cells (HPCs). After separation of paired-progenitor cells they cultured these cells individually and observed that in some cases siblings gave rise to colonies that significantly differed from each other (ie, they gave rise to different cell lineages and/or to colonies of different sizes).12-14 Recently, some evidence for the occurrence of asymmetric cell division of mouse HSCs was provided in a transplantation model.15,16
In studies using human HSC-enriched cells of the CD34+CD38- fraction we and others have shown that CD34+CD38- cells are heterogeneous in respect to their function and their proliferation kinetics (ie, the proliferation rate of more primitive cells is lower than that of committed ones).17-19 Furthermore, it was observed that approximately 30% of the CD34+CD38- cells gave rise to daughter cells with heterogeneous proliferation kinetics, perhaps the result of an asymmetric cell division.17,19,20
To increase the evidence for the occurrence of asymmetric cell divisions within the most primitive in vitro–detectable hematopoietic cell compartment we have separated the progenies of HSC candidates by micromanipulation and analyzed their developmental potential in primitive human progenitor assays.
Materials and methods
Cell source and preparation
Human umbilical cord blood (CB) was obtained from the umbilical vein after delivery of the placenta from mothers after informed consent according to the Declaration of Helsinki. Approval for CB was obtained from the Paul-Ehrlich Institute. Mononuclear cells were isolated from individual sources by Ficoll (Biocoll Separating Solution; Biochrom AG, Berlin, Germany) density gradient centrifugation. CD34+ cells were isolated by magnetic cell separation using the MidiMacs technique according to the manufacturer's instructions (Miltenyi Biotec, Bergisch Gladbach, Germany).
Immunofluorescence
For the proliferation kinetics, freshly isolated CD34+ cells were stained with PKH2 (Sigma-Aldrich Chemie, Taufkirchen, Germany) as described previously.19 Before the cells were analyzed by flow cytometry they were stained with AC133-phycoerythrin (PE; Miltenyi Biotec) and anti-CD34–PE/Cytochrome 5 (PCy5) antibodies (BD PharMingen, Heidelberg, Germany). Flow cytometric analyses were performed on a Cytomics FC 500 flow cytometer equipped with the RXP software (Beckman Coulter, Krefeld, Germany).
For functional analysis CD34+-enriched cells were stained with anti-CD34–PE (BD PharMingen) and anti-CD38–allophycocyanin (APC; BD PharMingen) antibodies. Individual CD34+CD38- cells were sorted into 96-well plates (NUNC, Roskilde, Denmark) using the Automated Cell Deposition Unit (ACDU) on a FACSvantage-SE flow cytometry system (BD Immunosystems, Heidelberg, Germany) equipped with an Apple G3 Power computer (Palo Alto, CA). To ensure that only a single cell was deposited, the ACDU was set up in a low-event “through-put” (200-500 events/second).
Cell culture (bulk proliferation assays)
Freshly enriched and PKH2-stained CD34+ cells were cultured in a humidified atmosphere at 37°C and 5% CO2 at a density of approximately 1 × 105 cells/mL in serum containing tissue-culture medium (Myelocult H5100; Stemcell Technologies, Vancouver, BC, Canada) supplemented with 1000 U/mL penicillin and 100 U/mL streptomycin (Invitrogen, Karlsruhe, Germany) in the presence of early-acting (recombinant human fetal liver tyrosine kinase 3 ligand [rhFLT3L], recombinant human stemcell factor [rhSCF], and recombinant human thrombopoietin [rhTPO], each at 10 ng/mL final concentration; PeproTech, Rocky Hill, NJ) or late-acting cytokines (10 ng/mL recombinant human interleukin-3 [rhIL-3], 500 U/mL rhIL-6, 10 ng/mL recombinant human granulocyte-macrophage colony-stimulating factor [rhGM-CSF], 2.5 ng/mL recombinant human basic fibroblast growth factor [rh-bFGF],10 ng/mL recombinant human insulin-like growth factor-1 [rhILGF; all Cell Systems, St Katharinen, Germany], 2.5 U/mL recombinant human erythropoietin [rhEpo; Roche Diagnostics, Mannheim, Germany], and 50 ng/mL rhSCF [R&D Systems, Minneapolis, MN]), respectively.
Functional assays (single-cell experiments)
Individual sorted CD34+CD38- cells were cultured for 10 days in Myelocult (Cell Systems) supplemented with 10 ng/mL rhIL-3, 500 U/mL rhIL-6, 10 ng/mL rhGM-CSF, 2.5 ng/mL rhbFGF, 10 ng/mL rhILGF (all Cell Systems), 2.5 U/mL rhEpo (Roche), 50 ng/mL rhSCF (R&D Systems), 1000 U/mL penicillin, and 100 U/mL streptomycin (Invitrogen) as described before.19
Before cell deposition the 96-well plates (Nunc) were precoated with bovine serum albumin (BSA) as described previously,19,21 and each well was subsequently filled with 150 μL culture medium as described. After single-cell deposition, cells were maintained in a humidified atmosphere at 37°C and 5% CO2 and fed every 48 hours with fresh medium. The proliferation of each cell was determined by light microscopy every 12 to 24 hours.
Immediately after single cells divided, the corresponding daughter cells were separated using a customized micromanipulation-system (Nikon-Germany, Düsseldorf, Germany) and individually transferred into the myeloid-lymphoid–initiating cell (ML-IC) assay.
ML-IC assay
This readout assay was described extensively in our previous reports.19,21 Briefly, separated daughter cells were individually deposited into 96-well plates (Nunc) containing a confluent, irradiated AFT024 feeder-cell layer. After 2 weeks of culture in a humidified atmosphere at 37°C and 5% CO2 in RPMI-medium containing 20% fetal bovine serum supplemented with 10 ng/mL rhFLT3L, 10 ng/mL rhSCF, 10 ng rhIL-7, and 10 ng/mL rhTPO (all Cell Systems), the content of each well was harvested by trypsination and split into 4 equal fractions. As described before, 2 fractions were transferred into duplicates of the lymphoid natural killer initiating cell (NK-IC) readout assay;21,22 the other 2 fractions were transferred into duplicates of the myeloid LTC-IC assay.19,21
Culture conditions in readout assays
Lymphoid differentiation readout (NK-IC assay). Transferred cells were cocultured with the murine cell line AFT024 in Dulbecco modified Eagle medium (DMEM) and Ham F12-medium (Invitrogen) mixed in a 2:1 (vol/vol) relation containing 20% heat-inactivated human AB serum (Cambrex, Taufkirchen, Germany), ascorbic acid (20 mg/mL; Invitrogen), selenium selenite (50 μM; Invitrogen), 2-mercaptoethanol (25 μM), and ethanolamine (50 μM; Invitrogen). The following cytokines were added to these cultures: rhIL-2 (1000 U/mL), rhIL-3 (5 ng/mL), rhIL-7 (20 ng/mL), rhSCF (10 ng/mL), and rhFlt3L (10 ng/mL). At weekly intervals half-media exchanges were performed using 10% instead of 20% human AB serum. Starting at week 2, the only cytokine added to the cultures was rhIL-2. After 5 to 7 weeks of culture, wells containing viable cells were harvested and cells were analyzed flow cytometrically using antibodies recognizing the NK cell–specific antigens CD16 and CD56 as well as CD3.
Confirming previous studies from our group and others the cells arising in the NK-IC assay express the NK cell–specific antigens CD16 and CD56 and are negative for the T-cell marker CD3. As such cells are able to kill cells of the commonly used NK cell target cell line K562, they are considered as functional NK cells.19,21-24 In agreement with this assumption, we found in our ongoing experiments that the arising cells express additional NK cell–specific antigens like NKp30/NKp44/NKp46 and NKG2A/CD94 as well as different killer immunoglobulin-like receptor (KIR) transcripts (M.P. and M. Uhrberg, manuscript in preparation).
Myeloid differentiation readout (LTC-IC assay). Transferred cells were cocultured with the murine cell line AFT024 in Iscove modified Dulbecco medium (IMDM; Invitrogen) supplemented with 12.5% fetal calf serum (FCS), 12.5% horse serum (Cell Systems), 2 mM l-glutamine (Invitrogen), 1000 U/mL penicillin, 100 U/mL streptomycin (Invitrogen), and 10-6 M hydrocortisone. Cultures were maintained for 5 weeks in a humidified atmosphere at 37°C and 5% CO2 and fed once a week. At week 5 all wells were overlaid with clonogenic methylcellulose medium (Methylcellulose [Fluka, Buchs, Switzerland] in a final concentration of 1.12% containing IMDM and supplemented with 30% FCS, 3 U/mL erythropoietin [Cell Systems], and supernatant of the bladder carcinoma cell line 5637 [10%]). Wells were scored for the occurrence of secondary colony-forming cells (CFCs) after an additional 2 weeks. As the initially deposited cells and their offspring were cultured for more than 8 weeks before reading out the secondary colony-forming cells as LTC-ICs, our myeloid readout reflects more primitive myeloid progenitors than those detected in conventional LTC-IC assays.
Statistics
Experimental results from different experiments were reported as mean ± standard deviation of the mean (SD). Significance analyses were performed with the paired Student t test.
Results
Experimental design
We and others have demonstrated that primitive human hematopoietic cells give rise to daughter cells following different proliferation kinetics.17,19,20 However, the cell fate of primitive hematopoietic cell siblings has not been analyzed at the single-cell level until now. Therefore, we decided to determine the developmental capacity of arising daughter cells individually. Because it is challenging to identify a single human primitive hematopoietic cell and subsequently separate its offspring, we compared the effect of 2 different culture conditions on the proliferation kinetics of primitive hematopoietic cells in bulk experiments first. For these analyses we assessed effects of 2 distinct cytokine cocktails, which we have successfully applied in our previous analysis,19,24 1 cocktail containing late-acting cytokines (LACs), and the other consisting of early-acting cytokines (EAC; ie, SCF, FLT3L, and TPO). The latter cocktail has been shown to be particularly effective in maintaining and slightly expanding human HSCs in suspension cultures up to 7 days.25-27 In order to compare the data presented here with our former results we performed our experiments in Dexter-type cultures.
Proliferation kinetics of primitive hematopoietic cells under different cytokine conditions
Isolated CD34+ cells (89.9% ± 5.9% purity; n = 3) were labeled with the fluorescent dye PKH2 and cultured up to a week in serum containing media supplemented either with EACs or LACs. Starting at day 3 cultured cells from individual aliquots were harvested every other day and simultaneously counterstained with antibodies recognizing the human stem-cell surrogate markers CD34 and CD133.28,29
Flow cytometric analysis of bulk experiments. Since the amount of cells depicted in the plots is normalized to the cell number initially used, plots can be compared semiquantitatively. Cells analyzed in panels B and C were gated according to the morphology depicted on the forward scatter/side scatter plots shown in panel A. Plots in panel B represent the distribution of CD34 and CD133 antigens in freshly isolated or in expanded CD34+ cells, respectively. A characteristic isotype control of expanded cells is shown as inlet in the right panels. To identify the individual subpopulations in panels A or C, CD34+CD133+ cells are labeled in red, CD34+CD133- cells in green, and CD34-CD133- cells in black. The PKH2 staining, representing the number of cell divisions single cells have performed, is plotted against the CD133 antigen distribution in panel C. PKH2bright cells are plotted within the right quadrants of the diagrams shown in panel C. See “Results” for more details.
Flow cytometric analysis of bulk experiments. Since the amount of cells depicted in the plots is normalized to the cell number initially used, plots can be compared semiquantitatively. Cells analyzed in panels B and C were gated according to the morphology depicted on the forward scatter/side scatter plots shown in panel A. Plots in panel B represent the distribution of CD34 and CD133 antigens in freshly isolated or in expanded CD34+ cells, respectively. A characteristic isotype control of expanded cells is shown as inlet in the right panels. To identify the individual subpopulations in panels A or C, CD34+CD133+ cells are labeled in red, CD34+CD133- cells in green, and CD34-CD133- cells in black. The PKH2 staining, representing the number of cell divisions single cells have performed, is plotted against the CD133 antigen distribution in panel C. PKH2bright cells are plotted within the right quadrants of the diagrams shown in panel C. See “Results” for more details.
By analyzing the influence of the culture conditions on CD34+ cells we observed that under both conditions cells increase in size and granularity (Figure 1A). Cells cultured in the presence of LACs expand more (45.2-fold ± 17.7-fold at day 5 compared with day 0; n = 3) than in the presence of EACs (14.7-fold ± 5.6-fold at day 5 compared with day 0; n = 3). Under both conditions most cells remain CD34+ until day 5 (Figure 1B) (LACs, 61.4% ± 20.2%; n = 3; EACs, 85.5% ± 10.4%; n = 3), while the percentage of CD133+ cells decreases over time (Figure 1B) (LACs, 14.6% ± 6.7%; n = 3; EACs, 33.0% ± 5.2%; n = 3 at day 5). However, the absolute numbers of CD133+ cells increase under both conditions (LACs, 8.6-fold ± 1.0-fold; n = 3; EACs, 5.4-fold ± 1.0-fold at day 5 compared with day 0; n = 3).
In addition, we show that under both conditions the average of the newly detected CD34+CD133- cells is less positive for PKH2 than CD34+CD133+ cells, suggesting that most arising CD34+CD133- cells have had a higher proliferation rate than the CD34+CD133+ cells (Figure 1C). Regarding their PKH2 staining, the LAC-stimulated CD34+CD133+ cell fraction is more heterogeneous than the corresponding EAC-stimulated fraction (Figure 1C), suggesting that EACs stimulate proliferation of CD34+CD133+ cells more homogeneously than LAC conditions. Within the LAC-stimulated fraction we observed more remaining PKHbright CD34+CD133+ cells than in the EAC-stimulated fraction (at day 5: LACs, 47.7% ± 24.1%; EACs, 39.4% ± 28.3% of the initially cultured PKHbright CD34+CD133+ cells, n = 3, P = .05; Figure 1C). Remarkably, the PKHbright fraction of LAC-stimulated cells contain less CD133- cells than the EAC-stimulated fraction, resulting in a sharp contrast of PKHbright CD34+CD133+ versus PKH+ CD34+CD133- cells within the LAC-stimulated fraction (Figure 1C).
According to our previous results primitive hematopoietic cells cultured under LAC conditions get highly enriched in the PKHbright or the so-called slow dividing fraction.19,24 This is most likely due to the fact that the most primitive hematopoietic cells do not immediately respond to LACs and therefore remain quiescent for the first few days under LAC conditions. Since the vast majority of the CD34+ cells get activated under EAC conditions and show similar cell division kinetics, the LAC conditions provide an opportunity to discriminate between primitive and more mature CD34+ cells according to just the way they initiate their cell division. Therefore, we concluded that LAC conditions were more suitable for our intended single-cell studies on primitive hematopoietic cells than EAC conditions.
Our conclusion was additionally supported by the finding that the percentage of the stem-cell surrogate marker CD133 within the slow dividing fraction (PKHbright cells) was significantly higher under LAC than under EAC conditions (at day 5: LAC, 61.1% ± 3.1%; EAC, 50.3% ± 3.9%; n = 3, P = .005).
Proliferation of individual primitive human hematopoietic cells
Due to the fact that freshly isolated CD34+ cells express similar amounts of CD133 (Figure 1B) and with the necessity to enrich the most primitive hematopoietic cells as much as possible, we decided to perform our single-cell analyses with CD34+CD38- cells.
In 4 independent experiments we sorted a total of 176 single CD34+CD38- cells per experiment into individual wells of 96-well plates. To determine the deposition frequency we analyzed each well 12 hours after finishing the sorting procedure by bright-field microscopy. We recovered a total of 556 single deposited cells, which corresponds to a deposition frequency of 79.0% ± 4.0%.
In previous experiments under LAC conditions, the most primitive hematopoietic cells remained quiescent for up to 5 days and underwent their first in vitro cell division between day 5 and day 10 after deposition.19 Therefore, we tracked the division process of each single cell within the first 10 days and grouped them into 3 different categories as shown in Table 1: (1) category I indicates cells that divided before day 5 (59% ± 12%); (2) category II, cells that performed their first cell division between day 5 and day 10 (31% ± 11%); and (3) category III, cells which did not perform any cell division within the first 10 days of culture (10% ± 4%).
Categorization of individual CD34+CD38- cells according to the occurrence of their initial in vitro cell division under LAC-stimulated culture conditions
. | Deposited cells . | Category I: first division before day 5 . | Category II: first division days 5-10 . | Category III: first division after day 10 . |
|---|---|---|---|---|
| Experiment 1 | 130 | 93 | 26 | 11 |
| Experiment 2 | 147 | 65 | 63 | 19 |
| Experiment 3 | 139 | 78 | 54 | 7 |
| Experiment 4 | 140 | 90 | 34 | 16 |
| Total cell no. (mean % ± SEM) | 556 | 326 (59.0 ± 11.7) | 177 (31.5 ± 11.1) | 53 (9.5 ± 3.5) |
. | Deposited cells . | Category I: first division before day 5 . | Category II: first division days 5-10 . | Category III: first division after day 10 . |
|---|---|---|---|---|
| Experiment 1 | 130 | 93 | 26 | 11 |
| Experiment 2 | 147 | 65 | 63 | 19 |
| Experiment 3 | 139 | 78 | 54 | 7 |
| Experiment 4 | 140 | 90 | 34 | 16 |
| Total cell no. (mean % ± SEM) | 556 | 326 (59.0 ± 11.7) | 177 (31.5 ± 11.1) | 53 (9.5 ± 3.5) |
Values in table are total numbers of cells except where indicated.
Analyses of the developmental potential of individually separated daughter cells
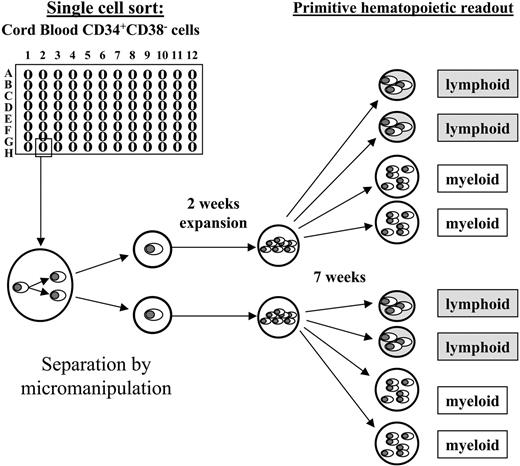
According to our focus on the most primitive human hematopoietic cells we only included cells from category II for the micromanipulation-based daughter cell analyses. Within 24 hours after performing their initial cell division, emerging daughter cells of category II cells were separated by micromanipulation and individually transferred into expansion cultures containing the murine fetal liver–derived stromal cell line AFT024.21 As shown in Figure 2, the expansion cultures were harvested after 2 weeks and split into 4 separate aliquots, which were transferred into secondary readout systems: 2 into primitive myeloid assays (LTC-IC) and 2 into primitive lymphoid assays (NK-IC). This enabled us to determine whether initially deposited cells gave rise to primitive progeny in any of the functional assays.
Experimental setup. Individual CD34+CD38- cells were deposited by fluorescent cell sorting into 96-well plates (1 cell/well), and observed at half-daily intervals. Initially deposited cells that performed their initial cell division between culture days 5 and 10 (category II) were considered for further analyses. Shortly after the first cell division, the arising daughter cells were separated by micromanipulation and individually transferred into secondary plates containing irradiated AFT024 cells as a stromal feeder. After 2 weeks of expansion the entire progeny of each individual daughter cell was split into 4 aliquots and transferred in equal amounts (in duplicates) into primitive myeloid (LTC-IC) or primitive lymphoid (NK-IC) readout assays, respectively. After an additional 7 weeks the assays were analyzed as described in “Materials and methods.” Originally deposited cells as well as singularized daughter cells that had both LTC-IC as well as NK-IC capacity were retrospectively considered ML-ICs.21
Experimental setup. Individual CD34+CD38- cells were deposited by fluorescent cell sorting into 96-well plates (1 cell/well), and observed at half-daily intervals. Initially deposited cells that performed their initial cell division between culture days 5 and 10 (category II) were considered for further analyses. Shortly after the first cell division, the arising daughter cells were separated by micromanipulation and individually transferred into secondary plates containing irradiated AFT024 cells as a stromal feeder. After 2 weeks of expansion the entire progeny of each individual daughter cell was split into 4 aliquots and transferred in equal amounts (in duplicates) into primitive myeloid (LTC-IC) or primitive lymphoid (NK-IC) readout assays, respectively. After an additional 7 weeks the assays were analyzed as described in “Materials and methods.” Originally deposited cells as well as singularized daughter cells that had both LTC-IC as well as NK-IC capacity were retrospectively considered ML-ICs.21
As shown in Table 2 we formed 3 groups, discriminating initially deposited cells which gave rise to (1) 2 colony forming daughter cells (category IIa; 55 pairs of daughter cells); (2) those in which only 1 daughter cell formed colonies (category IIb; 21 pairs); and (3) the ones in which none of the daughter cells formed any colonies (category IIc; 101 pairs).
Categorization of individual CD34+CD38– cells performing their initial in vitro cell division under LAC-stimulated culture conditions between days 5 and 10
. | Investigated daughter cell pairs . | Category IIa: 2 colony-initiating daughter cells . | Category IIb: 1 colony-initiating daughter cell . | Category IIc: No colony-initiating daughter cells . |
|---|---|---|---|---|
| Experiment 1 | 26 | 3 | 1 | 22 |
| Experiment 2 | 63 | 16 | 5 | 42 |
| Experiment 3 | 54 | 13 | 7 | 34 |
| Experiment 4 | 34 | 23 | 8 | 3 |
| Total cell no. (mean % ± SEM) | 177 | 55 (32.2 ± 24.5) | 21 (12.1 ± 8.5) | 101 (55.8 ± 32.7) |
. | Investigated daughter cell pairs . | Category IIa: 2 colony-initiating daughter cells . | Category IIb: 1 colony-initiating daughter cell . | Category IIc: No colony-initiating daughter cells . |
|---|---|---|---|---|
| Experiment 1 | 26 | 3 | 1 | 22 |
| Experiment 2 | 63 | 16 | 5 | 42 |
| Experiment 3 | 54 | 13 | 7 | 34 |
| Experiment 4 | 34 | 23 | 8 | 3 |
| Total cell no. (mean % ± SEM) | 177 | 55 (32.2 ± 24.5) | 21 (12.1 ± 8.5) | 101 (55.8 ± 32.7) |
Values in table are total numbers of cells except where indicated.
Category IIa and IIb cells, 76 cells in total, were further subgrouped regarding to the cell fate adopted by the individual daughter cells (Table 3). We used the following definitions: (1) initially plated cells that only gave rise to primitive myeloid hematopoiesis were defined as LTC-ICs; (2) those that gave rise only to NK cells were defined as NK-ICs; and (3) cells that were able to generate both LTC-ICs as well as NK-ICs were defined as ML-ICs, closely related to the most primitive human hematopoietic compartment.21
Cell fate classification of initially deposited colony-initiating CD34+CD38- cells
. | Category IIa and IIb cells . | ML-ICs . | LTC-ICs . | M-NK-ICs . | NK-ICs . | CFUs-M . |
|---|---|---|---|---|---|---|
| Experiment 1 | 4 | 1 | 0 | 1 | 2 | 0 |
| Experiment 2 | 21 | 5 | 2 | 5 | 2 | 7 |
| Experiment 3 | 20 | 4 | 0 | 5 | 8 | 3 |
| Experiment 4 | 31 | 5 | 1 | 13 | 5 | 7 |
| Total cell no. (mean % ± SEM) | 76 | 15 (21.2 ± 4.0) | 3 (3.2 ± 4.5) | 24 (28.9 ± 8.7) | 17 (28.9 ± 19.2) | 17 (17.7 ± 14.0) |
. | Category IIa and IIb cells . | ML-ICs . | LTC-ICs . | M-NK-ICs . | NK-ICs . | CFUs-M . |
|---|---|---|---|---|---|---|
| Experiment 1 | 4 | 1 | 0 | 1 | 2 | 0 |
| Experiment 2 | 21 | 5 | 2 | 5 | 2 | 7 |
| Experiment 3 | 20 | 4 | 0 | 5 | 8 | 3 |
| Experiment 4 | 31 | 5 | 1 | 13 | 5 | 7 |
| Total cell no. (mean % ± SEM) | 76 | 15 (21.2 ± 4.0) | 3 (3.2 ± 4.5) | 24 (28.9 ± 8.7) | 17 (28.9 ± 19.2) | 17 (17.7 ± 14.0) |
Values in table are total numbers of cells except where indicated.
In several cases colonies were found in the LTC-IC assays that did not fulfill the well-defined morphologic criteria of secondary colony formation. Since they resembled a macrophage-like morphology without clonogenic proliferation we did not categorize them as LTC-ICs but as macrophage colony-forming units (CFUs-M). Remarkably, we found many cases in which individual cells had the NK-IC capacity and gave rise to CFU-M–forming progeny. To discriminate these cells from ML-ICs we called them macrophage/NK cell–initiating cells (M-NK-ICs).
In total, 15 of the original deposited cells had the ML-IC capacity; 2 of them transmitted the ML-IC fate to both daughter cells, and in the remaining cases only 1 of the daughter cells adopted both, the LTC-IC as well as the NK-IC capacity (Table 4). Remarkably, in 6 (40%) cases the non–ML-IC daughter cell gave rise to NK-ICs and formed macrophage-like colonies in the LTC-IC assay, demonstrating that viable offspring of the corresponding daughter cells were transferred into the latter assay. These results suggest that in these cases the first-generation ML-IC daughter cells contained different developmental capacities. Surprisingly, we never found the constellation in which 1 ML-IC daughter cell adopted the myeloid and the other 1 the lymphoid potential.
Cell fate classification of separated offspring of initially deposited colony-initiating CD34+CD38- cells
. | DC no. 1 . | . | . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|
| DC no. 2 . | ML-ICs . | LTC-ICs . | M-NK-ICs . | NK-ICs . | CFUs-M . | ||||
| ML-ICs | 2 | — | — | — | — | ||||
| LTC-ICs | 3 | 2 | — | — | — | ||||
| M-NK-ICs | 6 | 0 | 11 | 0 | — | ||||
| NK-ICs | 1 | 1 | 0 | 7 | — | ||||
| CFUs-M | 2 | 0 | 8 | 0 | 13 | ||||
| Dead | 1 | 0 | 5 | 10 | 4 | ||||
| Identical cell fate, no. (%) | 2 (13.3) | 2 (66.7) | 11 (45.8) | 7 (41.2) | 13 (76.5) | ||||
| Different cell fate, no. (%) | 13 (86.7) | 1 (33.3) | 13 (54.2) | 10 (58.8) | 4 (23.5) | ||||
. | DC no. 1 . | . | . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|
| DC no. 2 . | ML-ICs . | LTC-ICs . | M-NK-ICs . | NK-ICs . | CFUs-M . | ||||
| ML-ICs | 2 | — | — | — | — | ||||
| LTC-ICs | 3 | 2 | — | — | — | ||||
| M-NK-ICs | 6 | 0 | 11 | 0 | — | ||||
| NK-ICs | 1 | 1 | 0 | 7 | — | ||||
| CFUs-M | 2 | 0 | 8 | 0 | 13 | ||||
| Dead | 1 | 0 | 5 | 10 | 4 | ||||
| Identical cell fate, no. (%) | 2 (13.3) | 2 (66.7) | 11 (45.8) | 7 (41.2) | 13 (76.5) | ||||
| Different cell fate, no. (%) | 13 (86.7) | 1 (33.3) | 13 (54.2) | 10 (58.8) | 4 (23.5) | ||||
The more primitive separated daughter cell is defined as DC no. 1, the other as DC no. 2. Values in table are total numbers of cells except where indicated.
— indicates not applicable.
Only 3 of the 76 originally deposited cells were determined exclusively as LTC-ICs; 2 of them gave rise to daughter cells, both containing the LTC-IC potential (Table 4). Originally deposited cells of the newly defined M-NK-IC group transmitted this cell fate to both daughter cells in 46% of the cases studied, and to only 1 daughter cell in 54% (Table 4). In the latter cases the non–M-NK-IC daughter cell died or gave rise to macrophage-like cells in LTC-IC assays. We never found any daughter cell that had the NK-IC potential only. Seventeen of the originally deposited cells had NK-IC potential only, which was transmitted in 7 cases to both daughter cells; in the remaining 10 cases only 1 of the siblings gave rise to NK cells in our assays (Table 4). Most of the cells (13 of 17) that were retrospectively named CFUs-M transmitted this fate to both daughter cells; in the remaining cases, only 1 daughter cell gave rise to the macrophage-like cells.
In summary, most of the originally deposited cells, retrospectively determined as ML-ICs, gave rise to daughter cells with different cell fates. In contrast, siblings of more-committed mother cells seem to inherit a higher ratio of identical cell fates in our assays (Table 4).
Discussion
Here, we analyzed primitive human hematopoietic cells in bulk cultures and at a single-cell level, and report 3 major findings: (1) we realized that upon cultivation CD34+ cells split up into a CD34+CD133+ and a CD34+CD133- fraction; (2) using our newly established single-cell separation approach we determined the cell fate of primitive human hematopoietic cells and their first-generation daughter cells individually and present evidence for the existence of progenitor cells containing the capacity to form NK cells and macrophages, but which lack more primitive myeloid capacities (LTC-ICs); and (3) we demonstrate that the most primitive ML-ICs have a high tendency (87%) to transmit their cell fate to only 1 of the arising daughter cells. In contrast, the ratio of more-committed progenitor cells giving rise to progeny adopting identical cell fates to those adopting different cell fates is more balanced.
Although there is increasing evidence that special hematopoietic niches are required to maintain HSCs,9-11 it is often suggested that hematopoietic stem cells can divide asymmetrically to form another HSC and a more-specified daughter cell.30 The latter hypothesis is highly supported by the finding that immediate progeny of primitive hematopoietic progenitor cells often adopt different cell fates in myeloid readout systems.12-14,16,18 However, as mentioned by Takano and colleagues, due to the lack of an appropriate assay, the lymphoid potential of separated cells was not analyzed in these studies.16 In this study we used the ML-IC assay, a sensitive and efficient assay for the detection of both the myeloid and lymphoid potentials of individual cells,19,21 and analyzed the myeloid and lymphoid potentials of progenies of primitive human hematopoietic cells at the single-cell level.
Supporting our previous data, we found that under LAC conditions approximately 30% of the deposited CD34+CD38- cells performed their initial cell division between culture day 5 and 10.19 We were able to determine retrospectively the cell fate of approximately 45% of the initially deposited cells that performed their first cell division between culture days 5 and 10. In 55% of the cases studied none of the separated daughter cells gave rise to any recognizable colony in our readout systems. Since similar frequencies were obtained in our previous and other studies, in which nonseparated CD34+CD38- cells were analyzed in ML-IC assays,19,31 we suggest that loss of cells during the micromanipulation procedure is more or less negligible. The efficiency of our micromanipulation procedure is additionally supported by the fact that we obtain similar ML-IC frequencies of nonseparated and separated CD34+CD38- cells performing their first cell division under LAC conditions between culture days 5 and 10 (15 [8.5%] cells of 177) or after day 5 (10.3% ± 3.5%),19 respectively.
In this context it should be mentioned that all but 1 initially deposited cell, which gave rise to only 1 colony-forming daughter cell, were retrospectively determined to be more mature cells (M-NK-ICs, NK-ICs, or CFUs-M), suggesting that the non–colony-forming daughter cell was more committed and terminally differentiated during the long-term culture period.
It is interesting to note that using the ML-IC assay we could identify a novel human progenitor cell that has not been described before. This progenitor, which we called M-NK-IC, has the capability to initiate NK cell development and also gives rise to macrophages but not to secondary clonogenic myeloid colonies in LTC-IC assays. Similar to our findings, murine fetal liver but not adult hematopoietic progenitors have been described as containing lymphoid and macrophage potential in short-term murine readout systems.32,33
According to the prevailing model of hematopoiesis, primitive hematopoietic cells give rise to common myeloid and common lymphoid progenitor cells, which have the developmental capacity to form all myeloid or lymphoid lineages, respectively.34 Because macrophages belong to the myeloid compartment, the discovery of progenitors containing the M-NK-IC capacity is not compatible with this model. In addition, the recent discovery of primitive hematopoietic cells in mice and humans which contain the lymphoid potential as well as the capacity to form granulocytes and macrophages but not cells of the erythromegakaryocytic lineage, is contrary to this model as well.35-37 In this context, Adolfsson and colleagues offered a so-called composite model in which primitive hematopoietic cells sequentially lose the capacity to form cells of the megakaryocyte/erythroid and then of the granulocyte/macrophage potential during lymphoid commitment.35 Our findings together with those of others32,33,38 support this hypothesis and further suggest that next to the megakaryocyte/erythroid potential the granulocytic developmental capacity is lost during early lymphoid commitment, leaving cells containing the capacity to initiate lymphoid and macrophage development.
In previous studies it was found that siblings of primitive hematopoietic cells often adopt different proliferation kinetics, whereas more-committed progenitor cells give rise to daughter cells dividing in a more uniform fashion. Differences in the proliferation kinetics are often interpreted as a result of an asymmetric cell division.17,19,20 The theory of asymmetric cell division is further supported by the finding that individual daughter cells of primitive hematopoietic cells frequently adopt different cell fates in myeloid progenitor assays.12-14,16 However, until now there was no evidence that the most primitive hematopoietic cells containing the lymphoid as well as the myeloid developmental potential give rise to daughter cells adopting different cell fates. Therefore, we demonstrate here for the first time evidence that the most primitive, in vitro–detectable hematopoietic cells, the ML-ICs, give rise to daughter cells adopting different cell fates, which could be the result of an asymmetric cell division. In addition, we provide evidence that offspring of more-committed progenitors that seem to contain similar proliferation kinetics also give rise to cells adopting different cell fates in approximately 50% of the cases studied. These findings demonstrate that proliferation kinetics of arising daughter cells cannot be interpreted as indication for the occurrence of asymmetric cell divisions.
Surprisingly, none of the deposited cells gave rise to siblings that both adopted only partial capacities of their mother cells. In all cases studied at least 1 of the arising daughter cells took over the developmental capacity of the mother cell, most likely resembling the predicted process of self-renewal. Committed progenitors have a high tendency to expand their cell fate, especially by forming 2 daughter cells containing the same developmental capacity. Cells of the most primitive compartment, the ML-ICs, have a very low tendency to transmit their cell fate to both of the arising daughter cells, which is consistent with the hypothesis that primitive hematopoietic cells have a high tendency to divide asymmetrically.30
However, can we really conclude from these data that there are asymmetric cell divisions within the primitive hematopoietic cell compartment? It depends how we define “asymmetric cell division.” If we define it from the point of the adopted cell fate, our data would fulfill these criteria. If we define asymmetric cell division as it is used in model organisms such as Drosophila or Caenorhabditis elegans, in which an asymmetric cell division describes the process in which 2 qualitatively different cells are formed by the different distribution of certain factors which might act as cell fate determinants,39 our data will not fulfill these criteria. In principle, it could be that even cells that form daughter cells adopting different cell fates divide in a symmetric way, giving rise to 2 equally specified daughter cells. Since the daughter cells in our as well as in other experiments had the ability to stay in close contact with each other before they were separated, they could theoretically have influenced each other's developmental capacity after mitosis. Developmental processes like that are well described. For example, the Notch-mediated process of lateral inhibition selects a single cell within a group of equivalent cells to adopt another cell fate than the remaining cells of that group.40,41 As Notch is required to maintain primitive hematopoietic cells in an undifferentiated state,42 it might be possible that 1 of the daughter cells activates the Notch signaling pathway in a process of lateral inhibition in its sister cell, resulting in the maintenance of the primitive cell fate in only 1 of the 2 cells.
Thus, we summarize that our data and data presented before are not sufficient to conclude that primitive hematopoietic cells can indeed divide asymmetrically. To unequivocally demonstrate that hematopoietic cells can divide asymmetrically, markers need to be defined which clearly segregate unequally within mitotic cells. We have learned from model organisms that all cells which divide asymmetrically are polarized during cell division and localize certain molecules to distinct regions of the cells, which then get transmitted in an unequal way.39 As we recently could show that several surface molecules, especially CD133, become distributed in a localized fashion in cultivated primitive hematopoietic cells,43 it will be interesting to analyze the distribution of these markers in dividing primitive hematopoietic cells. As we realized that the more primitive CD34+ cells of the slow-dividing fraction specifically express CD133, it might be possible that upon cell division CD133 segregates into one of the arising daughter cells, perhaps confirming the concept of asymmetric cell division within the primitive hematopoietic cell compartment.
Prepublished online as Blood First Edition Paper, October 25, 2005; DOI 10.1182/blood-2005-08-3139.
Supported by grants from the Deutsche Forschungsgemeinschaft (SPP1109 GI 336/1-2 to B.G. and P.W.; HO 914/2-1 to A.D.H.), from the Forschungskommission of the HHU-Düsseldorf (B.G., M.P.), and from the Faculty Research Program of the University of Heidelberg (F203532 to M.P.).
B.G. and T.Z. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The authors like to thank Rainer Saffrich and Martin Peschel (Nikon-Germany) for their excellent support in establishing the micromanipulation system, as well as Volker Eckstein, Johannes Fischer, and Katrin Miesala for their technical support. Umbilical cord blood samples were kindly provided by the Department of Gynecology and Obstetrics of the University of Heidelberg and by Gesine Kögler of the Heinrich-Heine University, Düsseldorf.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal