Abstract
Decreased major histocompatibility class II (MHCII) expression is associated with poor survival in diffuse large B-cell lymphoma (DLBCL). Immune-privileged site DLBCL (IP-DLBCL) patients reportedly have frequent large deletions at the MHCII locus whereas the mechanism of decreased expression in non-IP-DLBCL is unknown. Gene expression profiling data were used for correlation analyses between expression levels of MHCII genes with each other and their transcriptional regulator, CIITA. Comparative genomic hybridization (CGH) assessed chromosomal alterations at MHCII-related loci. Finally, a map was created of expression of genes that are telomeric, within, or centromeric to the MHCII locus. Correlation coefficients among MHCII genes ranged from 0.73 to 0.92, whereas those between adjacent and intervening genes were lower (-0.12 to 0.49). Correlations between MHCII and CIITA expression were higher (0.53 to 0.60) than between CIITA and neighboring genes (-0.05 to 0.22). In 23 MHCII- cases, CGH detected 2 losses and 2 gains at MHCII loci. Expression of genes telomeric, within, and centromeric to MHCII loci were near normal in most MHCII- cases. Large deletions of the MHCII locus are uncommon in non-IP-DLBCL, implicating altered transcription as the operative mechanism for decreased expression.
Introduction
Diffuse large B-cell lymphoma (DLBCL) is a lethal disease and the most common lymphoma diagnosed in the United States. Patient survival is highly variable, spurring recent efforts to further classify the disease and identify prognostically important parameters.1-3 The Leukemia and Lymphoma Molecular Profiling Project (LLMPP) reported results from multi-institutional gene expression microarray studies of 240 DLBCL cases using the Lymphochip.4 This consortium described 4 gene expression signatures that were associated with outcome in DLBCL including the proliferation, major histocompatibility (MHC) class II, lymph node (host response), and germinal center signatures.5 We recently reported on a reanalysis of the LLMPP microarray data that demonstrated that decreasing MHC class II expression was highly predictive of worse patient survival. We correlated the gene expression profiling data with protein immunohistochemistry and demonstrated that the presence or absence of MHC class II protein correlated with high or low percentages of tumor-infiltrating CD8+ T cells, respectively.6 However, the mechanisms of lost MHC class II expression in most cases of DLBCL are unknown.
The classical MHC class II proteins are expressed constitutively on antigen-presenting cells including monocytes, macrophages, dendritic cells, and B cells and are responsible for presenting peptide antigens derived from exogenous proteins to T cells. The classical MHC class II proteins, HLA-DR, HLA-DQ, and HLA-DP, are almost always expressed concurrently on the surface of antigen-presenting cells. The nonclassical molecules, HLA-DM, HLA-DO, and invariant chain (Ii), are expressed within antigen-presenting cells and modulate the antigen presentation pathway. All of the classical and nonclassical MHC class II proteins, except for Ii which is monomeric, consist of α and β peptide chains encoded by different A and B genes.7 All of the genes encoding classical and nonclassical MHC class II proteins, except Ii, are expressed from a single locus located on chromosome 6.8 Thus, decreases in expression of these MHC class II molecules imply either large genetic deletions covering the entire MHC class II region or altered transcriptional activity.
MHC class II transcription including all of the classical molecules, nonclassical molecules (HLA-DM and HLA-DO), and Ii is controlled by the master transactivator CIITA, which works in conjunction with other elements of the transcriptosome, RFX, NF-Y, and CREB. The binding of these various proteins appears highly cooperative. While RFX, NF-Y, and CREB bind directly to DNA sequences at the MHC class II promoters, CIITA binds to these proteins and not to the DNA directly.7 RFX and CIITA are specific activators of the MHC class II genes and have few other known functions, whereas NF-Y and CREB are less specific activators implicated in the control of numerous genes.7 RFX is a protein consisting of 3 subunits, RFX5, RFXB, and RFXAP. Mutation or loss of expression of any of the 3 RFX subunits or of CIITA can completely eliminate all MHC class II expression, as evidenced in the MHC class II deficiency syndromes.9
Loss of MHC class II antigens in lymphomas is a frequent event in a subset of DLBCL involving immune-privileged sites (IP-DLBCL) such as testes or brain.10 Lymphomas that arise in these locations occur more often in patients with immunodeficiencies such as AIDS or who are on immunosuppressive drug therapy following solid organ transplantation or who have congenital immune disorders. In addition, these lymphomas are nearly always of B-cell lineage (rarely T cell), nearly always intermediate to high grade, and often associated with Epstein-Barr virus infection.11 Thus, they are different in many ways from DLBCL originating in other anatomic sites. Using fluorescence in situ hybridization (FISH) analysis, large homozygous or hemizygous losses at the MHC class II locus with consequent loss of MHC class II antigen expression were detected in 11 (61%) of 18 IP-DLBCLs in contrast to 0 of 7 DLBCLs from other locations.10 The loss of MHC class II expression in the IP-DLBCLs was later determined to result from deletions rather than chromosome loss or mitotic recombination.12 These deletions are very large, ranging from 100 to 900 kb (kilobases).10 These results were in contrast to the mechanism of loss in spontaneously mutated lymphoma cell lines or nonhematopoietic malignancies such as melanoma or colonic or laryngeal carcinoma, where chromosome loss or mitotic recombination were reportedly more common.13,14 Proposed mechanisms for non-IP-DLBCL have included point mutations or small deletions undetectable by FISH or methylation of the promoter region, although none of these mechanisms have yet been demonstrated.10 Whether loss of MHC class II molecules confers a survival advantage to tumors arising in or metastasizing to immune-privileged sites is unknown. In our previous work, we analyzed data from the 240 cases of DLBCL studied in the LLMPP. Of these, only 3 were from an immune-privileged site (testes) and none were included in the cases that made up the lowest 10% of MHC class II expression. Thus, the results of our previous paper relating loss of MHC class II to poor patient survival were applicable only to non-IP-DLBCL.6
The purpose of this study was to determine whether the loss of MHC class II expression in DLBCL cases in the LLMPP study group resulted from large chromosomal deletions, as is the reported mechanism in IP-DLBCL, or if other mechanisms are responsible. We present gene expression profiling and comparative genomic hybridization (CGH) evidence that large genetic deletions are infrequent in MHC class II- non-IP-DLBCL and that a more likely explanation is decreased transcription as directed by the master transactivator molecule CIITA.
Materials and methods
We used the publicly available data from the LLMPP published on the internet for DLBCL.15 The data were derived from microarray gene expression analysis using the Lymphochip,4 a type 2 spotted microarray specifically designed to include genes preferentially expressed in lymphoid cells or relevant to immunology or cancer. All of the patient samples were compared with a pooled control sample made from multiple cell lines. The publicly available data were a subset of the microarray data selected for high variability between samples and had been preanalyzed, normalized to median, and log 2 transformed after elimination of excessively low values. We first searched the details of the Lymphochip4 library for array elements annotated as specific for the MHC class II classical and nonclassical and activator genes. These genes included HLA-DRA, HLA-DRB, HLA-DQA, HLA-DQB, HLA-DPA, HLA-DPB, HLA-DMA, HLA-DMB, HLA-DOA, HLA-DOB, Ii, CIITA, and the RFX, NF-Y, and CREB genes. We then independently verified the specificity of these expressed sequence tag (EST) sequences (for those which had a specific EST sequence) as described previously.6 RFX, NF-Y, CREB, and HLA-DO each did not have EST sequences for all of the proteins composing them in the published data set. All of the microarray elements for each gene that passed our criteria were averaged to give a single set of expression values for each gene, which were used in subsequent analysis. We then determined the MHC class II expression, which we defined as the average of the expression of all the classical and nonclassical MHC class II genes present in the dataset: HLA-DRA, -DRB, -DQA, -DQB, -DPA, -DPB, -DMA, -DMB, and Ii. We next identified those cases in the lowest 10% of MHC class II expression, which we defined as MHC class II- cases. These cases account for 24 of the 240 cases in the DLBCL database. Cases in the upper 90% of expression are designated as MHC class II+. Of the 240 cases in the LLMPP data set, only 3 were from immune-privileged sites (all from the testes) so that nearly 99% of cases were non-IP-DLBCL. None of the 3 testicular cases were part of the lowest 10% of MHC class II expressers. Therefore, the data on correlation analysis, positional expression profiling, and CGH presented here were all from non-IP-DLBCL cases.
Chromosome CGH from genomic DNA samples from the lowest-10% and highest-10% MHC class II expressers in the LLMPP project was used to evaluate the frequency of chromosomal losses, gains, and amplifications according to our previously published method.16-18 Briefly, nick translation was used to label normal and tumor DNA with different fluorescent dyes. This mixture of labeled DNA was then hybridized against normal metaphase spreads. Binding of the 2 DNAs was evaluated by image analysis. Differences in fluorescence ratios were evaluated with Cytovision software (Applied Imaging, San Jose, CA). This technique has a resolution of approximately 10 Mb (megabases). The rate of chromosomal losses, gains, and amplifications were examined at several loci: 6p21-31 (MHC class II), 5q32 (Ii), 16p13 (CIITA), 1q21 (RFX5), and 13q13 (RFXAP). Overall chromosomal alteration rate was compared between the lowest-10% and highest-10% MHC class II expressers. One of the lowest-10% cases was not evaluated due to lack of available DNA.
Next, using the National Center for Biotechnology Information website,19 we searched for genes that were located within or closely adjacent to the MHC class II complex on chromosome 6p21-25 and were represented in the expression data. We found 6 genes that were up to 600 kb telomeric, 4 genes within, and 6 genes that were up to 450 kb centromeric from the beginning and end of the MHC class II locus. These genes and their relative positions are listed in Table 1. EST sequences for these genes were verified when possible for sequence specificity as previously described.6
Genes in the vicinity of the MHC class II locus, found in microarray data
Gene . | Position in relation to MHC class II loci* . |
|---|---|
| HSPAIL | –597 kb from HLA-DRA |
| HSPAIA | –594 kb from HLA-DRA |
| BAT8 | –532 kb from HLA-DRA |
| RDBP | –453 kb from HLA-DRA |
| CREBLI | –317 kb from HLA-DRA |
| PBX2 | –255 kb from HLA-DRA |
| TAP2 | Between HLA-DQA and DMB |
| TAP1 | Between HLA-DQA and DMB |
| PSMB9 | Between HLA-DQA and DMB |
| BRD2 | Between HLA-DMA and HLA-DPA |
| RXRB | +77 kb from HLA-DPB |
| RING1 | +85 kb from HLA-DPB |
| RPSI8 | +148 kb from HLA-DPB |
| TAPBP | +186 kb from HLA-DPB |
| DAXX | +195 kb from HLA-DPB |
| BAK1 | +452 kb from HLA-DPB |
Gene . | Position in relation to MHC class II loci* . |
|---|---|
| HSPAIL | –597 kb from HLA-DRA |
| HSPAIA | –594 kb from HLA-DRA |
| BAT8 | –532 kb from HLA-DRA |
| RDBP | –453 kb from HLA-DRA |
| CREBLI | –317 kb from HLA-DRA |
| PBX2 | –255 kb from HLA-DRA |
| TAP2 | Between HLA-DQA and DMB |
| TAP1 | Between HLA-DQA and DMB |
| PSMB9 | Between HLA-DQA and DMB |
| BRD2 | Between HLA-DMA and HLA-DPA |
| RXRB | +77 kb from HLA-DPB |
| RING1 | +85 kb from HLA-DPB |
| RPSI8 | +148 kb from HLA-DPB |
| TAPBP | +186 kb from HLA-DPB |
| DAXX | +195 kb from HLA-DPB |
| BAK1 | +452 kb from HLA-DPB |
Positional differences calculated from the 5′ ends of the genes compared with the 5′ end of the nearest MHC class II molecule; positive numbers indicate centromeric direction
Pearson correlations were performed with Splus 6.1 (Insightful, Seattle, WA). A heat map of the pairwise correlations, including all MHC class II genes, their neighboring and intervening genes, and CIITA, was created to examine both spatial and group correlations. Tests of average correlations between groups of genes used the bootstrap method to correctly accommodate covariances between the estimated correlation coefficients.20 Exact binomial confidence intervals (CIs) were calculated for the chromosomal loss and gain data.
Results
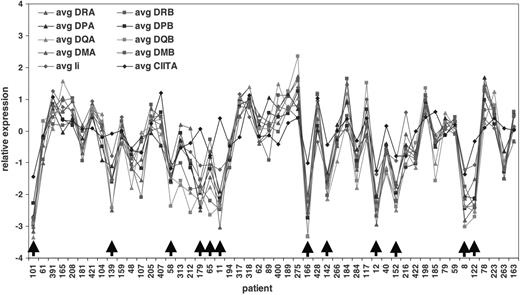
We first investigated whether there were any differences in expression patterns of the MHC class II genes. Initially, we plotted the relative gene expression of the individual MHC class II genes and CIITA for each DLBCL patient, with the patients ranked by increasing time under observation. For Figure 1, we chose to show the first 50 patients from this larger plot, those with the shortest survival after diagnosis (all 50 of these patients are dead), because this part of the plot showed both MHC class II+ and a number of MHC class II- cases. This analysis demonstrates a substantial coordination of MHC class II expression on all cases including those cases identified as MHC class II-. Visual inspection of this graph indicates that when expression of one MHC class II gene is reduced, expression is almost always decreased in the remaining genes including Ii. Importantly, expression of the transactivator CIITA is also decreased in many of the MHC class II- cases, implying that down-regulated transcription, rather than genetic deletion, may be responsible for diminished MHC class II expression in these cases.
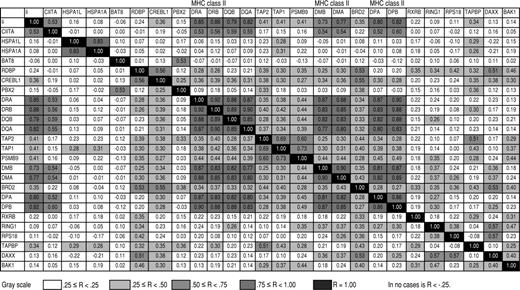
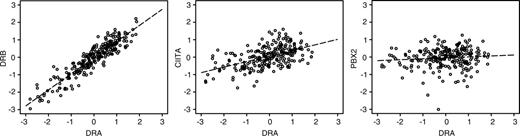
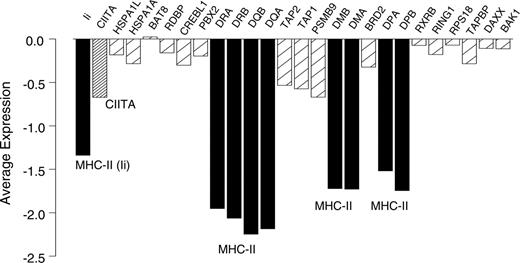
We next produced specific Pearson correlations between the MHC class II genes, their chromosomal neighbors, Ii, and CIITA (Figure 2). These analyses demonstrated high correlation coefficients within the classical and nonclassical MHC class II genes (range, 0.77 to 0.92) and between Ii (which is on a separate chromosome) and MHC class II genes (range, 0.73 to 0.88). However, correlations between MHC class II genes (including Ii) and genes adjacent to or within the MHC class II locus were significantly lower (range, -0.12 to 0.49; bootstrap P < .001). In addition, correlation between the transactivator gene CIITA and any of the MHC class II genes including Ii was significantly higher (range, 0.52 to 0.60) than between CIITA and the adjacent and intervening genes (range, -0.05 to 0.22; bootstrap P < .001). Figure 3 illustrates these different correlations, showing representative scatterplots within MHC class II genes (DRA vs DRB), between an MHC class II gene (DRA) and the transactivator gene CIITA, and between an MHC class II gene and an adjacent gene (DRA vs PBX2). Coordinate expression of MHC class II genes with themselves but not with neighboring or intervening genes, and the correlation between CIITA and MHC class II genes but not with adjacent or intervening genes, implies that loss of transcriptional control may be involved in the loss of MHC class II expression. Figure 4 shows average expression for cases in the lowest 10% of average MHC class II expression, again indicating low expression of the MHC class II genes, a corresponding low expression of CIITA, and higher expression of the adjacent and intervening genes.
Highly coordinated expression of MHC class II classical and nonclassical molecules, invariant chain, and the master transactivator CIITA in individual DLBCL patients including those with the overall 10% lowest expression (arrows). The 50 patients with the shortest survival times of the 240 in the LLMPP DLBCL data set are shown from left to right. The x-axis shows each individual patient by study ID number, whereas the y-axis shows relative expression of genes (log 2).
Highly coordinated expression of MHC class II classical and nonclassical molecules, invariant chain, and the master transactivator CIITA in individual DLBCL patients including those with the overall 10% lowest expression (arrows). The 50 patients with the shortest survival times of the 240 in the LLMPP DLBCL data set are shown from left to right. The x-axis shows each individual patient by study ID number, whereas the y-axis shows relative expression of genes (log 2).
Because large genetic deletions of the MHC class II loci are a frequent cause of decreased expression in IP-DLBCL, we wanted to evaluate this possibility in our cases. Chromosome CGH was performed on the lowest 10% and highest 10% of the MHC class II expressers in the DLBCL cases from the LLMPP study. Chromosomal gains or losses at the MHC class II-related loci were quantified and the results summarized in Table 2. Of the 23 lowest-expressing MHC class II cases that were assayed, CGH found alterations at 6p21 (which includes as a small part the MHC class II locus) in 4 cases (17%; 95% CI, 5%-37%), including 2 losses (9%; 95% CI, 1%-27%) and 2 gains (9%; 95% CI, 1%-27%). Neither of the 2 cases showing loss of chromosomal material at 6p21 was from an immune-privileged site. No losses (0%; 95% CI, 0%-14%) were detected near the Ii locus (5q32), CIITA locus (16p13), RFX5 locus (1q21), or RFXAP locus (13q14). Each of these loci showed 1 case with 1 amplification each, except the RFX5 locus, which showed 3 gains. These results indicate that large deletions identifiable by CGH were not a frequent cause of MHC class II loss in our group of 24 lowest expressers. We also explored whether chromosome alterations were more common in MHC class II- cases, which would imply that there was more frequent large-scale DNA damage. However, we found that the average number of chromosome alterations per patient was nearly identical between the lowest-10% and highest-10% MHC class II expressers (4.3 ± 3.7 vs 4.3 ± 3.9, respectively).
Comparative genomic hybridization results from the lowest 10% MHC class II–expressing cases, n = 23
Case no. . | MHCII, 6p21 . | Ii, 5q32 . | CIITA, 16p13 . | RFX5, 1q21 . | RFXAP, 13q14 . |
|---|---|---|---|---|---|
| 8 | No alt | No alt | No alt | No alt | No alt |
| 11 | ND | ND | ND | ND | ND |
| 12 | Loss | No alt | No alt | No alt | No alt |
| 15 | No alt | No alt | No alt | Gain | No alt |
| 19 | No alt | No alt | No alt | No alt | No alt |
| 58 | No alt | No alt | No alt | Gain | No alt |
| 64 | No alt | No alt | No alt | No alt | No alt |
| 65 | No alt | No alt | No alt | No alt | No alt |
| 80 | Loss | No alt | No alt | No alt | No alt |
| 101 | No alt | No alt | No alt | No alt | No alt |
| 119 | No alt | Gain | No alt | No alt | No alt |
| 122 | No alt | No alt | No alt | No alt | No alt |
| 132 | High gain | No alt | No alt | No alt | No alt |
| 139 | Gain | No alt | Gain | No alt | No alt |
| 142 | No alt | No alt | No alt | Gain | No alt |
| 152 | No alt | No alt | No alt | No alt | No alt |
| 166 | No alt | No alt | No alt | No alt | No alt |
| 179 | No alt | No alt | No alt | No alt | No alt |
| 203 | No alt | No alt | No alt | No alt | No alt |
| 272 | No alt | No alt | No alt | No alt | Gain |
| 277 | No alt | No alt | No alt | No alt | No alt |
| 284 | No alt | No alt | No alt | No alt | No alt |
| 285 | No alt | No alt | No alt | No alt | No alt |
| 322 | No alt | No alt | No alt | No alt | No alt |
| 423 | No alt | No alt | No alt | No alt | No alt |
Case no. . | MHCII, 6p21 . | Ii, 5q32 . | CIITA, 16p13 . | RFX5, 1q21 . | RFXAP, 13q14 . |
|---|---|---|---|---|---|
| 8 | No alt | No alt | No alt | No alt | No alt |
| 11 | ND | ND | ND | ND | ND |
| 12 | Loss | No alt | No alt | No alt | No alt |
| 15 | No alt | No alt | No alt | Gain | No alt |
| 19 | No alt | No alt | No alt | No alt | No alt |
| 58 | No alt | No alt | No alt | Gain | No alt |
| 64 | No alt | No alt | No alt | No alt | No alt |
| 65 | No alt | No alt | No alt | No alt | No alt |
| 80 | Loss | No alt | No alt | No alt | No alt |
| 101 | No alt | No alt | No alt | No alt | No alt |
| 119 | No alt | Gain | No alt | No alt | No alt |
| 122 | No alt | No alt | No alt | No alt | No alt |
| 132 | High gain | No alt | No alt | No alt | No alt |
| 139 | Gain | No alt | Gain | No alt | No alt |
| 142 | No alt | No alt | No alt | Gain | No alt |
| 152 | No alt | No alt | No alt | No alt | No alt |
| 166 | No alt | No alt | No alt | No alt | No alt |
| 179 | No alt | No alt | No alt | No alt | No alt |
| 203 | No alt | No alt | No alt | No alt | No alt |
| 272 | No alt | No alt | No alt | No alt | Gain |
| 277 | No alt | No alt | No alt | No alt | No alt |
| 284 | No alt | No alt | No alt | No alt | No alt |
| 285 | No alt | No alt | No alt | No alt | No alt |
| 322 | No alt | No alt | No alt | No alt | No alt |
| 423 | No alt | No alt | No alt | No alt | No alt |
No alt indicates no alteration at that locus; ND, not done.
Pearson correlation coefficients between average gene expression of MHC class II genes, adjacent and intervening genes, CIITA, and Ii. -.25 ≤ R < .25; .25 ≤ R < .50; .50 ≤ R < .75; .75 ≤ R < 1.00; R = 1.00. In no case is R < -.25.
Pearson correlation coefficients between average gene expression of MHC class II genes, adjacent and intervening genes, CIITA, and Ii. -.25 ≤ R < .25; .25 ≤ R < .50; .50 ≤ R < .75; .75 ≤ R < 1.00; R = 1.00. In no case is R < -.25.
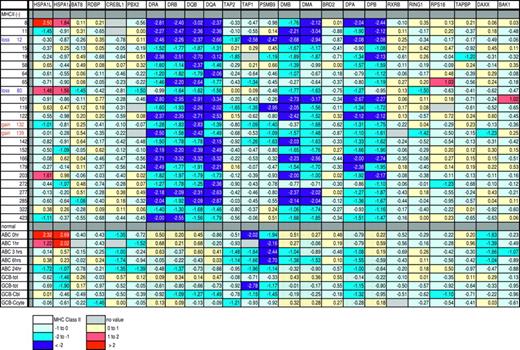
The resolution of the CGH technique used in our study was 10 Mb; however, previous reports have described MHC class II deletions of 100 to 900 kb (10- to 100-fold smaller) in IP-DLBCL.10 We therefore performed positional expression profiling, creating a map of color-coded gene expression of selected genes within approximately 500 kb of the MHC class II locus, for the 24 MHC class II- cases (Figure 5). Relative locations of these genes are detailed in Table 1. Our reasoning was that if we could identify genes within a few hundred kb of the MHC class II genes and these were expressed at normal or near-normal levels, then the large deletions described in IP-DLBCL could not exist in our cases. We could also see if the deletions found by CGH were localized to the MHC class II locus or might have occurred outside our area of interest. As can be seen, although the expression of MHC class II molecules is very low as expected in all of the MHC class II- cases, expression of the preceding, intervening, and following genes is not suppressed in 22 (92%) of the 24 cases. These results imply that the MHC class II genes are under selective transcriptional repression rather than a regional effect on gene expression (such as a large deletion or regional chromatin condensation), which would be expected to affect all adjacent genes.
In the remaining 2 cases (8%), there was very low expression of the intervening genes (case no. 12 and no. 119). In both cases no. 12 and no. 119, all 3 genes that were between HLA-DQA and HLA-DMB were expressed at or below 2-fold less than median with at least one below 4-fold less expression. One other case, no. 19, had low expression of 2 of the 3 genes between HLA-DQA and HLA-DMB, although there was no value for the third intervening gene, TAP1. Case no. 12 was one of the same cases in which CGH identified chromatin loss at 6p21, which includes the MHC class II locus. This case was particularly interesting, since it demonstrated that deletions occurring somewhere in a large region can be tentatively localized using CGH and gene expression analysis techniques. The other case showing loss by CGH, no. 80, showed no loss of expression of neighboring genes in our map, implying that the deletion at 6p21 occurred outside the MHC class II locus. It is possible that case no. 119 is an example of a smaller deletion not visible by CGH.
Representative scatterplots of intra-MHC correlation (DRA vs DRB, 0.92), correlation between an MHC gene and CIITA (DRA vs CIITA, 0.53), and correlation between an MHC gene and an adjacent gene (DRA vs PBX2, 0.09). A least-square regression line of the fit of the data is included, indicating the trend of the correlation.
Representative scatterplots of intra-MHC correlation (DRA vs DRB, 0.92), correlation between an MHC gene and CIITA (DRA vs CIITA, 0.53), and correlation between an MHC gene and an adjacent gene (DRA vs PBX2, 0.09). A least-square regression line of the fit of the data is included, indicating the trend of the correlation.
Discussion
Loss of the MHC class II molecules has emerged as one of the most important factors associated with a poor prognosis in DLBCL. Earlier single-institutional reports using protein immunohistochemistry had demonstrated this effect in a small number of patients and linked it to loss of tumor immunosurveillance.21-24 This information received little attention until recent large, multi-institutional, multinational gene expression profiling studies found that loss of MHC class II molecules in non-IP-DLBCL was a prominent predictor of patient outcome.5 We immediately pursued this lead, demonstrating that loss of MHC class II molecules was associated with poor patient outcome independent of international prognostic index (IPI) score, occurred in all histologic subtypes, and had a dosage effect on patient survival. We further linked loss of MHC class II molecules to decreased percentages of tumor-infiltrating lymphocytes, implicating diminished tumor immunosurveillance in the mechanism of action, as had previous studies.6 While documentation of this association is of scientific and biologic interest, medically, it is important to determine the reasons for MHC class II loss so that corrective therapeutic strategies can be explored.
Average gene expression of MHC class II and adjacent and intervening genes for cases in the lowest 10% of MHC class II expression. ▪ indicates MHC class II genes;  , adjacent and intervening genes; and
, adjacent and intervening genes; and  , CIITA.
, CIITA.
Average gene expression of MHC class II and adjacent and intervening genes for cases in the lowest 10% of MHC class II expression. ▪ indicates MHC class II genes;  , adjacent and intervening genes; and
, adjacent and intervening genes; and  , CIITA.
, CIITA.
Searching the literature, we found several elegant articles from a single laboratory which have explored the mechanism of lost MHC class II expression in a particular subset of DLBCLs, those arising in immune-privileged sites. This group has reported that there are large hemizygous or homozygous deletions involving the MHC class II locus, ranging in size from 100 kb to 900 kb. While these investigators used non-IP-DLBCL as controls, there were very few of these cases included in their studies since their primary focus was on IP-DLBCL. Our intent was to determine whether non-IP-DLBCL cases followed the same pattern of large genetic deletions as the underlying cause of lost MHC class II. Correction of expression in these types of cases would require gene therapy strategies to reinsert the correct DNA sequences. Thus, determining the differences in mechanism of lost MHC class II expression between IP and non-IP-DLBCL is a previously unrecognized distinction that may have significant clinical implications.
Our studies using the gene expression data from the LLMPP group demonstrated high correlation between the MHC class II genes (including Ii, which is under some of the same transcriptional control) and the transcriptional regulator CIITA, and low correlation between the adjacent and intervening genes and CIITA. In addition, CGH studies found that large genetic deletions were not frequent in our cases. We therefore hypothesized that transcriptional activity regulates MHC expression in our non-IP-DLBCL cases. The fact that the MHC class II genes were highly correlated with each other but not with the adjacent or intervening genes further supported this hypothesis.
Reasoning that large genetic deletions such as those described in IP-DLBCL would be nonselective and would necessarily involve adjacent genes as well as the MHC class II genes, we undertook an analysis called positional expression profiling.25 To perform this analysis, we used published information to determine genes that were in the region of the MHC class II locus within the distances involved in large deletions described in IP-DLBCL. Analysis of expression of these neighboring genes demonstrated that there was a specific pattern of repressed gene expression exclusively involving the MHC class II genes and none of the adjacent genes in most cases. This further confirmed our correlation analyses and CGH results.
Positional expression profiling map showing relative expression of MHC class II genes and those genes physically located telomeric, within, and centromeric to the MHC class II loci. Color coding is used to indicate the relative expression of genes. Each row indicates one of the MHC class II- cases by individual patient study number. The last 9 rows are activated B cells (ABC; with hours of activation indicated) and germinal center B cells (GCB). tot indicates total from a normal lymph node; Cbl, centroblastic cells from healthy individuals; and Ccyte, centrocytes from healthy individuals. MHC class II genes are indicated with dots. Gene expression (e) is colored as follows: dark blue, e < -2; medium blue, -2 ≤ e < -1; light blue, -1 ≤ e < 0; yellow, 0 ≤ e < 1; orange, 1 ≤ e < 2; red, e ≥ 2; and light gray, no value.
Positional expression profiling map showing relative expression of MHC class II genes and those genes physically located telomeric, within, and centromeric to the MHC class II loci. Color coding is used to indicate the relative expression of genes. Each row indicates one of the MHC class II- cases by individual patient study number. The last 9 rows are activated B cells (ABC; with hours of activation indicated) and germinal center B cells (GCB). tot indicates total from a normal lymph node; Cbl, centroblastic cells from healthy individuals; and Ccyte, centrocytes from healthy individuals. MHC class II genes are indicated with dots. Gene expression (e) is colored as follows: dark blue, e < -2; medium blue, -2 ≤ e < -1; light blue, -1 ≤ e < 0; yellow, 0 ≤ e < 1; orange, 1 ≤ e < 2; red, e ≥ 2; and light gray, no value.
In this study, we have excluded large genetic deletions as a frequent cause of lost MHC class II expression; however, other mechanisms for lost expression exist. These include small deletions or point mutations of CIITA or other elements of the transcriptosome; physiologic down-regulation of the CIITA via transcriptional control mechanisms involving BCL-6, BLIMP-1, or other upstream regulators; or epigenetic changes such as hypermethylation or hypoacetylation of the MHC class II or CIITA promoter regions. Since CIITA is substantially down-regulated in many of our samples, it would be inadvisable to try sequencing its cDNA, as residual CIITA mRNA could come from nontumor cells included in the sample. Looking for mutations in the coding sequence will therefore involve sequencing genomic DNA, which will be less susceptible to contamination from nontumor cells. Sequencing the complete CIITA gene is complicated due to the large size of the gene that includes 20 coding exons with over 3000 bases. We have initiated this work in a few cell lines and cases. However, the results are not yet completed or confirmed. The latter possibilities involving epigenetic changes are particularly interesting, since demethylating agents and histone deacetylase inhibitors are classes of drugs that have shown activity in up-regulating MHC class II expression in mouse B-cell lymphoma lines, human T-cell lymphoma lines, and primary patient tumor samples.26-28 In addition, CIITA is known to influence MHC class II expression via acetylation and is itself controlled via acetylation.29,30 Thus, chromatin accessibility may be a particularly relevant mechanism of transcriptional control in the MHC class II system, which presents possibilities for new therapeutic strategies in MHC class II- DLBCL. Although we demonstrated that the worst outcome was seen in DLBCL patients with complete loss of MHC class II expression, enhancing expression may be a rational therapeutic strategy for most DLBCL cases since in our previous analysis, the hazard ratio of death went steeply upward for all cases below the mean.6
Finally, our findings beg the intriguing question of why there are different mechanisms of MHC class II expression based on anatomic origin of the tumor (large genetic deletions in IP-DLBCL vs transcriptional disruption in non-IP tumors). At this time, we can only speculate on this issue. It is known that MHC class II expression, in particular, the expression of Ii, can be differently regulated in immune-privileged versus non-immune-privileged sites. Lack of invariant chain expression, which frequently occurs in immune-privileged sites, favors the presentation of endogenous as opposed to exogenous peptides that may alter details of the inflammatory response.31,32 Investigation of this matter will be the focus of future research that may have implications regarding different mechanisms of lymphomagenesis at different anatomic locations, a previously unexplored area of DLBCL biology.
Prepublished online as Blood First Edition Paper, October 20, 2005; DOI 10.1182/blood-2005-04-1510.
Supported by Spanish Ministry of Science CICYT SAF 02/3261, Instituto Nacional de Salud Carlos III Red Temática de Genómica de Cáncer 03/10, Estudio de Linfomas 03/179, and National Institutes of Health Grants U10-CA-84967 and U10-CA-32102.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal