Abstract
In chronic lymphocytic leukemia (CLL), chromosomes usually evade detailed cytogenetic analyses because cells poorly respond to the traditionally used set of mitogens. We applied novel technologies, such as stimulation of CLL cells either with CD40 ligand or with a combination of CpG-oligodeoxynucleotides and IL-2, to increase the freequncy of metaphase spreads for detailed chromosome analysis in 96 patients with CLL. This approach revealed that translocations occurred in 33 of 96 (34%) of our patients with CLL. The presence of translocations defined a new prognostic subgroup because these patients have significantly shorter median treatment-free survival (24 months vs 106 months; P < .001) and significantly inferior overall survival (OS; median, 94 months) than patients without translocations (346 months; P < .001). In multivariate analysis—including Binet stage, complex karyotype, CD38 expression, and 17p deletions—translocation proved to be the prognostic marker with the highest impact for an unfavorable clinical outcome (P < .001). In summary, we identified a new subgroup of patients with CLL defined by chromosomal trans-locations and poor prognosis. Our data may facilitate the identification of molecular events crucial for transforming activity in this disease and should have implications for risk-adapted clinical management of patients with CLL. (Blood. 2006;107:742-751)
Introduction
B-cell chronic lymphocytic leukemia (B-CLL) is the most common form of leukemia in adults and has a highly variable clinical course. At present, in addition to Binet stage, the mutational status of the VH genes, molecular markers such as ZAP-70 and CD38, and chromosomal aberrations 11q-, 13q-, 17p-, and +12 are used to predict survival.1-5
Cytogenetic analyses in CLL were performed in the 1980s evaluating metaphases that were obtained after stimulation with B-cell mitogens such as TPA.6-13 Although chromosomal aberrations were detected by this approach in 40% to 50% of patients, this technique and its prognostic implications were mostly replaced when interphase fluorescence in situ hybridization (FISH) became available. Nowadays, chromosome analysis is usually performed by interphase FISH using a probe set that, according to our current knowledge, covers regions often involved in numerical or structural rearrangements. Because of these technical limitations, it is more or less impossible to assess the entire chromosome complement and to identify new cytogenetic markers with prognostic value. In particular, new structural rearrangements, such as translocations or inversions, are not visible by FISH with a defined probe set, which underscores the necessity for novel cytogenetic approaches in this disease entity.
Here we used such novel technologies—stimulation of CLL cells with CD40 ligand (CD40L) or with a combination of CpG-oligodeoxynucleotides (CpG-ODN) and IL-2—for the preparation of metaphase spreads from CLL cells.14-16 Applying these approaches to CLL cells from 96 patients, we unexpectedly identified translocations in 34% of patients. We confirmed the presence of these translocations by interphase FISH on unstimulated cells and correlated the cytogenetic data with other prognostic markers and clinical parameters.
Patients and methods
Patients
Between January 1999 and May 2003, 109 consecutive patients with B-CLL (35 women, 74 men; Binet stage A, 31%; Binet stage B, 30%; Binet stage C, 39%; age range, 35-86 years; median age, 60 years) were enrolled in the study.17 After informed consent, peripheral blood was obtained for cytogenetic analysis, and patients were observed with regard to treatment-free survival (TFS) and overall survival (OS). Sixty percent of the patients were untreated before cytogenetic analysis, 29% of the patients received one chemotherapeutic regimen, and 11% received 2 or more treatments before cytogenetic assessment at least 3 months preceding the study. Median time from diagnosis to cytogenetic analysis was 36 months. In a pilot phase of our study, 7 of the first 13 samples for which we failed to induce enough metaphases had WBC counts of less than 20 × 109/L; therefore, for all other patients, WBC counts of at least 20 × 109/L were required. The diagnosis of CLL in the 2 patients with t(11;14) was confirmed by lymph node biopsy to exclude mantle cell lymphoma. The study was approved by the ethics committee of the Ludwig-Maximilians-University in Munich.
Metaphase cytogenetic analysis
Mononuclear cells were isolated on a Ficoll-Hypaque (Seromed, Berlin, Germany) density gradient. Stimulation of B-CLL cells coexpressing CD5 and CD19 was performed as previously described.14,18 Briefly, γ-irradiated CD40L-expressing HeLa cells were plated in 6-well plates and incubated overnight. B-CLL cells were added at 2.5 × 106 cells/mL in Iscove medium (Biochrome, Berlin, Germany) supplemented with 10% heat-inactivated fetal calf serum (FCS), 100 U/mL penicillin, and 100 μg/mL streptomycin (Sigma-Aldrich, Steinheim, Germany), incubated for 72 hours in the presence of IL-4 (2 ng/mL; Cellconcepts, Umkirch, Germany), and purified by Ficoll-Hypaque density gradient. In 14 patients another stimulation protocol was applied using CpG-ODN, as previously described.15,16 CLL cells were incubated for 72 hours in the presence of 1 μM CpG-ODN DSP30 (TCGTCGCTGTCTCCGCTTCTTCTTGCC) (TibMolBiol, Berlin, Germany) and 50 U/mL IL-2.15 After washing, Colcemid (Sigma-Aldrich) was added at 0.15 μg/mL, and chromosome preparation, staining, and classification were performed according to standard protocols, as previously described.14,19-21 Translocations were counted if the same aberration was detectable in at least 2 metaphases. To assess the number of aberrations in a given patient, a translocation, interstitial deletions, and inversions all counted as one aberration though 2 breakpoints were involved. A median of 20 metaphases per patient were analyzed, and a median of 10 metaphases showed the same aberration.
In patients determined by metaphase analysis to have complex aberrant karyotypes, additional 24-color FISH analyses (multiplex FISH [M-FISH]) with commercially available probes (Metasystems, Altlussheim, Germany) were performed.22 FISH analysis was performed using an axioscope (Zeiss, Jena, Germany) with filters for 4,6-diamidino-phenyl-indole (DAPI), fluorescein isothiocyanate (FITC), Texas red, cyanine 3 (Cy3), Cy5, and diethyl amino methyl coumarin (DEAC) (Feuerbacher, Tübingen, Germany) using ISIS software (Metasystems).
Interphase cytogenetic analysis
A comprehensive set of commercially available probes (Vysis, Downers Grove, IL) was used, as follows: del(11q22-q23), ATM; +12, centromeric probe for chromosome 12; del(13q14), D13S319, D13S25, and D13S272; del(17p13), p53 and the subtelomeric probes for chromosomes 5, 17, and 18 (Vysis).3 A minimum of 100 interphase nuclei were evaluated per probe for each patient.
Immunophenotyping
Monoclonal antibodies (mAbs) CD3, CD5, CD19 (Beckman Coulter, Krefeld, Germany), and CD38 (BD PharMingen, Heidelberg, Germany) conjugated with FITC, phycoerythrin (PE), or PE-Cy5 were used. We used a cut-off level of 30% to identify CD38+ samples. Fluorescence was measured with a Coulter Epics XL-MCL (Beckman Coulter, Kre-feld, Germany).
Statistical analysis
End points of the study were TFS and OS. TFS was defined as the interval from initial diagnosis to the start of first-line treatment in previously untreated patients. In patients with advanced disease, TFS referred to the end of one treatment to retreatment if cytogenetic analysis was performed in this time interval. TFS times were plotted using Kaplan-Meier estimates, and 95% confidence intervals (95% CIs) for hazard ratios were computed. The proportional-hazards regression model of Cox with a limited backward-selection procedure was used to identify differences with respect to TFS attributed to prognostic factors. The following variables were included: Binet stage, presence of a complex karyotype (3 or more aberrations per sample), CD38 expression, 17p deletions, and occurrence of translocations. Analysis was stratified according to the variable previous treatment because previous treatment had a significant influence on the length of TFS. OS was calculated from the initial diagnosis until death. OS times were plotted using Kaplan-Meier estimates, and 95% CIs for hazard ratios were computed. The proportional hazards regression model of Cox, with a limited backward-selection procedure, was used to identify differences with respect to OS attributed to prognostic factors. The 3 variables with the highest log rank value in the univariate analysis concerning OS (occurrence of translocations, deletion of 11q, complex karyotype) were included in the backward-selection procedure. Groupwise comparison of the distributions of clinical and laboratory variables at the time of genetic analysis were performed with 2-sided Kruskal-Wallis, Mann-Whitney U, or Fisher exact test. An effect was considered statistically significant if P was .05 or lower. Calculations were determined using SPSS version 13 (SPSS, Chicago, IL).
Results
Interphase cytogenetic analysis
Interphase FISH was performed in 93 patients; 81% of patients exhibited abnormalities. Sixty patients had 1 aberration, 14 patients had 2 aberrations, and 1 patient had 3 aberrations, with 11q-, trisomy 12, 13q-, and 17p-considered chromosomal aberrations. Eighteen patients had normal interphase patterns. Forty-four (47%) patients had the 13q deletion as a single aberration. Fifty-nine (63%) patients had 13q-, 14% had +12, 13% had 11q-, and 7.5% had 17p-aberrations.
Metaphase cytogenetic analysis
We obtained metaphases after CD40L stimulation in 96 of 109 patients with CLL. We analyzed them for the presence of structural rearrangements using banding analyses and M-FISH. In addition to the known aberrations found by interphase FISH, we observed balanced and unbalanced translocations at a high frequency in 33 of 96 (34%) patients. In fact, in these 33 patients, we discovered 62 translocations altogether, 29 of which were balanced and 33 unbalanced (Figure 1). Most of these translocations were not previously described in CLL and were not recurrently observed in our patient cohort.23 However, most (70%) breakpoints were recurring and were previously reported, though with different partner chromosomes.23 Thirty-four percent of the breakpoints were clustered in regions previously described as deleted in CLL, such as in 13q14, 11(q21q25), and 14q32, or in regions often involved in other types of lymphoma, such as 1(p32p36), 1(q21q25), 2(p11p13), 6(p11p12), 6(p21p25), and 18q21 (Table 1; Figure 1).
Patients with translocations according to their pretreatment status, Binet stage, CD38 expression, and chromosomal aberrations detected by interphase FISH and metaphase cytogenetics
Patient no. . | Treatment before analysis . | Binet stage . | CD38, % . | Interphase FISH del11q . | Interphase FISH + 12 . | Interphase FISH del13q . | Interphase FISH del17p . | Gains, losses . | Balanced translocations . | Unbalanced translocations . | No. metaphases analyzed . | No. aberrations . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Yes | C | ND | 0 | 0 | 0 | 0 | del(6)(q21)[2], | t(7;15)(q35;q12)[2], | der(1)t(1;6)(p33;?)[2], | 10 | 9 |
| −9[2],−17[2] | t(13;14)(q14;q32)[2] | det(3)t(3;9)(q21;?)[2], | ||||||||||
| der(4)t(1;4)(p33;p16)[2], | ||||||||||||
| der(12)t(12;17)(p11;q11)[2] | ||||||||||||
| 2 | No | B | 0 | 0 | 0 | 1 | 0 | − | t(2;16)(p11;p13)[2] | der(2)t(2;16)(p11;p13)[16] | 30 | 6 |
| der(4)t(2;4)(p21;q13)[16] | ||||||||||||
| der(7)t(4;7)(q13;q22)[16] | ||||||||||||
| der(13)t(7;13)(q22;q14)[16] | ||||||||||||
| der(16)t(2;16)(p11;p13)[16] | ||||||||||||
| 3 | No | B | 30.6 | 1 | 0 | 1 | 0 | del(13)(q12q14)[1] | — | der(1)t(1;6)(q25;p11)[4] | 5 | 7 |
| del(5)(q13)[4] | der(3)t(3;5)(q21;q13)[4] | |||||||||||
| del(6)(p11)[4] | der(6)t(1;6)(q25;p25)[4] | |||||||||||
| del(11)(q13)[4] | ||||||||||||
| 4 | No | B | 2.0 | 0 | 0 | 1 | 0 | del(13)(q14)[10] | — | der(11)t(4;11)(q11;p13)[2] | 18 | 5 |
| ins(11;13)(q12;q21q34)[2] | ||||||||||||
| der(8)(ins8;15)(p12;qq)[2] | ||||||||||||
| der(18)t(3;18)(?;q21)[3] | ||||||||||||
| 5 | Yes | C | 15.0 | 0 | 1 | 0 | 0 | +12[15] | t(4;7)(q23;q11.2)[2] | der(4)t(4;7)(q23;q11.2)[2] | 20 | 5 |
| der(6)t(6;7)(p21;q11.1)[2] | ||||||||||||
| der(7)t(6;7)(p21;q11.1)[2] | ||||||||||||
| 6 | Yes | C | 1.8 | 1 | 0 | 1 | 0 | del(11)(q13)[11] | t(1;15;19)(p36;q15;p11)[11] | der(3)(3;11)(p25;q22)[11] | 12 | 5 |
| del(13)(q12q14)[11] | t(12;14)(q13;q32)[11] | |||||||||||
| 7 | No | C | 30.4 | 1 | 0 | 1 | 0 | trp(8)(q23q24)[7] | t(3;6)(p21;p22)[4] | — | 20 | 5 |
| del(11)(q13q22)[11] | t(15;17)(q22;q25)[4] | |||||||||||
| del(13)(q12q22)[7] | ||||||||||||
| 8 | Yes | C | ND | 0 | 0 | 1 | 1 | del(5)(p)[10] | t(1;1)(p36;q10) [18] | der(17)(1;13;17) | 20 | 4 |
| t(10;18)(q21;q21)[18] | (q36;q14;q21)[18] | |||||||||||
| 9 | Yes | C | ND | 0 | 0 | 0 | 1 | −13[3]−16[3] | t(12;18)(q23;q21)[3] | der(17)t(13;17)(q11;p11)[3] | 22 | 4 |
| 10 | No | B | 7.0 | 0 | 0 | 1 | 0 | — | der(1)t(1;6)(q21;?)[2] | 10 | 3 | |
| der(6)(1;6)(q21;p11)[2] | ||||||||||||
| der(20)t(2;20)(p11;p13)[3] | ||||||||||||
| 11 | No | B | 7.0 | 0 | 0 | 1 | 0 | del(13)(q12q14)[1] | inv(4)(p16q12)[9] | — | 12 | 4 |
| del(11)(q13q21)[2] | ||||||||||||
| dup(8)(q11q23)[9] | ||||||||||||
| 12 | Yes | C | 47.0 | 0 | 0 | 0 | 0 | del(11)(q14)[10] | — | der(20)t(8;20)(q13;p13)[15] | 15 | 4 |
| dup(1)(p22p32)[5] | ||||||||||||
| del(13)(q13q22)[5] | ||||||||||||
| 13 | Yes | C | 49.6 | 0 | 1 | 0 | 0 | +12[30] | t(1;22)(p32;q11)[30] | — | 30 | 3 |
| t(2;18)(p12;q21)[30] | ||||||||||||
| 14 | Yes | B | 41.0 | 1 | 0 | 0 | 0 | del(11)q(13)[8]-Y[8] | — | der(4)(2;4)(p13;p15)[8] | 23 | 2 |
| del(13)(q12q21)[2] | ||||||||||||
| 15 | Yes | C | 3.7 | 0 | 0 | 1 | 0 | del(3)(q21)[2] | — | der(14)(3;14)(q21;q11)[2] | 11 | 3 |
| del(13)(q12q14)[2] | ||||||||||||
| 16 | Yes | C | 2.5 | 0 | 0 | 1 | 0 | del(11)(q22q23)[14] | t(1;7)(q21;p22)[3] | — | 20 | 3 |
| t(1;11)(q21;q24)[6] | ||||||||||||
| 17 | Yes | B | 65.0 | 0 | 0 | 1 | 0 | del(13)q(q12q14)[6] | t(4;20)(q32;p13)[6] | — | 12 | 3 |
| +12[5] | ||||||||||||
| 18 | Yes | B | 4.6 | 0 | 1 | 1 | 0 | del(13)(q12q14)[5] | t(6;14)(q25;q32)[5] | — | 5 | 3 |
| del(11)(q22q24)[9] | ||||||||||||
| 19 | Yes | C | 57.8 | 1 | 0 | 1 | 0 | del(13)(q12q14)[9] | t(6;22)(p21;q11)[9] | — | 25 | 3 |
| del(11)(q13q21)[5] | ||||||||||||
| 20 | No | C | 2.6 | 0 | 0 | 0 | 0 | i(1)(q10)[2] | t(11;16)(p15;p13.1)[13] | — | 21 | 3 |
| t(5;13)(q32;q14)[3] | ||||||||||||
| 21 | Yes | C | ND | 0 | 0 | 1 | 0 | — | t(8;13)(q12;q14)[12] | — | 15 | 2 |
| 22 | Yes | C | 25.0 | 0 | 0 | 0 | 1 | del(6)q12[13] | — | der(17;18)(q10;q10)[13] | 13 | 2 |
| 23 | Yes | C | 1.1 | 0 | 0 | 0 | 1 | del(10)q24)[10] | — | der(17)t(10;17)(q24;p12)[10] | 10 | 2 |
| 24 | Yes | C | 15.4 | 0 | 0 | 0 | 1 | del(14)(q22q32)[5] | — | der(14)t(14;17)(q10;q10)[3] | 18 | 2 |
| 25 | Yes | C | 3.0 | 0 | 0 | 0 | 1 | −17[18] | — | der(6)(6;17)(q11;q11)[18] | 20 | 2 |
| 26 | No | A | 16.1 | 0 | 0 | 1 | 0 | del(13)(q12q14)[3] | t(1;6)(p36;p12)[5] | — | 15 | 2 |
| 27 | Yes | C | 2.7 | 0 | 0 | 0 | 0 | +5[4] | t(8;10)(p23;q25)[4] | — | 11 | 2 |
| 28 | Yes | B | 2.7 | 0 | 0 | 1 | 0 | del(13)(q12q14)[3] | t(14;18)(q32;q21)[3] | — | 20 | 2 |
| 29 | No | A | 1.0 | 0 | 0 | 1 | 0 | del(13)(q11q14)[10] | t(14;18)(q32;q21)[10] | — | 10 | 2 |
| 30 | No | C | 0 | 0 | 0 | 0 | 0 | — | t(3;7)(p25;q22)[14] | — | 18 | 1 |
| 31 | Yes | B | 56.0 | 0 | 0 | 0 | 0 | — | t(5;8)(q11;q24)[6] | — | 15 | 1 |
| 32 | Yes | C | 1.0 | 0 | 0 | 0 | 0 | — | t(11;14)(q13;q32)[16] | — | 18 | 1 |
| der(11)t(11;14) | ||||||||||||
| 33 | Yes | C | 32.9 | 1 | 0 | 0 | 0 | — | — | (q13;q24)[20] | 20 | 1 |
Patient no. . | Treatment before analysis . | Binet stage . | CD38, % . | Interphase FISH del11q . | Interphase FISH + 12 . | Interphase FISH del13q . | Interphase FISH del17p . | Gains, losses . | Balanced translocations . | Unbalanced translocations . | No. metaphases analyzed . | No. aberrations . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Yes | C | ND | 0 | 0 | 0 | 0 | del(6)(q21)[2], | t(7;15)(q35;q12)[2], | der(1)t(1;6)(p33;?)[2], | 10 | 9 |
| −9[2],−17[2] | t(13;14)(q14;q32)[2] | det(3)t(3;9)(q21;?)[2], | ||||||||||
| der(4)t(1;4)(p33;p16)[2], | ||||||||||||
| der(12)t(12;17)(p11;q11)[2] | ||||||||||||
| 2 | No | B | 0 | 0 | 0 | 1 | 0 | − | t(2;16)(p11;p13)[2] | der(2)t(2;16)(p11;p13)[16] | 30 | 6 |
| der(4)t(2;4)(p21;q13)[16] | ||||||||||||
| der(7)t(4;7)(q13;q22)[16] | ||||||||||||
| der(13)t(7;13)(q22;q14)[16] | ||||||||||||
| der(16)t(2;16)(p11;p13)[16] | ||||||||||||
| 3 | No | B | 30.6 | 1 | 0 | 1 | 0 | del(13)(q12q14)[1] | — | der(1)t(1;6)(q25;p11)[4] | 5 | 7 |
| del(5)(q13)[4] | der(3)t(3;5)(q21;q13)[4] | |||||||||||
| del(6)(p11)[4] | der(6)t(1;6)(q25;p25)[4] | |||||||||||
| del(11)(q13)[4] | ||||||||||||
| 4 | No | B | 2.0 | 0 | 0 | 1 | 0 | del(13)(q14)[10] | — | der(11)t(4;11)(q11;p13)[2] | 18 | 5 |
| ins(11;13)(q12;q21q34)[2] | ||||||||||||
| der(8)(ins8;15)(p12;qq)[2] | ||||||||||||
| der(18)t(3;18)(?;q21)[3] | ||||||||||||
| 5 | Yes | C | 15.0 | 0 | 1 | 0 | 0 | +12[15] | t(4;7)(q23;q11.2)[2] | der(4)t(4;7)(q23;q11.2)[2] | 20 | 5 |
| der(6)t(6;7)(p21;q11.1)[2] | ||||||||||||
| der(7)t(6;7)(p21;q11.1)[2] | ||||||||||||
| 6 | Yes | C | 1.8 | 1 | 0 | 1 | 0 | del(11)(q13)[11] | t(1;15;19)(p36;q15;p11)[11] | der(3)(3;11)(p25;q22)[11] | 12 | 5 |
| del(13)(q12q14)[11] | t(12;14)(q13;q32)[11] | |||||||||||
| 7 | No | C | 30.4 | 1 | 0 | 1 | 0 | trp(8)(q23q24)[7] | t(3;6)(p21;p22)[4] | — | 20 | 5 |
| del(11)(q13q22)[11] | t(15;17)(q22;q25)[4] | |||||||||||
| del(13)(q12q22)[7] | ||||||||||||
| 8 | Yes | C | ND | 0 | 0 | 1 | 1 | del(5)(p)[10] | t(1;1)(p36;q10) [18] | der(17)(1;13;17) | 20 | 4 |
| t(10;18)(q21;q21)[18] | (q36;q14;q21)[18] | |||||||||||
| 9 | Yes | C | ND | 0 | 0 | 0 | 1 | −13[3]−16[3] | t(12;18)(q23;q21)[3] | der(17)t(13;17)(q11;p11)[3] | 22 | 4 |
| 10 | No | B | 7.0 | 0 | 0 | 1 | 0 | — | der(1)t(1;6)(q21;?)[2] | 10 | 3 | |
| der(6)(1;6)(q21;p11)[2] | ||||||||||||
| der(20)t(2;20)(p11;p13)[3] | ||||||||||||
| 11 | No | B | 7.0 | 0 | 0 | 1 | 0 | del(13)(q12q14)[1] | inv(4)(p16q12)[9] | — | 12 | 4 |
| del(11)(q13q21)[2] | ||||||||||||
| dup(8)(q11q23)[9] | ||||||||||||
| 12 | Yes | C | 47.0 | 0 | 0 | 0 | 0 | del(11)(q14)[10] | — | der(20)t(8;20)(q13;p13)[15] | 15 | 4 |
| dup(1)(p22p32)[5] | ||||||||||||
| del(13)(q13q22)[5] | ||||||||||||
| 13 | Yes | C | 49.6 | 0 | 1 | 0 | 0 | +12[30] | t(1;22)(p32;q11)[30] | — | 30 | 3 |
| t(2;18)(p12;q21)[30] | ||||||||||||
| 14 | Yes | B | 41.0 | 1 | 0 | 0 | 0 | del(11)q(13)[8]-Y[8] | — | der(4)(2;4)(p13;p15)[8] | 23 | 2 |
| del(13)(q12q21)[2] | ||||||||||||
| 15 | Yes | C | 3.7 | 0 | 0 | 1 | 0 | del(3)(q21)[2] | — | der(14)(3;14)(q21;q11)[2] | 11 | 3 |
| del(13)(q12q14)[2] | ||||||||||||
| 16 | Yes | C | 2.5 | 0 | 0 | 1 | 0 | del(11)(q22q23)[14] | t(1;7)(q21;p22)[3] | — | 20 | 3 |
| t(1;11)(q21;q24)[6] | ||||||||||||
| 17 | Yes | B | 65.0 | 0 | 0 | 1 | 0 | del(13)q(q12q14)[6] | t(4;20)(q32;p13)[6] | — | 12 | 3 |
| +12[5] | ||||||||||||
| 18 | Yes | B | 4.6 | 0 | 1 | 1 | 0 | del(13)(q12q14)[5] | t(6;14)(q25;q32)[5] | — | 5 | 3 |
| del(11)(q22q24)[9] | ||||||||||||
| 19 | Yes | C | 57.8 | 1 | 0 | 1 | 0 | del(13)(q12q14)[9] | t(6;22)(p21;q11)[9] | — | 25 | 3 |
| del(11)(q13q21)[5] | ||||||||||||
| 20 | No | C | 2.6 | 0 | 0 | 0 | 0 | i(1)(q10)[2] | t(11;16)(p15;p13.1)[13] | — | 21 | 3 |
| t(5;13)(q32;q14)[3] | ||||||||||||
| 21 | Yes | C | ND | 0 | 0 | 1 | 0 | — | t(8;13)(q12;q14)[12] | — | 15 | 2 |
| 22 | Yes | C | 25.0 | 0 | 0 | 0 | 1 | del(6)q12[13] | — | der(17;18)(q10;q10)[13] | 13 | 2 |
| 23 | Yes | C | 1.1 | 0 | 0 | 0 | 1 | del(10)q24)[10] | — | der(17)t(10;17)(q24;p12)[10] | 10 | 2 |
| 24 | Yes | C | 15.4 | 0 | 0 | 0 | 1 | del(14)(q22q32)[5] | — | der(14)t(14;17)(q10;q10)[3] | 18 | 2 |
| 25 | Yes | C | 3.0 | 0 | 0 | 0 | 1 | −17[18] | — | der(6)(6;17)(q11;q11)[18] | 20 | 2 |
| 26 | No | A | 16.1 | 0 | 0 | 1 | 0 | del(13)(q12q14)[3] | t(1;6)(p36;p12)[5] | — | 15 | 2 |
| 27 | Yes | C | 2.7 | 0 | 0 | 0 | 0 | +5[4] | t(8;10)(p23;q25)[4] | — | 11 | 2 |
| 28 | Yes | B | 2.7 | 0 | 0 | 1 | 0 | del(13)(q12q14)[3] | t(14;18)(q32;q21)[3] | — | 20 | 2 |
| 29 | No | A | 1.0 | 0 | 0 | 1 | 0 | del(13)(q11q14)[10] | t(14;18)(q32;q21)[10] | — | 10 | 2 |
| 30 | No | C | 0 | 0 | 0 | 0 | 0 | — | t(3;7)(p25;q22)[14] | — | 18 | 1 |
| 31 | Yes | B | 56.0 | 0 | 0 | 0 | 0 | — | t(5;8)(q11;q24)[6] | — | 15 | 1 |
| 32 | Yes | C | 1.0 | 0 | 0 | 0 | 0 | — | t(11;14)(q13;q32)[16] | — | 18 | 1 |
| der(11)t(11;14) | ||||||||||||
| 33 | Yes | C | 32.9 | 1 | 0 | 0 | 0 | — | — | (q13;q24)[20] | 20 | 1 |
n = 33 patients.
Interphase FISH: 0, aberration not detected; 1, aberration detected.
Metaphase analysis: Numbers in parentheses are those of metaphases of a given aberration.
ND indicates not detected;—, none.
Verification of the presence of translocations in unstimulated CLL cells
We addressed whether the translocations might have been caused by stimulation with CD40L. Based on our metaphase spread analyses, we selected clones derived from bacterial artificial chromosomes (BACs) that mapped into regions of imbalances and hybridized them to unstimulated, uncultured CLL cells of the same patient. These analyses were performed for 13 of the 33 translocation carriers.
In 5 of these 13 patients, a translocation involving band 13q14 was observed. In these patients, we identified a deletion of 13q14 in unstimulated interphase nuclei and CD40L metaphase spreads. Similarly, we detected a 17p13 loss in stimulated and unstimulated cells in 6 patients. In addition, analysis of metaphase spreads in our patients revealed that the 17p13 deletions were not interstitial deletions but were caused by unbalanced translocations. Most important, we confirmed in unstimulated cells that regions not commonly known to be involved in CLL were indeed lost. For example, in one patient we found, among other abnormalities, a deletion of the short arm of chromosome 5 in approximately 50% of all metaphases (Figure 2A). In 60% of unstimulated nuclei from this patient, only one signal was detectable with a 5p probe (Figure 2B). In another patient, der(17;18)(q10;q10) resulted in losses of 17p and 18p (Figure 2C). These losses were confirmed in unstimulated cells with probes for 17p13 and subtelomeric probes for 17p and 18p (Figure 2D-G). In summary, in all the analyzed patients, interphase FISH findings correlated well with the imbalances observed on metaphase spreads of the same patient after CD40L stimulation.
Distribution of breakpoints identified by metaphase analysis in 33 patients with CLL. In 33 of 96 evaluable patients, 62 translocations were detected. Three patients (patients 4, 6, 8) had translocations involving 3 breakpoints (Table 1). This resulted in 127 breakpoints, but in 6 of 127 breakpoints, the exact band was not assignable; therefore, 121 breakpoints are displayed. Each dot represents a single breakpoint.
Distribution of breakpoints identified by metaphase analysis in 33 patients with CLL. In 33 of 96 evaluable patients, 62 translocations were detected. Three patients (patients 4, 6, 8) had translocations involving 3 breakpoints (Table 1). This resulted in 127 breakpoints, but in 6 of 127 breakpoints, the exact band was not assignable; therefore, 121 breakpoints are displayed. Each dot represents a single breakpoint.
To further confirm that the chromosomal aberrations were not artificially caused by the CD40L protocol, we applied an additional stimulation protocol in 14 patients using CpG-ODN and IL-2, which is not known to induce double-strand breaks, and we again observed the same aberrations observed with the CD40L stimulation (data not shown).
Evolution of karyotypes over time
To address whether the observed aberrations were early events or were markers of progression or genetic instability, we studied 12 patients with CLL undergoing metaphase analyses at 2 different time points with a median interval of 29 months (range, 12-60 months). Interestingly, most (8 of 12) patients analyzed had a stable karyotype regardless of whether they underwent chemotherapy or had progressive disease. In 4 patients, karyotypic evolution was detectable over time. In one patient, the trisomy 12 subclone expanded during treatment with chlorambucil. Another patient with a deletion of 13q was found to have a biallelic deletion after 3 years of observation without treatment. One patient who already showed a translocation gained further translocations within 31 months of treatment with alemtuzumab and allogeneic bone marrow transplantation. Finally, a fourth patient with trisomy 12 was found to have additional translocations after 12 months of observation without treatment.
Correlations with clinical and laboratory parameters
Data regarding TFS were available for 92 patients. As can be seen in Table 2, we were able to confirm the prognostic significance regarding TFS of many previously known important factors, such as Binet stage, β2-microglobulin concentration, thymidine kinase (TK) levels, CD38 expression, and cytogenetic subgroups identified by interphase FISH. New classifiers such as complex karyo-type (3 or more aberrations) or occurrence of translocations had even higher influence on TFS in univariate analysis using the log rank test. Because of the significant influence of previous treatment on the TFS interval, we stratified our analyses with respect to the number of previous treatments, but the translocations still determined the TFS in each subgroup.
Univariate analysis of TFS
. | . | . | Patients with feature . | . | Patients without feature . | . | ||
|---|---|---|---|---|---|---|---|---|
. | Log rank . | P . | Median TFS (95% Cl) . | No. . | Median TFS (95% Cl) . | No. . | ||
| Binet stage | 18.05 | < .001 | — | — | — | — | ||
| A | — | — | 132 | 27 | — | — | ||
| B | — | — | 42 (21-63) | 27 | — | — | ||
| C | — | — | 36 (24-48) | 37 | — | — | ||
| β2-microglobulin level 3.5 mg/L or greater | 4.98 | .026 | 32 (15-49) | 26 | 58 (32-84) | 55 | ||
| TK level 10 IU/L or greater | 6.332 | .012 | 36 (17-55) | 39 | 77 (48-106) | 33 | ||
| Complex karyotype* | 25.72 | < .001 | 26 (15-37) | 24 | 106 (61-151) | 68 | ||
| CD38 | 17.31 | < .001 | 20 (7-33) | 13 | 88 (40-136) | 61 | ||
| 13q- single | 17.06 | < .001 | 106 (54-158) | 43 | 30 (18-42) | 46 | ||
| 11q- | 2.78 | .095 | 36 (0-80) | 12 | 58 (38-78) | 75 | ||
| +12 | 1.54 | .215 | 36 (23-49) | 11 | 48 (28-68) | 78 | ||
| 17p- | 7.35 | .007 | 28 (2-54) | 7 | 58 (34-82) | 82 | ||
| Balanced translocations | 19.59 | < .001 | 24 (18-30) | 21 | 77 (40-114) | 71 | ||
| Unbalanced translocations | 18.21 | < .001 | 26 (12-40) | 17 | 77 (39-115) | 75 | ||
| Translocations | 35.45 | < .001 | 24 (17-31) | 32 | 106 (74-138) | 60 | ||
| One translocation vs more than one translocation | 0.84 | .359 | 24 (14-34) | 18 | 24 (9-39) | 13 | ||
| Translocations stratified according to no. previous treatments | 17.95 | < .001 | — | — | — | — | ||
| Untreated patients | — | — | 26 (6-46) | 11 | 110 (NC) | 44 | ||
| Patients who had received one chemotherapeutic regimen | — | — | 24 (14-34) | 12 | 48 (22-74) | 13 | ||
| Patients who had received more than one chemotherapeutic regimen | — | — | 13 (0-41) | 8 | 228 (NC) | 2 | ||
. | . | . | Patients with feature . | . | Patients without feature . | . | ||
|---|---|---|---|---|---|---|---|---|
. | Log rank . | P . | Median TFS (95% Cl) . | No. . | Median TFS (95% Cl) . | No. . | ||
| Binet stage | 18.05 | < .001 | — | — | — | — | ||
| A | — | — | 132 | 27 | — | — | ||
| B | — | — | 42 (21-63) | 27 | — | — | ||
| C | — | — | 36 (24-48) | 37 | — | — | ||
| β2-microglobulin level 3.5 mg/L or greater | 4.98 | .026 | 32 (15-49) | 26 | 58 (32-84) | 55 | ||
| TK level 10 IU/L or greater | 6.332 | .012 | 36 (17-55) | 39 | 77 (48-106) | 33 | ||
| Complex karyotype* | 25.72 | < .001 | 26 (15-37) | 24 | 106 (61-151) | 68 | ||
| CD38 | 17.31 | < .001 | 20 (7-33) | 13 | 88 (40-136) | 61 | ||
| 13q- single | 17.06 | < .001 | 106 (54-158) | 43 | 30 (18-42) | 46 | ||
| 11q- | 2.78 | .095 | 36 (0-80) | 12 | 58 (38-78) | 75 | ||
| +12 | 1.54 | .215 | 36 (23-49) | 11 | 48 (28-68) | 78 | ||
| 17p- | 7.35 | .007 | 28 (2-54) | 7 | 58 (34-82) | 82 | ||
| Balanced translocations | 19.59 | < .001 | 24 (18-30) | 21 | 77 (40-114) | 71 | ||
| Unbalanced translocations | 18.21 | < .001 | 26 (12-40) | 17 | 77 (39-115) | 75 | ||
| Translocations | 35.45 | < .001 | 24 (17-31) | 32 | 106 (74-138) | 60 | ||
| One translocation vs more than one translocation | 0.84 | .359 | 24 (14-34) | 18 | 24 (9-39) | 13 | ||
| Translocations stratified according to no. previous treatments | 17.95 | < .001 | — | — | — | — | ||
| Untreated patients | — | — | 26 (6-46) | 11 | 110 (NC) | 44 | ||
| Patients who had received one chemotherapeutic regimen | — | — | 24 (14-34) | 12 | 48 (22-74) | 13 | ||
| Patients who had received more than one chemotherapeutic regimen | — | — | 13 (0-41) | 8 | 228 (NC) | 2 | ||
To convert β2-microglobulin from milligrams per liter to nanomoles per liter, multiply milligrams per liter by 85.
TK indicates thymidine kinase; —, none; NC, not calculated.
Three or more chromosomal aberrations per sample.
In multivariate analysis using the Cox regression model, the following variables were included: Binet stage, presence of complex karyotype, CD38 expression, 17p deletions, and occurrence of translocations. Analysis was stratified according to previous treatment. The presence of translocations in CLL patients proved to be an independent factor and had the highest influence on TFS in our cohort (P < .001) (Table 3).
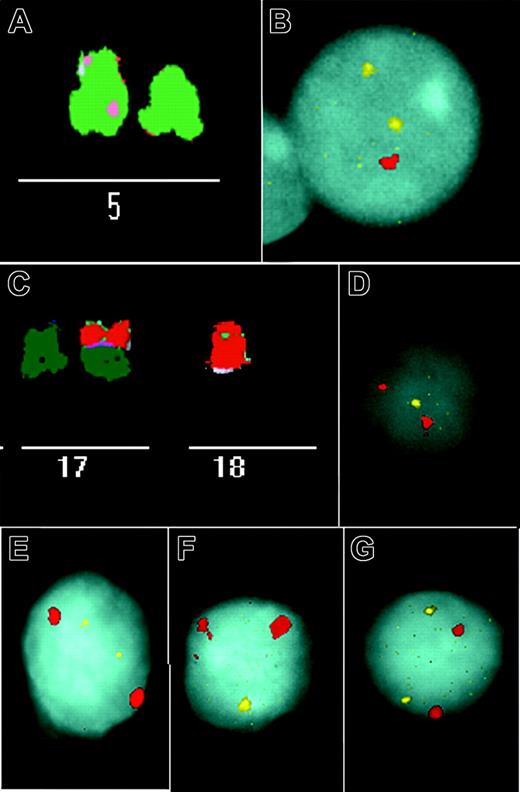
Examples of imbalances identified in stimulated and unstimulated CLL cells. (A) Partial M-FISH metaphase spread showing a deletion of 5p in patient 8 (Table 1). (B) An unstimulated interphase nucleus of this patient shows one signal for 5p (red) but 2 signals for 5q (yellow). (C) M-FISH identified der(17;18)(q10;q10) in patient 22 (Table 1). This rearrangement resulted in losses of 17p and 18p. (D-E) Interphase FISH was performed with 3 chromosome 17 and 18 probe sets. There was only one signal for a 17p subtelomere probe (yellow) (D), but there were 2 for 17q (yellow) (E), and in each hybridization there were 2 signals for a chromosome 17 centromere probe (red). (F-G) Similarly, we observed only one 18p subtelomere signal (yellow) (F), 2 18q signals (yellow) (G), and in each case 2 signals for 18 centromere (red). Images were obtained using a Leica DMRXA microscope (Leica, Wetzlar, Germany) with a 63 × 1.32 numeric aperture Plan Apo objective. Imaging medium was P-phenylenediamine dihydrochloride antifade solution. Images were captured using a Sensys CCD camera (Photometrics, Ottobrunn, Germany) and a Kodak KAF 1400 chip (Eastman Kodak, Rochester, NY), and were acquired using Leica Q-FISH and processed using Leica MCK software.
Examples of imbalances identified in stimulated and unstimulated CLL cells. (A) Partial M-FISH metaphase spread showing a deletion of 5p in patient 8 (Table 1). (B) An unstimulated interphase nucleus of this patient shows one signal for 5p (red) but 2 signals for 5q (yellow). (C) M-FISH identified der(17;18)(q10;q10) in patient 22 (Table 1). This rearrangement resulted in losses of 17p and 18p. (D-E) Interphase FISH was performed with 3 chromosome 17 and 18 probe sets. There was only one signal for a 17p subtelomere probe (yellow) (D), but there were 2 for 17q (yellow) (E), and in each hybridization there were 2 signals for a chromosome 17 centromere probe (red). (F-G) Similarly, we observed only one 18p subtelomere signal (yellow) (F), 2 18q signals (yellow) (G), and in each case 2 signals for 18 centromere (red). Images were obtained using a Leica DMRXA microscope (Leica, Wetzlar, Germany) with a 63 × 1.32 numeric aperture Plan Apo objective. Imaging medium was P-phenylenediamine dihydrochloride antifade solution. Images were captured using a Sensys CCD camera (Photometrics, Ottobrunn, Germany) and a Kodak KAF 1400 chip (Eastman Kodak, Rochester, NY), and were acquired using Leica Q-FISH and processed using Leica MCK software.
Multivariate analysis of TFS, stratified according to previous treatment before cytogenetic analysis
Variable . | Hazard ratio . | 95% Cl . | P . |
|---|---|---|---|
| Translocations | 5.78 | 2.26-14.71 | < .001 |
Variable . | Hazard ratio . | 95% Cl . | P . |
|---|---|---|---|
| Translocations | 5.78 | 2.26-14.71 | < .001 |
Cox regression; final model. Variables included Binet stage, presence of complex karyotype, CD38 expression, 17p deletions, and occurrence of translocations.
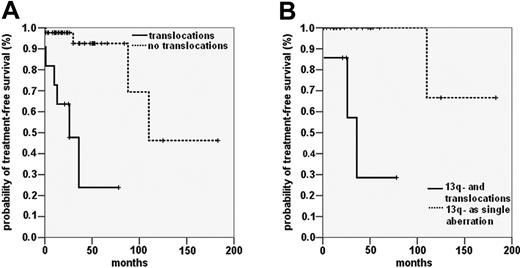
Patients with unbalanced translocations and patients with balanced translocations showed significantly shorter time to treatment than patients without unbalanced or balanced translocations (unbalanced translocations: median TFS, 26 months [95% CI, 12-40 months] vs 77 months [39-115 months]; log rank, 18.21; P < .001; balanced translocations: median TFS, 24 months [18-30 months] vs 77 months [40-114] months; log rank, 19.59; P < .001). There was no significant difference between the TFS rates of patients with unbalanced translocations and those with balanced translocations (log rank, 0.30; P = .860). Comparison of the combined group of translocation carriers with the group without translocations revealed median TFS times of 24 months (95% CI, 17-31 months) and 106 months (95% CI, 74-138 months; log rank, 35.45; P < .001) (Figure 3A).
We also analyzed whether other chromosomal aberrations with an established influence on prognosis affected the reduced TFS of CLL patients with translocations. Deletions of 13q as single aberrations in interphase FISH have been associated with favorable prognosis. In our cohort, 11 of 43 patients with the 13q deletion, detected as a single aberration in interphase FISH, had additional translocations when metaphase spreads were analyzed. These patients had significantly shorter TFS times than patients with the 13q deletion as a sole aberration (median TFS, 41 months vs 132 months; log rank, 14.37; P < .001) (Figure 3B).
Deletions of 11q and 17p have been associated with poor prognosis. In our study, patients with 11q- and translocations (n = 6) had significantly shorter TFS (median, 13 months) than patients with 11q- without translocations (n = 6; median TFS, 48 months; log rank, 5.05; P = .025). Loss of 17p is known to have a negative prognostic value. All patients with 17p- had translocations. The TFS for patients with translocations but without 17p- was similar to that for patients with 17p- (median, 24 months vs 18 months; P = .567).
We investigated the impact of CD38 expression and the presence of translocations on prognosis. Using a cut-off value of 30% positive cells, 13 of 82 (16%) patients were positive for CD38 expression. Median TFS was 20 months for patients with high CD38 expression levels compared with median TFS of 88 months (log rank, 17.31; P < .001) for patients with low CD38 expression levels. Nine of 13 patients with high CD38 values also had translocations. Nevertheless, when we compared the TFS of translocation carriers with low CD38 and those with high CD38, the TFS was short for both groups without a statistically significant difference (median, 26 months vs 14 months; P = .198).
Clinical and laboratory data at the time of cytogenetic analysis for the study population are given in Table 4. Patients with unbalanced or balanced translocations (34% of all patients) are shown as one group because there was no significant difference in any of the parameters examined. There was no difference in the time from diagnosis to the cytogenetic study between patients with or without translocations (P = .528). Previous treatment with chlorambucil had no influence on the incidence of translocations (P = .355). Patients with translocations were older (P = .021), had more advanced disease (higher Binet [P < .001] or Rai stage [P = .001]), higher β2-microglobulin levels (P < .001), higher thymidine kinase levels (P = .004), lower hemoglobulin levels (P = .024), and significantly higher CD38 expression levels (P = .003).
Comparison of clinical and laboratory data among patients with and without translocations at cytogenetic analysis
Parameter . | With translocations . | Without translocations . | P . |
|---|---|---|---|
| Median age, y (range) | 63 (39-86) | 59 (35-83) | .020* |
| No. male/no. total (% male) | 24/33 (73) | 39/63 (62) | .371† |
| Median time from diagnosis to investigation, mo (range) | 53 (0-228) | 35 (0-324) | .491‡ |
| Disease stage at enrollment, no. patients/no. total (% of patients) | |||
| Binet A | 2/33 (6) | 26/60 (43) | < .001† |
| Binet B | 10/33 (30) | 18/60 (30) | |
| Binet C | 21/33 (64) | 16/60 (27) | |
| Rai 0 | 1/33 (3) | 8/59 (14) | .002† |
| Rai 1 | 2/33 (6) | 13/59 (22) | |
| Rai 2 | 8/33 (24) | 21/59 (35) | |
| Rai 3 | 7/33 (21) | 0/59 (0) | |
| Rai 4 | 15/33 (46) | 17/59 (29) | |
| Median WBC count, × 109/L (range) | 43.4 (5.2-210.0) | 34.7 (5.6-254.0) | .851* |
| Median hemoglobin level, g/dL (range) | 11.5 (8.2-17.0) | 13.5 (7.4-17.2) | .003* |
| Median platelet count, × 109/L (range) | 131 (9-238) | 148 (7-311) | .079* |
| Median lactate dehydrogenase level, IU/L (range) | 238 (126-622) | 192 (128-581) | .112* |
| Median albumin level, g/dL (range) | 3.95 (3.3-5.4) | 4.50 (4.0-5.5) | .061* |
| Median β2-microglobulin level, mg/L (range) | 4.27 (1.4-14.3) | 2.47 (1.1-7.4) | < .001* |
| Median thymidine kinase level, IU/L (range) | 19.2 (4.9-58.5) | 8.8 (2.6-45.0) | .004* |
| High CD38 expression, no. patients/no. total (% of patients) | 9/15 (60) | 4/52 (8) | .003† |
| Lymphocyte doubling time below 12 months, no. patients/no. total (% of patients) | 13/28 (46) | 15/56 (27) | .089† |
| Splenomegaly, no. patients/no. total (% of patients) | 20/26 (77) | 31/55 (56) | .089† |
| Peripheral lymphadenopathy, no. patients/no. total (% of patients) | 20/25 (80) | 42/56 (75) | .779† |
| Median time from diagnosis to first treatment, mo (range) | 24 (17-31) | 110 (74-138) | < .001‡ |
| Previous treatment with chlorambucil, no. patients/no. total (% of patients) | 15/21 (71) | 16/17 (94) | .104† |
Parameter . | With translocations . | Without translocations . | P . |
|---|---|---|---|
| Median age, y (range) | 63 (39-86) | 59 (35-83) | .020* |
| No. male/no. total (% male) | 24/33 (73) | 39/63 (62) | .371† |
| Median time from diagnosis to investigation, mo (range) | 53 (0-228) | 35 (0-324) | .491‡ |
| Disease stage at enrollment, no. patients/no. total (% of patients) | |||
| Binet A | 2/33 (6) | 26/60 (43) | < .001† |
| Binet B | 10/33 (30) | 18/60 (30) | |
| Binet C | 21/33 (64) | 16/60 (27) | |
| Rai 0 | 1/33 (3) | 8/59 (14) | .002† |
| Rai 1 | 2/33 (6) | 13/59 (22) | |
| Rai 2 | 8/33 (24) | 21/59 (35) | |
| Rai 3 | 7/33 (21) | 0/59 (0) | |
| Rai 4 | 15/33 (46) | 17/59 (29) | |
| Median WBC count, × 109/L (range) | 43.4 (5.2-210.0) | 34.7 (5.6-254.0) | .851* |
| Median hemoglobin level, g/dL (range) | 11.5 (8.2-17.0) | 13.5 (7.4-17.2) | .003* |
| Median platelet count, × 109/L (range) | 131 (9-238) | 148 (7-311) | .079* |
| Median lactate dehydrogenase level, IU/L (range) | 238 (126-622) | 192 (128-581) | .112* |
| Median albumin level, g/dL (range) | 3.95 (3.3-5.4) | 4.50 (4.0-5.5) | .061* |
| Median β2-microglobulin level, mg/L (range) | 4.27 (1.4-14.3) | 2.47 (1.1-7.4) | < .001* |
| Median thymidine kinase level, IU/L (range) | 19.2 (4.9-58.5) | 8.8 (2.6-45.0) | .004* |
| High CD38 expression, no. patients/no. total (% of patients) | 9/15 (60) | 4/52 (8) | .003† |
| Lymphocyte doubling time below 12 months, no. patients/no. total (% of patients) | 13/28 (46) | 15/56 (27) | .089† |
| Splenomegaly, no. patients/no. total (% of patients) | 20/26 (77) | 31/55 (56) | .089† |
| Peripheral lymphadenopathy, no. patients/no. total (% of patients) | 20/25 (80) | 42/56 (75) | .779† |
| Median time from diagnosis to first treatment, mo (range) | 24 (17-31) | 110 (74-138) | < .001‡ |
| Previous treatment with chlorambucil, no. patients/no. total (% of patients) | 15/21 (71) | 16/17 (94) | .104† |
P values for Binet A-C and Rai 0-4 are calculated collectively.
To convert hemoglobin from grams per deciliter to grams per liter, multiply grams per deciliter by 10.
To convert albumin from grams per deciliter to grams per liter, multiply grams per deciliter by 10.
To convert β2-microglobulin from milligrams per liter to nanomoles per liter, multiply milligrams per liter by 85.
P values according to Mann-Whitney U test for continuous variables.
P values according to ϰ2 for categorical variables.
P values according to log rank test.
Chromosomal aberrations in previously untreated patients
Fifty-five patients were untreated at the time of cytogenetic analysis. In 11 (20%) of these patients, translocations were detected (balanced translocations, 6 patients; unbalanced translocations, 5 patients). Seven of these 11 patients had complex translocations (ie, more than 2 chromosomes were involved). As these data indicate that the occurrence of translocations is not treatment related, we reanalyzed TFS for the subgroup of untreated patients. Untreated patients with translocations showed significantly lower TFS than did patients without translocations (n = 44); median TFS was 26 months compared with 110 months (log rank, 15.62; P < .001) (Figure 4A). Again, patients with 11q deletion and translocations (n = 2) had significantly shorter TFS than did patients with 11q- without translocations (n = 4; log rank, 4.90; P = .027). Of particular interest were untreated patients with the 13q deletion as a single aberration in FISH. Usually such patients are expected to have TFS of 10 years or longer. However, patients with 13q-deletion who had additional translocations (n = 7) had significantly shorter TFS than patients with a single 13q deletion (n = 23; median TFS, 36 months vs not yet reached; log rank, 11.71; P = .001) (Figure 4B).
Overall survival
Overall survival data were available in 85 patients. After a median follow-up of 75 months, 23 of 85 evaluated patients died. Median survival time of the entire group was 201 months (95% CI, 61-341 months). Patients with translocations had significantly inferior overall survival (median OS, 94 months [95% CI, 53-135 months]) compared with patients without translocations (346 months [95% CI, not calculated (NC)]); log rank, 18.23; P < .001) (Figure 3C). In addition, the prognostic factors defined by interphase FISH had a significant influence on OS. Patients with or without deletions of 11q (median OS, 86 months [95% CI, 82-90] vs 234 months [95% CI, 172-296 months]; log rank, 11.94; P = .001), patients with or without deletions of 13q as a single aberration (median OS, 346 months [95% CI, NC] vs 113 months [95% CI, 82-144 months]; log rank, 10.87; P = .001), and patients with or without deletions of 17p (median OS, 84 months [95% CI, 67-101 months] vs 201 months [95% CI, 113-289 months]; log rank, 8.89; P = .003) could be distinguished. Additionally, the presence of a complex karyo-type had a significant influence on the OS (median OS, 107 months [95% CI, 57-157 months] vs 346 months [95% CI, NC]; log rank, 15.44; P < .001). In multivariate analysis including complex karyotype, deletion of 11q, and translocations, the occurrence of translocations (P = .002; hazard ratio, 5.13; 95% CI, 1.81-14.5) and deletion of 11q (P = .048; hazard ratio, 2.54 [95% CI, 1.01-6.41]) had the strongest influence on OS.
Discussion
We present a new strategy for the cytogenetic analysis of CLL cells that outperforms previous methods because it allows a more detailed analysis of the entire chromosomal complement, which was thought for a long time to be inaccessible for analysis. More than 20 years ago, several studies used B-cell mitogens for the analysis of metaphase spreads in CLL. However, these time-consuming metaphase analyses were completely replaced by interphase FISH and fell into oblivion.6-13 After stimulation with CD40L or CpG-ODN, we are now able to detect chromosomal aberrations in approximately 90% of patients with CLL. In fact, aberrations can be identified twice as frequently as they are by chromosome banding after stimulation with classic B-cell mitogens and at even higher frequency (82%) compared with the routinely used interphase FISH.3,6-13,24
Most important, we observed translocations in approximately one third of studied patients, though those aberrations were considered rare events in CLL.25,26 Half the translocations appeared to be balanced in the banding analysis; however, the presence of small deletions or duplications at the breakpoints cannot be ruled out with certainty. Three lines of evidence indicate that these translocations are not an artifact caused by our stimulation technique. First, it has been reported that 14-day stimulation of CLL cells with CD40L and IL-4 induces activation-induced cytidine deaminase (AID) and thus may cause double-strand breaks.27,28 However, in our cell culture system, CLL cells were grown and stimulated only for 3 days. Second, we found in each tested patient the same translocations when we used another stimulation protocol in parallel with CpG-oligodeoxynucleotides, which activates the NFκB signaling pathway but does not induce AID.15,16,29 Third, and most important, interphase FISH on unstimulated cells identified the same imbalances observed on metaphase spreads of the same patient after CD40L stimulation. This demonstrates unequivocally that the observed translocations are represented in in vivo conditions and are not caused by our preparation method.
We did not find a CLL-specific translocation but found instead many different translocations, confirming observations made more than a decade ago.6-13 However, recurring breakpoints cluster in regions that are also affected in other types of lymphomas, such as 1(p32p36), 1(q21q25), 2(p11p13), 6(p11p12), 6(p21p25), 13q14, 14q32, and 18q21, which definitely could harbor genes crucially involved in tumorigenesis or disease progression.23 High-resolution breakpoint mapping should facilitate the identification of those genes in the future.
We also tried to establish whether the presence of translocations has any impact on prognosis. At present, established prognostic factors in CLL include mutational status of the IgVH genes, molecular markers such as ZAP-70 and CD38, and chromosomal losses of 13q, 11q, 17p, and trisomy 12.1-3,30,31 Because of the usually indolent and long-lasting clinical course of CLL, data for OS are in general only available after very long observation times. Therefore, we and others32 have focused on TFS as an additional parameter to evaluate prognosis in CLL patients, but we also evaluated OS. Regarding OS, the presence of translocations was an independent prognostic factor that had the highest influence on OS in our cohort. For patients with advanced disease, TFS times in general decrease with every additional therapy. Therefore, we split our patient cohort into 3 groups—untreated patients, patients treated with one chemotherapeutic regimen, and patients treated with more than one chemotherapeutic protocol. In all groups, and when all patients were taken together, the occurrence of translocations critically determined the prognosis.
TFS and OS in the entire study cohort. Probability of disease progression, as indicated by the TFS interval, including all patients with available clinical data. (A) TFS of patients with translocations (n = 32) (solid line) and without translocations (n = 60) (dashed line) detected by metaphase analysis (P < .001). (B) TFS in patients with 13q deletion as the sole abnormality observed by FISH with (n = 11) (solid line) and without (n = 32) (dashed line) additional translocations (P < .001). (C) Probability of death, as indicated by OS including all patients with (n = 31) (solid line) and without (n = 54) (dashed line) translocations (P < .001).
TFS and OS in the entire study cohort. Probability of disease progression, as indicated by the TFS interval, including all patients with available clinical data. (A) TFS of patients with translocations (n = 32) (solid line) and without translocations (n = 60) (dashed line) detected by metaphase analysis (P < .001). (B) TFS in patients with 13q deletion as the sole abnormality observed by FISH with (n = 11) (solid line) and without (n = 32) (dashed line) additional translocations (P < .001). (C) Probability of death, as indicated by OS including all patients with (n = 31) (solid line) and without (n = 54) (dashed line) translocations (P < .001).
Using multivariate regression analysis, including factors such as Binet stage, complex karyotype, CD38 expression, 17p deletions, and translocations, we showed that the occurrence of translocations was an independent prognostic factor with the highest impact on TFS.
As was recently shown for patients with acute myeloid leukemia (AML), we were similarly able to demonstrate that patients with a complex karyotype in CLL (3 or more aberrations per patient) had inferior clinical outcomes compared with patients with fewer aberrations.33,34 The complex rearrangements may reflect some underlying genetic instability in this patient group. Therefore, it may be speculated that a putative genetic instability or the occurrence of translocations determines the clinical outcome. However, 92% of our patients with complex karyotypes had unbalanced or balanced translocations, and we achieved better distinction between prognostic subgroups using aberration type as a parameter for separation. The importance of translocations for determining prognosis is best reflected by the fact that patients with more than one translocation had the same TFS rates as patients with only one translocation.
Interestingly, all patients with 17p deletions or with high CD38 expression constituted subgroups within the group of patients with translocations. When patients with 17p- or high CD38 expression were omitted from the group of translocations, however, the remaining patients with translocations still had comparably short TFS times, proving that the negative impact on TFS is not caused by the known adverse factors of 17p deletion or high CD38 expression. Furthermore, it has been found that CLL patients with a translocation involving 14q32 have poorer prognosis.7,35,36 This subgroup had the same poor prognosis as other patients with translocations in our analysis.
TFS in previously untreated patients. Probability of disease progression, as indicated by TFS. (A) TFS of patients with translocations (n = 11) and without translocations (n = 44) detected by metaphase analysis (P < .001). (B) TFS in patients with 13q deletion as the sole abnormality observed by FISH with (n = 7) and without (n = 23) additional translocations (P = .001).
TFS in previously untreated patients. Probability of disease progression, as indicated by TFS. (A) TFS of patients with translocations (n = 11) and without translocations (n = 44) detected by metaphase analysis (P < .001). (B) TFS in patients with 13q deletion as the sole abnormality observed by FISH with (n = 7) and without (n = 23) additional translocations (P = .001).
One important benefit of our metaphase cytogenetic analysis is that the prognostic subgroups defined by interphase FISH can be further subdivided if translocation status is included. For example, 13q deletion is associated with good prognosis. However, in 26% of patients with the 13q deletion as a single aberration in the interphase FISH screen, we also identified translocations, and this changed the prognosis significantly. In addition, the group of patients with an 11q deletion can be further subdivided into patients with and patients without translocations and significantly different TFS rates. Our observations merit further analysis and confirmation in a large prospective clinical trial in early-stage CLL, as planned by the German CLL Study Group.
Several arguments suggest that the observed translocations are not treatment-associated secondary events. First, we and others8 observed simple and even complex translocations in untreated patients and at the time of initial diagnosis. Second, it was previously shown that the karyotypes of most (73%) CLL patients remained stable during the course of the disease, regardless of progression or even of chemotherapeutic treatment.8 We can confirm this by our own observations. Karyotypes in 8 of 12 patients for whom serial cytogenetic analyses were performed remained stable. Third, not all patients who were previously treated had translocations. Thus, observed translocations in our study seem not to have been treatment-associated secondary events.
The occurrence of translocations is associated with higher Binet stages. The distribution of Binet stages in our cohort was slightly shifted toward more advanced stages. Therefore, the proportion of patients with translocations might have been overestimated in our study. However, in our group of untreated patients, the distribution of Binet stages (Binet stage A, 52%; Binet stage B, 33%; Binet stage C, 15%) was similar to that seen in the cohort described by Döhner et al.3 Although half of the latter cohort presented with early-stage disease, the proportion of patients with translocations reached 20%. Finally, it seems that translocations indicate an advanced disease stage, but a delayed time point of study can be ruled out as bias in our study because the time from diagnosis to cytogenetic study was comparable in both groups.
Our data suggest a revised assessment of prognosis in patients with CLL if future studies confirm our results. In summary, with the use of CD40L stimulation, we show that translocations can be detected in a high proportion of CLL patients and can serve as a prognostic marker for TFS and OS. Nevertheless, it will be necessary to assess the definitive prognostic value of translocations in CLL in large, prospective clinical trials to define whether they have significant impact on risk-adapted therapy for patients with CLL.
Prepublished online asBlood First Edition Paper, September 22, 2005; DOI 10.1182/blood-2005-05-2093.
C.M., M.H., and C.M.W. conceived and designed the present work. C.M. and M.R.S. performed the research. D.M.K., R.B., and J.S. contributed analytical tools. R.B. conducted statistical analysis of the data. C.M. and C.M.W. analyzed the data. C.M., M.R.S., and C.M.W. wrote the paper.
Supported by the Deutsche Forschungsgemeinschaft (grant SFB 455) (C.M., M.H., C.M.W.), Deutsche Krebshilfe (C.M.), Wilhelm-Sander-Stiftung (grant 95-056-2), and Bundesministerium für Bildung und Forschung (grant BMBF/NGFN-1).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank the many physicians who unconditionally referred patients or contributed patient material. We also thank Stefan Bohlander and Manfred Gabriel for critical reading of the manuscript and for stimulating discussions.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal