Abstract
The reduced folate carrier (RFC) is the dominant influx transporter for antifolates. A major mechanism of antifolate resistance is loss of RFC (SLC19A1) gene expression due to decreased GC-box-dependent transcription. However, despite the poor GC-box binding in multiple antifolate-resistant cell lines, normal Sp1 levels were retained. Here we explored the post-translational modifications that may disrupt Sp1 function. Phospho-affinity purification of nuclear proteins revealed that resistant cells contained approximately 8-fold more phosphorylated Sp1 than parental cells; treatment of nuclear proteins from these cells with alkaline phosphatase restored GC-box binding. As protein kinase A phosphorylates Sp1, resistant cells were treated with various cAMP-reactive agents, revealing no apparent effect on GC-box binding except for the general phosphodiesterase inhibitor IBMX. As cGMP levels also may be affected by IBMX, resistant cells were treated with 8-pCPT-cGMP, resulting in the complete restoration of GC-box binding, luciferase reporter activity, and RFC mRNA levels. This restoration was abolished in the presence of the protein phosphatase 2A inhibitor (PP2A) okadaic acid. Importantly, whereas resistant cells showed multiple phosphorylated Sp1 forms barely detectable in parental cells, treatment with 8-pCPT-cGMP resulted in their elimination; this disappearance, however, was prevented by the copresence of okadaic acid. These findings provide the first evidence that loss of RFC gene expression in antifolate-resistant cells is associated with an inhibitory Sp1 phosphorylation that can be eliminated by a cGMP-dependent activation of PP2A. (Blood. 2006;107:708-715)
Introduction
Reduced folates are essential nutrients necessary for one-carbon transfer reactions, resulting in the de novo biosynthesis of purine and pyrimidine nucleotides and amino acids.1 Since mammalian cells are auxotrophic for folates, these vitamin cofactors are transported into cells from exogenous sources primarily via the reduced folate carrier (RFC).2,3 RFC also is the primary uptake route for hydrophilic antifolates used in chemotherapy of various human cancers, including acute lymphoblastic leukemia (ALL). These antifolates include the dihydrofolate reductase inhibitors methotrexate (MTX)4 and edatrexate (EDX),5 the thymidylate synthase (TS) inhibitor tomudex (raltitrexed),6 as well as the glycinamide ribonucleotide transformylase inhibitors pemetrexed (alimta)7 and AG2034.8
The human RFC (SLC19A1) promoter contains several consensus elements for the binding of various transcription factors, some of which appear to be essential for gene expression.9-11 Like many housekeeping genes, the human RFC promoter region contains, among several consensus elements, essential constitutive GC-box elements. The latter were shown to be physically occupied by Sp1 and Sp3,12 thereby substantially contributing to RFC gene expression in various cancer cell lines, including human CCRF-CEM T-cell leukemia.9,10 Specifically, by transiently transfecting various tumor cell lines with a series of 5′- and 3′- deletions in the human RFC promoters B and A, the minimal promoters were localized within 46 and 47 base pairs.12 Gel mobility shift assays and antibody-mediated supershift analysis with this human RFC basal promoter region confirmed specific DNA protein complexes involving a highly conserved GC-box and Sp1 or Sp3 binding sites.9,10,12 Furthermore, transient introduction of Sp1 and the long Sp3 isoform into Drosophila SL2 cells that are devoid of Sp1 family members potently transactivated basal RFC promoter activity.12
Sp1 was originally identified as a nuclear factor that binds to and activates transcription from multiple GC-box elements in the early promoter of simian virus 40 (SV40).13 The cloning of SP1 paralogous genes uncovered the existence of 8 nuclear proteins termed Sp1 through Sp8.14 While SP1 and SP3 are ubiquitously expressed, SP4 and SP7 exhibit a restricted pattern of expression in certain tissues. Members of the Sp family contain a highly conserved DNA-binding domain composed of 3 zinc fingers close to the C-terminus, as well as serine/threonine-rich domains. However, only Sp1, Sp3, and Sp4 contain 2 glutamine-rich domains in their N-terminal regions that are essential for transcriptional transactivation.14 Sp1 can act as a strong activator as it harbors glutamine-rich regions. Although the cellular level of Sp1 is regulated to a large extent at the mRNA level,15 there are additional regulatory mechanisms that occur at the protein level. For instance, Sp1 is known to undergo various posttranslational modifications that alter its DNA binding and/or transactivation capabilities; these include phosphorylation,16-21 O-linked glycosylation,22,23 and ubiquitin-independent proteasome degradation.24 Whereas various Sp1 phosphorylations have been documented to increase its DNA binding and transactivation activity,21,25,26 a few reports suggested that certain phosphorylations compromise Sp1 binding and transactivation activity.17,18 Furthermore, Sp1 has been shown to undergo phosphorylation by cAMP-dependent protein kinase (PKA),27,28 in which case the phosphorylation increased its GC-box-binding capability but rendered it a repressor of gene expression. Consistently, alternative phosphorylations also have been reported to render Sp1 a transcriptional repressor.29,30
An important mechanism of antifolate resistance in ALL31 and other malignancies32 is impaired antifolate transport due to loss of RFC function,2,33 resulting from mutational inactivation34 and/or transcriptional silencing.9-11 This loss of function, as we have recently shown in multiple antifolate-resistant human leukemia cell lines with defective antifolate uptake, is due either to mutations in the RFC gene34 or decreased RFC mRNA levels caused by a methylation-independent transcriptional silencing of the RFC gene.9-11 This was specifically achieved by the loss of expression and/or function of various transcription factors, thereby resulting in a significant decrease in their binding to various consensus elements in the RFC promoter region including GC-box, CRE, Mzf-1, AP-1, and E-box.9-11 Although approximately 50% of these antifolate-resistant leukemia cell lines displayed a marked decrease in their GC-box-binding capability, the vast majority surprisingly retained wild-type Sp1 protein levels.10 As these results suggested that Sp1 undergoes inhibitory posttranslational modifications that impair its function, we explored this hypothesis in the current paper. Hence, we herein provide the first evidence that loss of RFC gene expression in multiple antifolate-resistant cells is associated with an inhibitory phosphorylation of Sp1 that abolishes its GC-box-binding and transcriptional transactivation capability. We further show that this inhibitory Sp1 phosphorylation can be eliminated by a cGMP-dependent activation of PP2A.
Materials and methods
Drugs
1,4-dihydro-5-(2-propoxyphenyl)-7H-1,2,3-triazolo(4,5-d)pyrimidin-7-one (zaprinast), sodium nitroferricyanide (SNP), forskolin, 8-(4-chlorophenylthio)-guanosine 3′,5′-cyclic monophosphate (pCPT-cGMP), 3-isobuty-1-methylxanthine (IBMX), N,6 2′-O-dibutyryladenosine 3′,5′-cyclic monophosphate (Bt2-cAMP), and cholera toxin were purchased from Sigma Chemical (St Louis, MO). Okadaic acid was from Alomone Laboratories (Jerusalem, Israel). Antifolate drugs were generous gifts from the following sources: AG2034, Dr T. Boritzki (Agouron Pharmaceuticals, San Diego, CA); PT523, Dr W. T. McCulloch (Sparta Pharmaceuticals, Durham, NC), and ZD9331, Dr A. Jackman (Institute of Cancer Research, Sutton, United Kingdom), and Edatrexate (EDX), Dr J. H. Schornagel (Netherlands Cancer Institute, Amsterdam, The Netherlands).
Cell lines and tissue culture
CCRF-CEM, a human T-cell leukemia line, and its antifolate-resistant sublines,34 as well as the human ovarian carcinoma cell line 2008, were maintained in RPMI-1640 medium containing 2.3 μM folic acid (GIBCO, Carlsbad, CA), supplemented with 10% fetal calf serum, 2 mM glutamine, 100 units/mL penicillin G, and 100 μg/mL streptomycin sulfate (Biological Industries, Beth-Haemek, Israel). The cell lines were established by stepwise antifolate selection of parental CCRF-CEM cells as previously described.34
Purification of nuclear phosphoproteins
Nuclear extracts were prepared from exponentially growing cells (2 × 108 cells) as previously described.35 The purification of phosphorylated nuclear proteins was performed by phospho-affinity chromatography using a PhosphoProtein Purification Kit according to the instructions of the manufacturer (Qiagen, Hilden, Germany). Protein concentration was determined by the colorimetric method of Bradford.36
Western blot analysis
For one-dimensional gels, nuclear proteins or phosphorylated/nonphosphorylated purified proteins (20 μg) were resolved by electrophoresis on 10% polyacrylamide gels containing sodium dodecyl sulfate (SDS), electroblotted onto Protran BA83 cellulose nitrate membranes (Schleicher & Schuell, Dassel, Germany), and reacted with anti-Sp1 antibodies according to the instructions of the manufacturer (Serotec, Raleigh, NC). Following 3 10-minute washes in TBST (TBS containing 0.5% Tween 20) at room temperature, blots were reacted with a secondary antibody (Jackson Immunoresearch Labs, Baltimore, PA), rewashed, and enhanced chemiluminescence (ECL) detection was performed according to the manufacturer's instructions (Biological Industries, Beth-Haemek, Israel). ECL was recorded on x-ray films using several exposure times.
Electrophoretic mobility shift and antibody-mediated supershift assays
Exponentially growing cells (2 × 107 cells) were incubated for 2 to 3 hours in growth medium containing or lacking 1 mM IBMX, 1 mM Bt2-cAMP, 20 μM forskolin, 0.5 μg/mL cholera toxin, 1 mM pCPT-cGMP, or 1 μM okadaic acid. Nuclear extracts were prepared as previously described.35 DNA protein complexes were formed by incubating nuclear extract proteins (6 μg) with [α-32P]dCTP end-labeled GC-box double-stranded oligonucleotide (5′-AGT CGA TCG GGG CGG GGC GAG C-3′) as detailed elsewhere.37 For supershift analysis, nuclear proteins (6 μg) were incubated for 1 hour at 4°C with 4 μg anti-Sp1 (Serotec) or Sp3 (Santa Cruz Biotechnology, Santa Cruz, CA) antibodies, prior to the addition of radiolabeled oligonucleotides.
Dephosphorylation of nuclear extracts
The dephosphorylation of nuclear proteins was performed with calf intestinal alkaline phosphatase (Promega, Madison, WI) as previously described.18 The dephosphorylated proteins were used immediately in electrophoretic mobility shift assays (EMSA) as described in “Electrophoretic mobility shift and antibody-mediated supershift assays.”
Transient transfections with expression constructs
Exponentially growing suspension cells (2 × 107) were harvested by centrifugation and transiently transfected by electroporation (1000 μF, 234 V) with 10 μg of the GC-box-luciferase expression plasmid (pGL3-Luciferase, Promega). Cells were then seeded at a density of 2 × 106/mL in prewarmed growth medium, incubated for 24 hours at 37°C, and harvested for extraction of total proteins.
GC-box-luciferase activity assay
Twenty-one hours after transient transfection with the GC-box-luciferase expression vector, cells were incubated for 3 hours in growth medium containing or lacking 1 mM IBMX, 1 mM pCPT-cGMP, 1 μM okadaic acid, 100 μM zaprinast, or 100 μM SNP. Cells were then harvested by centrifugation, washed with phosphate-buffered saline (PBS), lysed, and firefly luciferase activity was assayed using a luciferase kit (Promega) and a luminometer. Results were obtained from at least 3 independent transient transfections performed in duplicate cultures.
High-resolution 2-dimensional gel electrophoresis
The 2-dimensional gel electrophoresis was performed as previously described.11 The nylon membranes were then reacted with the Sp1 antibody described in “Western blot analysis.”
Semiquantitative RT-PCR analysis of cGMP-dependent protein kinase (PKG) gene expression
Cells (107) from CCRF-CEM, a human T-cell leukemia line, and 2008, a human ovarian carcinoma cell line, were harvested for RNA isolation; reverse transcription and PCR reactions were performed as previously described.11 The primers used for the semiquantitative reverse transcriptase-polymerase chain reaction (RT-PCR) of PKG were as follows: PKG Iα, 5′-TGG AGA AGC GGC TGT CAG AG-3′; PKG Iβ, 5′-CAA GAG CCC ACA GTC CAA GG-3′; and 5′-GCT CTG GAA TGT TGG AAC GC-3′ for both; PKG II, 5′-TGT GGA CCA CAT TTG GGG AG-3′ and 5′-CAT CAC GGT TCA GGT TTG CC-3′.
Semiquantitative RT-PCR analysis of RFC gene expression
Cells (107) were incubated in growth medium containing or lacking 1 mM pCPT-cGMP for 3 hours, following which the medium was replaced and the cells were either incubated for 3 more hours or harvested for RNA isolation; reverse transcription and PCR reactions were performed as previously described.11 The primers used for the semiquantitative RT-PCR of RFC and GAPDH were described previously.9
Treatment of cells with an inhibitor of O-linked glycosylation
To rule out the possibility that Sp1 underwent some O-glycosylation that disrupts its DNA binding activity, parental and antifolate-resistant cells were treated with the potent O-glycosylation inhibitor benzyl-2-acetamido-2-deoxy-α-D-galactopyranoside (BenzylGalNac, 2 mM) for 72 hours. Nuclear extracts were then isolated, and EMSA was performed.
Results
Sp1 phosphorylation abolishes its GC-box binding in antifolate-resistant cells
Since phosphorylation is an established mechanism of regulation of Sp1 activity,16-21 we first explored the status of Sp1 phosphorylation in antifolate-resistant cells. Toward this end, total nuclear extract proteins from parental CCRF-CEM cells and their various antifolate-resistant sublines were first purified by phospho-affinity chromatography. Equal amounts of proteins were then analyzed by Western blot analysis using monoclonal antibodies to Sp1, revealing that purified nuclear phosphoproteins from antifolate-resistant cell lines contained approximately 8-fold more phosphorylated Sp1 than wild-type cells (Figure 1A, compare lanes 6 and 9 with 3), whereas nuclear extracts contained equal amounts of unphosphorylated Sp1 (Figure 1A, compare lanes 4 and 7 with 1). EMSA with a [32P]GC-box consensus oligonucleotide confirmed that all unpurified nuclear proteins from antifolate-resistant cells displayed a dramatic loss of GC-box binding (Figure 1B, compare lane 1 with 5). Furthermore, despite the marked phospho-affinity enrichment of phosphorylated Sp1 (Figure 1A), antifolate-resistant cells still displayed approximately 8-fold decreased GC-box binding, thereby suggesting that the Sp1 phosphorylation was inhibitory to GC-box binding (Figure 1C, compare lane 1 with 5). Antibody-mediated supershift analysis with nuclear proteins from parental cells before (Figure 1B, lanes 2-4) and after phospho-affinity purification (Figure 1C, lanes 2-4) demonstrated that GC-box binding was mediated by Sp1 and Sp3. In contrast, Sp1- and Sp3-binding to [32P]labeled GC-box oligonucleotide was essentially undetectable in nuclear proteins from antifolate-resistant cells (Figure 1B, lanes 6-8). The minimal restoration of GC-box binding after Phospho-affinity purification of nuclear proteins from antifolate-resistant cells (compare lanes 5 in Figure 1B,C) allowed for the detection of low Sp1- and Sp3-binding to the GC-box oligonucleotide upon supershift analysis (Figure 1C, lanes 6-8).
Treatment of nuclear proteins from antifolate-resistant cells with alkaline phosphatase restores GC-box binding
As these results suggested the presence of an inhibitory Sp1 phosphorylation that disrupts its GC-box binding, nuclear proteins from antifolate-resistant cells were treated with calf intestinal alkaline phosphatase (Figure 2A). Dephosphorylation of nuclear proteins from various antifolate-resistant cell lines with alkaline phosphatase resulted in restoration of GC-box binding. A small increase in GC-box binding also was observed with phosphatase-treated nuclear proteins from parental cells. Antibody-mediated supershift analysis confirmed that only parental cells had a prominent binding of Sp1 and Sp3 to the radiolabeled GC-box oligonucleotide (Figure 2B, lanes 2-4), whereas antifolate-resistant cells had only a marginal Sp1- and Sp3-binding to the GC-box oligonucleotide (Figure 2B, lanes 6-8). Consistently, antibody-mediated supershift analysis demonstrated the restoration of Sp1- and Sp3-binding to the radiolabeled GC-box oligonucleotide in antifolate-resistant cell lines after treatment with alkaline phosphatase (Figure 2C, lanes 6-8).
Phospho-affinity purification of nuclear proteins and functional examination by supershift analysis. (A) Western blot analysis of Sp1 expression and phosphorylation in parental and antifolate-resistant cell lines. Nuclear proteins from parental and antifolate-resistant sublines were purified by phospho-affinity chromatography, thereby revealing phosphorylated and unphosphorylated fractions. These proteins were then resolved by polyacrylamide gels containing SDS, transferred to a cellulose nitrate Protran membrane, and reacted with antibodies against human Sp1 as detailed in “Materials and methods.” (B, C) Antibody-mediated supershift analysis. Nuclear proteins, 6 μg of untreated extracts (B) and 4 μg of phospho-affinity-purified extracts (C), from parental and a representative antifolate-resistant cell line, EDXR0.03, with loss of binding to the GC-box element, were first incubated with antibodies (4 μg) against Sp1 and/or Sp3 for 1 hour at 4°C. Then, binding to [32P]-labeled GC-box oligonucleotide was determined by a Phosphorimager. The supershifted antibody-nuclear protein(s)-oligonucleotide complexes are denoted on the left by a, b, and c.
Phospho-affinity purification of nuclear proteins and functional examination by supershift analysis. (A) Western blot analysis of Sp1 expression and phosphorylation in parental and antifolate-resistant cell lines. Nuclear proteins from parental and antifolate-resistant sublines were purified by phospho-affinity chromatography, thereby revealing phosphorylated and unphosphorylated fractions. These proteins were then resolved by polyacrylamide gels containing SDS, transferred to a cellulose nitrate Protran membrane, and reacted with antibodies against human Sp1 as detailed in “Materials and methods.” (B, C) Antibody-mediated supershift analysis. Nuclear proteins, 6 μg of untreated extracts (B) and 4 μg of phospho-affinity-purified extracts (C), from parental and a representative antifolate-resistant cell line, EDXR0.03, with loss of binding to the GC-box element, were first incubated with antibodies (4 μg) against Sp1 and/or Sp3 for 1 hour at 4°C. Then, binding to [32P]-labeled GC-box oligonucleotide was determined by a Phosphorimager. The supershifted antibody-nuclear protein(s)-oligonucleotide complexes are denoted on the left by a, b, and c.
Pulse-treatment of antifolate-resistant cells with cGMP reactive agents restores GC-box binding and GC-box-driven transcriptional activity
Recently, we have shown that protein kinase A (PKA)-dependent phosphorylation markedly contributes to alteration of the activity of transcription factors, including CREB-1, resulting in dramatic changes in RFC gene expression.11 Since it has been shown that Sp1 undergoes a PKA-dependent phosphorylation,19,27,28 the possibility that the PKA pathway is involved in the generation of the inhibitory phosphorylation of Sp1 was explored. Treatment of antifolate-resistant cells with various cAMP-reactive agents, including cholera toxin, forskolin (data not shown), and dibutyryl cAMP (Bt2cAMP) (Figure 3A) had no effect on the DNA binding capability of Sp1. In contrast, the general phosphodiesterase inhibitor IBMX that increases cellular cAMP levels induced a substantial restoration of GC-box binding (Figure 3A). These results suggested that elevation in cAMP levels is not the mechanism underlying the IBMX-dependent restoration of GC-box binding in antifolate-resistant cells. Since cGMP is a substrate for certain phosphodiesterases, some of which are inhibited by IBMX, including phosphodiesterase 5 (PDE5),38 the possibility arose that IBMX induces an increase in the cellular levels of cGMP via inhibition of certain phosphodiesterases, thereby resulting in restoration of GC-box binding in antifolate-resistant cells. Pulse-treatment of the latter cells with the cell-permeable cGMP analog 8-pCPT-cGMP resulted in the complete restoration of GC-box binding (Figure 3A,B); this restoration was prevented in the presence of okadaic acid, a protein phosphatase 2A (PP2A) inhibitor (Figure 3B).
Dephosphorylation of nuclear proteins using calf intestinal alkaline phosphatase. (A) Restoration of promoter element binding after treatment with calf intestinal alkaline phosphatase in antifolate-resistant cell lines lacking GC-box binding. Drug-resistant cell lines with decreased (or loss of) binding to the cis-acting element, due to specific modifications of specific transcription factors, were treated with alkaline phosphatase. The binding to [32P]-labeled GC-box was then determined. (B, C) Antibody-mediated supershift analysis. Nuclear proteins, 6 μg of untreated extracts (B) and 3 μg of alkaline phosphatase-treated extracts (C) from parental and a representative antifolate-resistant cell line EDXR0.03, with loss of binding to the GC-box element, were first incubated with antibodies (4 μg) against Sp1 and/or Sp3 for 1 hour at 4°C. Binding to [32P]-labeled GC-box oligonucleotide was then determined by a Phosphorimager. The supershifted antibody-nuclear protein(s)-oligonucleotide complexes are denoted on the left by a, b, and c.
Dephosphorylation of nuclear proteins using calf intestinal alkaline phosphatase. (A) Restoration of promoter element binding after treatment with calf intestinal alkaline phosphatase in antifolate-resistant cell lines lacking GC-box binding. Drug-resistant cell lines with decreased (or loss of) binding to the cis-acting element, due to specific modifications of specific transcription factors, were treated with alkaline phosphatase. The binding to [32P]-labeled GC-box was then determined. (B, C) Antibody-mediated supershift analysis. Nuclear proteins, 6 μg of untreated extracts (B) and 3 μg of alkaline phosphatase-treated extracts (C) from parental and a representative antifolate-resistant cell line EDXR0.03, with loss of binding to the GC-box element, were first incubated with antibodies (4 μg) against Sp1 and/or Sp3 for 1 hour at 4°C. Binding to [32P]-labeled GC-box oligonucleotide was then determined by a Phosphorimager. The supershifted antibody-nuclear protein(s)-oligonucleotide complexes are denoted on the left by a, b, and c.
Effect of various compounds on GC-box binding. EMSA with [32P]-labeled GC-box oligonucleotide was performed as follows: nuclear proteins (6 μg) from parental cells and their antifolate-resistant subline before and after treatment with Bt2-cAMP, IBMX, or pCPT-cGMP (A), pCPT-cGMP, pCPT-cGMP plus okadaic acid, or okadaic acid alone (B), were first incubated with [32P]-labeled GC-oligonucleotide, resolved by electrophoresis on polyacrylamide gels, and analyzed by a Phosphorimager.
Effect of various compounds on GC-box binding. EMSA with [32P]-labeled GC-box oligonucleotide was performed as follows: nuclear proteins (6 μg) from parental cells and their antifolate-resistant subline before and after treatment with Bt2-cAMP, IBMX, or pCPT-cGMP (A), pCPT-cGMP, pCPT-cGMP plus okadaic acid, or okadaic acid alone (B), were first incubated with [32P]-labeled GC-oligonucleotide, resolved by electrophoresis on polyacrylamide gels, and analyzed by a Phosphorimager.
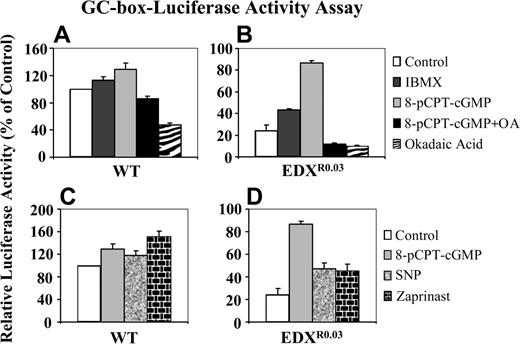
GC-box-driven luciferase reporter gene activity. A GC-box-luciferase construct was transfected by electroporation into parental CCRF-CEM wild-type (WT) cells (A, C) and their antifolate-resistant subline EDXR0.03 (B, D), after which a portion of cells was treated with the various agents for 3 hours: IBMX, pCPT-cGMP, pCPT-cGMP plus okadaic acid or okadaic acid alone (A, B), as well as pCPT-cGMP, SNP, or zaprinast (C, D). Cells were then lysed, and reporter gene activities were determined as detailed in “Materials and methods.” Results presented are normalized mean reporter activities (relative to untreated parental cells) ± SD obtained from 3 independent experiments.
GC-box-driven luciferase reporter gene activity. A GC-box-luciferase construct was transfected by electroporation into parental CCRF-CEM wild-type (WT) cells (A, C) and their antifolate-resistant subline EDXR0.03 (B, D), after which a portion of cells was treated with the various agents for 3 hours: IBMX, pCPT-cGMP, pCPT-cGMP plus okadaic acid or okadaic acid alone (A, B), as well as pCPT-cGMP, SNP, or zaprinast (C, D). Cells were then lysed, and reporter gene activities were determined as detailed in “Materials and methods.” Results presented are normalized mean reporter activities (relative to untreated parental cells) ± SD obtained from 3 independent experiments.
Consistently, pulse-treatment of antifolate-resistant cells with 8-pCPT-cGMP resulted in a marked increase in the GC-box-driven luciferase reporter gene activity upon transient transfection into antifolate-resistant cells (Figure 4B), whereas the same treatment of parental cells had only a slight increase in reporter gene activity (Figure 4A). Moreover, inclusion of the PP2A inhibitor okadaic acid during the pulse-treatment with 8-pCPT-cGMP resulted in ablation of the transcriptional stimulatory effect of the latter (Figure 4B). As these results were suggestive of cGMP-dependent transcriptional activation, antifolate-resistant cells were treated with the nitric oxide (NO) donor sodium nitroferricyanide (SNP), which activates soluble guanylyl cyclase, as well as with zaprinast, a specific PDE5 inhibitor: these resulted in a prominent restoration of GC-box-driven luciferase reporter activity (Figure 4D), whereas these treatments had only a slight stimulatory effect on parental cells (Figure 4C).
Pulse-treatment of antifolate-resistant cells with 8-pCPT-cGMP eliminates the inhibitory Sp1 phosphorylation
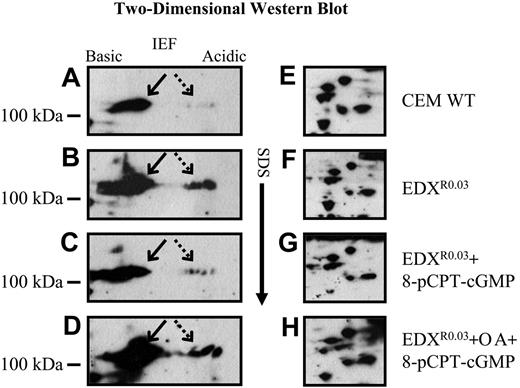
High-resolution 2-dimensional gel electrophoresis (2-DGE) followed by Western analysis with an anti-Sp1 antibody revealed that antifolate-resistant cells displayed high levels of multiple phosphorylated forms of Sp1 (Figure 5B, dashed arrow) that were barely detectable in parental cells (Figure 5A). As these phosphorylated Sp1 forms were shown in Figures 1, 2, and 4 to be inhibitory to GC-box binding and to GC-box-driven transcription, and since treatment with 8-pCPT-cGMP restored these activities, antifolate-resistant cells were pulse-treated with 8-pCPT-cGMP after which nuclear proteins were prepared and examined by Western blots with Sp1 antibodies after 2-DGE. Treatment of antifolate-resistant cells with this cGMP-reactive agent resulted in a marked dephosphorylation of Sp1 (Figure 5C), thereby restoring the poorly phosphorylated pattern of Sp1 in parental cells (Figure 5A); however, when the PP2A inhibitor okadaic acid was included during the treatment with 8-pCPT-cGMP, a complete retention of the phosphorylated forms of Sp1 was observed (Figure 5D). A cluster of well-resolved proteins is shown in Figure 5E-H to confirm that equal amounts of total nuclear proteins from antifolate-resistant and parental cells were being analyzed.
Exploration of PKG expression in leukemia cell lines
The pathway for cellular cGMP signaling occurs primarily via PKG39 ; we therefore explored the gene expression status of PRKG1A, PRKG1B, and PRKG2. RT-PCR analysis with specific primers revealed that none of these PKG variants was expressed in parental cells and their antifolate-resistant cells, whereas a control group of human ovarian carcinoma 2008 cells expressed low levels of all 3 PKG transcripts (data not shown).
Western blot analysis after high-resolution 2-D gel electrophoresis (2-DGE) of Sp1 and its phosphorylated forms. Nuclear proteins (75 μg) isolated from untreated parental (A, E) and antifolate-resistant EDXR0.03 cells (B, F), as well as pCPT-cGMP-treated EDXR0.03 cells (C, G) and pCPT-cGMP plus okadaic acid (OA)-treated EDXR0.03 cells (D, H) were subjected to high-resolution 2-DGE. The blots were then reacted with an antibody to Sp1 to detect nonphosphorylated (arrow) as well as phosphorylated forms of Sp1 (dashed arrow).
Western blot analysis after high-resolution 2-D gel electrophoresis (2-DGE) of Sp1 and its phosphorylated forms. Nuclear proteins (75 μg) isolated from untreated parental (A, E) and antifolate-resistant EDXR0.03 cells (B, F), as well as pCPT-cGMP-treated EDXR0.03 cells (C, G) and pCPT-cGMP plus okadaic acid (OA)-treated EDXR0.03 cells (D, H) were subjected to high-resolution 2-DGE. The blots were then reacted with an antibody to Sp1 to detect nonphosphorylated (arrow) as well as phosphorylated forms of Sp1 (dashed arrow).
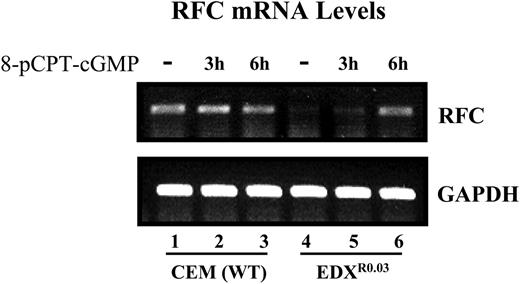
Restoration of RFC gene expression in parental and antifolate-resistant cells upon treatment with pCPT-cGMP. The effect of pCPT-cGMP on the restoration of RFC mRNA levels in parental and antifolate-resistant cells was examined by RT-PCR analysis (30 cycles) using internal GAPDH (glyceraldehyde-3-phosphate dehydrogenase) normalization as detailed in “Materials and methods.”
Restoration of RFC gene expression in parental and antifolate-resistant cells upon treatment with pCPT-cGMP. The effect of pCPT-cGMP on the restoration of RFC mRNA levels in parental and antifolate-resistant cells was examined by RT-PCR analysis (30 cycles) using internal GAPDH (glyceraldehyde-3-phosphate dehydrogenase) normalization as detailed in “Materials and methods.”
Restoration of RFC gene expression
Since decreased Sp1 DNA binding has been shown to contribute to RFC down-regulation,9,10 we examined whether the dephosphorylation of Sp1 could restore RFC gene expression. Treatment of antifolate-resistant cells with 8-pCPT-cGMP resulted in a marked restoration of RFC mRNA levels (Figure 6). These results provide the first evidence that loss of RFC gene expression is associated with an inhibitory phosphorylation of Sp1 that can be eliminated with a cGMP-dependent phosphatase that is independent of PKG, as the latter was not expressed at all in parental and antifolate-resistant cells.
Sp1 from antifolate-resistant cells does not harbor inhibitory O-linked glycosylation
To rule out the possibility that Sp1 from antifolate-resistant cells underwent some O-linked glycosylation that may disrupt its DNA binding, parental and antifolate-resistant cells were treated with the potent O-glycosylation inhibitor benzyl-2-acetamido-2-deoxy-α-D-galactopyranoside for 72 hours. Nuclear extracts were then isolated, and GC-box binding was assessed by EMSA; this O-glycosylation inhibitor did not alter the GC-box binding in either parental or antifolate-resistant cells, thereby suggesting that Sp1 from these cells did not harbor any inhibitory O-glycosylation.
Discussion
Several lines of evidence suggest that Sp1 undergoes an inhibitory phosphorylation that disrupts its GC-box binding and consequently its transactivation capability. First, purification of nuclear proteins by phospho-affinity chromatography followed by Western analysis revealed that antifolate-resistant cells had 8-fold more phosphorylated Sp1 levels than their parental cells; however, this phosphorylated Sp1 conversely showed an 8-fold decrease in the GC-box binding. This implies that a certain inhibitory phosphorylation of Sp1 dramatically decreases its GC-box binding by a factor of approximately 64-fold. This finding is consistent with previous studies suggesting that certain phosphorylations resulted in disruption of Sp1 binding to GC-box.17,18 Second, nuclear proteins from antifolate-resistant cells contained highly phosphorylated states of Sp1, as revealed by the increased acidity of the Sp1 forms observed upon high-resolution 2-D gel electrophoresis followed by Western blotting. Surprisingly, however, these phosphorylated Sp1 forms displayed poor GC-box binding. Third, dephosphorylation of nuclear proteins from antifolate-resistant cells with alkaline phosphatase resulted in a marked restoration of GC-box binding by Sp1, as revealed by antibody-mediated supershift analysis.
Various Sp1 phosphorylations have been documented to increase both Sp1 binding to GC-box and transactivation activity; these include serine phosphorylation by protein kinase C-γ (PKC-γ),21 Ser59 phosphorylation by cyclin A-dependent kinase 2,25 as well as Thr453 and Thr739 phosphorylation by p42/p44 mitogen-activated protein kinase.26 Furthermore, Sp1 also has been shown to undergo phosphorylation by PKA27,28 ; however, although this phosphorylation increased its GC-box binding, it converted Sp1 to a repressor of gene expression. This is consistent with studies in which certain phosphorylations conferred upon Sp1 a transcriptional repression activity.29,30 However, little is known about inhibitory phosphorylations that disrupt the DNA-binding capability of Sp1.17,18 Casein kinase II, which is the only kinase known to inhibit Sp1 activity,40 phosphorylates the latter on a Thr579 in the second zinc finger domain; this phosphorylation results in a reduced binding affinity of Sp1 for GC-box.17 Therefore, it is likely that in our antifolate-resistant cell lines, Sp1 contains a similar inhibitory phosphorylation that impairs its GC-box binding, thereby resulting in abolished transactivation capability including that of the RFC gene.
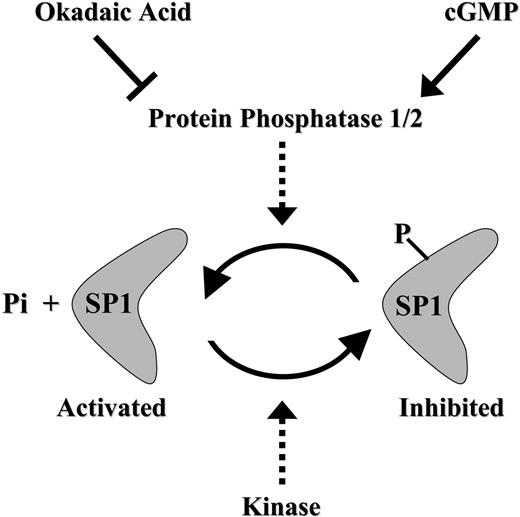
Schematic model summarizing the cGMP-dependent restoration of Sp1 activity via Sp1 dephosphorylation in antifolate-resistant cells.
Schematic model summarizing the cGMP-dependent restoration of Sp1 activity via Sp1 dephosphorylation in antifolate-resistant cells.
Our results suggest that antifolate-resistant cells apparently suffer from an imbalance in Sp1 inhibitory hyperphosphorylation when compared with the poorly phosphorylated state present in parental cells. Importantly, this inhibitory phosphorylation could be readily reversed (ie, by dephosphorylation) by pulse treatment with the cell-permeable cGMP-reactive agent pCPT-GMP. We therefore propose here that Sp1 function can be restored in antifolate-resistant cells via a cGMP-dependent pathway that presumably activates PP2A, resulting in dephosphorylation of Sp1 (Figure 7); this in turn resulted in restoration of GC-box binding and GC-box-driven transcriptional activity, including that of the RFC gene. This suggestion is supported by the following evidence: (1) Coincubation of antifolate-resistant cells with 8-pCPT-GMP and the efficient PP2A inhibitor okadaic acid ablated the cGMP-dependent restoration of GC-box binding and GC-box-driven transcriptional activity. These findings are consistent with previous studies: Armstrong et al17 showed that upon terminal differentiation in vivo, Sp1 undergoes a casein kinase II-dependent phosphorylation at Thr579 in the second zinc finger domain, thereby resulting in decreased DNA binding; and PP1 was identified as the endogenous nuclear protein that dephosphorylates Sp1. Furthermore, it was demonstrated that transcriptional activity of Sp1 is up-regulated in dividing primary T lymphocytes by a PP2A-dependent dephosphorylation.41 It should be noted that 1 μM okadaic acid is an efficient inhibitor of PP2A without affecting PP1 from various cells,42 including the T-cell line CCRF-CEM that was used in the current study. Furthermore, PP2A but not PP1 was found to eliminate the inhibitory phosphorylation of Sp1 in T lymphocytes.41 Hence, it is likely that PP2A is the protein phosphatase that eliminates the inhibitory phosphorylation of Sp1 in antifolate-resistant cell lines. (2) The general PDE inhibitor IBMX, as well as zaprinast, a specific inhibitor of PDE 5 that has high specificity for cGMP,43 both of which are known to elevate cGMP levels, increased GC-box-driven transcriptional activity of a reporter gene. Moreover, this IBMX-dependent restoration effect appears to be due to induction of cGMP levels and not due to elevation of cAMP levels; this conclusion is based upon the finding that various cAMP-reactive agents along the PKA pathway, including cholera toxin, Bt2cAMP, and forskolin, failed to restore Sp1 activity. (3) Direct dephosphorylation of Sp1 by alkaline phosphatase restored Sp1 binding to GC-box.
In the current paper we note that treatment of antifolate-resistant cells with the membrane-permeable cGMP-reactive agent 8-pCPT-GMP resulted in the loss of the inhibitory phosphorylation of Sp1, thereby resulting in restoration of GC-box binding and GC-box-driven transcriptional activity. cGMP is a key secondary messenger involved in crucial cellular signal transduction pathways and is generated either by soluble guanylyl-cyclases (sGC), which are activated by nitric oxide, or by receptor guanylyl cyclases (rGCs), activated by natriuretic peptides.44 However, we find here that PKGI and PKGII transcripts were completely absent from both parental CCRF-CEM and their antifolate-resistant cells. The finding that PKG is not expressed in the T-cell line CCRF-CEM used in this study is consistent with previous reports that upon in vitro culturing of primary T-cells, PKGI expression is lost,45 whereas PKGII is not expressed at all in blood cells.38 Hence, since cGMP exerts its biologic effect by modulating various proteins, including cGMP-dependent PKGI and PKGII,38 PDEs,43 and cyclic nucleotide-regulated ion channels,46 it is clear that the putative cGMP-dependent activation of PP2A is independent of PKG. Further studies are warranted to delineate the cGMP-mediated yet PKG-independent pathway of PP2A activation in antifolate-resistant cells. It is tempting to speculate that cGMP activates PP2A via cyclic nucleotide-regulated Ca++ or K+-channels.
High-resolution 2-dimensional gel electrophoresis followed by Western analysis revealed multiple phosphorylated Sp1 forms (ie, more acidic) in antifolate-resistant cells when compared to the more discrete unphosphorylated complex of Sp1 (ie, basic) from parental cells. These findings are in agreement with a previous report that Sp1 from adult rat liver is phosphorylated at multiple sites,18 thereby resulting in loss of DNA-binding activity. Sp1 is known to undergo various Ser/Thr phosphorylations, the vast majority of which increase its DNA-binding capability and consequent transcriptional transactivation capacity.21,25,26 In contrast, at least one phosphorylation at Thr579 at the second zinc finger domain has been shown to disrupt its GC-box-binding activity.17 It appears that Sp1 from antifolate-resistant cells harbors multiple phosphorylations, some of which may be stimulatory phosphorylations; however, the co-presence of a single inhibitory phosphorylation such as Thr579 at the second zinc finger domain is presumably sufficient to disrupt its DNA binding capability. Furthermore, since much of the Sp1 protein remained in the unphosphorylated form in antifolate-resistant cells, it is possible that the inhibitory phosphorylation results in a negative dominant effect exerted on DNA binding and transactivation capability.
The possibility that Sp1 undergoes some inhibitory O-glycosylation in antifolate-resistant cells also was explored. Thus, drug-resistant cells were subjected to a long-term treatment (72 hours) with benzylGalNac, a potent O-glycosylation inhibitor. However, since no alteration in the GC-box binding could be observed in both parental and drug-resistant cells, we conclude that Sp1 from these cells appears to lack any inhibitory Sp1 O-glycosylation.
The potential clinical implication of our present study for the possible future exploitation of the cGMP signaling pathway in the reversal of antifolate-resistance as well as in cancer chemotherapy is as follows. Currently there is a growing interest in the exploitation of the cGMP signaling pathway as a novel means of anticancer drug treatment. For example, exisulind (sulindac sulfone) is a novel proapoptotic drug that induces regression and prevents recurrence of adenomatous polyps in patients with familial adenomatous polyposis.47 Furthermore, exisulind and its analogs exert their proapoptotic activity in cancer cells by blocking the catalytic activities of cGMP PDE5, 2 and 1, thereby resulting in a sustained increase in the intracellular levels of cGMP in colon cancer cells.48 Consistently, benzylamide sulindac analogs also were found to induce apoptosis in chronic lymphocytic leukemia (CLL) cells via loss of microtubules and cell cycle (G2-M) arrest.49 Similarly, membrane-permeant acetoxymethyl esters of cGMP analogs (ie, bioactivatable via intracellular esterase activity) also induced cell death in promyelocytic leukemia cells via apoptosis.50 Moreover, long-acting natriuretic peptides and related peptides also were shown to inhibit cancer cell proliferation via inhibition of DNA synthesis mediated by cGMP.51 Based on these reports and our present findings, it is reasonable to suggest that future treatment of antifolate-resistant tumors due to impaired drug uptake with membrane-permeant cGMP analogs could restore antifolate sensitivity and at the same time induce programmed cell death, thereby resulting in an enhanced eradication of tumor cells. Hence, further in vivo studies are warranted that evaluate the susceptibility of antifolate-resistant tumor xenografts to membrane-permeable cGMP analogs.
In summary, in the current study we show for the first time that impaired antifolate transport in multiple antifolate-resistant cell lines resulting from transcriptional silencing of the RFC gene is associated with an inhibitory phosphorylation of Sp1. We provide further evidence that this inhibitory Sp1 phosphorylation can be reversed by a cGMP-dependent, yet PKG-distinct, activation of a protein phosphatase, most likely PP2A, thereby resulting in restoration of RFC gene expression.
Prepublished online as Blood First Edition Paper, September 13, 2005; DOI 10.1182/blood-2005-07-2743.
Supported by the Star Foundation (Y.G.A.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

![Figure 1. Phospho-affinity purification of nuclear proteins and functional examination by supershift analysis. (A) Western blot analysis of Sp1 expression and phosphorylation in parental and antifolate-resistant cell lines. Nuclear proteins from parental and antifolate-resistant sublines were purified by phospho-affinity chromatography, thereby revealing phosphorylated and unphosphorylated fractions. These proteins were then resolved by polyacrylamide gels containing SDS, transferred to a cellulose nitrate Protran membrane, and reacted with antibodies against human Sp1 as detailed in “Materials and methods.” (B, C) Antibody-mediated supershift analysis. Nuclear proteins, 6 μg of untreated extracts (B) and 4 μg of phospho-affinity-purified extracts (C), from parental and a representative antifolate-resistant cell line, EDXR0.03, with loss of binding to the GC-box element, were first incubated with antibodies (4 μg) against Sp1 and/or Sp3 for 1 hour at 4°C. Then, binding to [32P]-labeled GC-box oligonucleotide was determined by a Phosphorimager. The supershifted antibody-nuclear protein(s)-oligonucleotide complexes are denoted on the left by a, b, and c.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/107/2/10.1182_blood-2005-07-2743/4/m_zh80020689610001.jpeg?Expires=1768415713&Signature=pg2eE~ielrhSybvcjcfj73Qyx6xhvr-2JgR0FNKV91l-5jaTZV-sZ1QywDgJcrt2-vbNcmlWUYCSMsFjpHGzw3LxialzMK6-bCqZUjvK-5OhSMueNDlWAqgP-DeoQXoUTAuIa4XRAZwufBBBK-E94D8L-ybYog12bM8SdpuioIz1O5oIIL13WQ7sIX-HnqG0PaCRYRMw-vSDvwWmIOTUVYSYyoGMRPtcYcjiT3F5gbSNWYAM5aZLYbZ4naf4fK80DVWEpMRLa~aSYgaobK8Hn-b7vsJMJZbTt6TB7cK9l-weNhAYNiLyt-ruEdRed926azds0UG4bkBrNv0D8aBOqg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 2. Dephosphorylation of nuclear proteins using calf intestinal alkaline phosphatase. (A) Restoration of promoter element binding after treatment with calf intestinal alkaline phosphatase in antifolate-resistant cell lines lacking GC-box binding. Drug-resistant cell lines with decreased (or loss of) binding to the cis-acting element, due to specific modifications of specific transcription factors, were treated with alkaline phosphatase. The binding to [32P]-labeled GC-box was then determined. (B, C) Antibody-mediated supershift analysis. Nuclear proteins, 6 μg of untreated extracts (B) and 3 μg of alkaline phosphatase-treated extracts (C) from parental and a representative antifolate-resistant cell line EDXR0.03, with loss of binding to the GC-box element, were first incubated with antibodies (4 μg) against Sp1 and/or Sp3 for 1 hour at 4°C. Binding to [32P]-labeled GC-box oligonucleotide was then determined by a Phosphorimager. The supershifted antibody-nuclear protein(s)-oligonucleotide complexes are denoted on the left by a, b, and c.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/107/2/10.1182_blood-2005-07-2743/4/m_zh80020689610002.jpeg?Expires=1768415713&Signature=brWFciPRjSc1Z~t9nkEuCADqvYwAZYV-ryW3qTpVzZb1y9KMZD3LkKHeuE-MP8JxYOPRxO6Y9aCoY8yyPyHTJZbitT1vpxWP7EFMTEjY~k2My90~5xuM8qm3zfOvvxmA4VmsQo64z6qusjoKMoW6nWWemxtn4n50ioV~1MyLhMAv1lZsiMLHJvZH9VG3~gzLukC9kdE1TRWRw7keqtztc2-rZ3nupmWLVRtn1ylMmgPJNPYMjZn3Hh70wNn074ReYifj2etjq6nzFfbDhvQG6xNlvh3g2ozq9tiW2SPIDFPoxVTBwtyVnMFtURQi12LLFHZNdzm8GwOf4NZXlz4-vw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 3. Effect of various compounds on GC-box binding. EMSA with [32P]-labeled GC-box oligonucleotide was performed as follows: nuclear proteins (6 μg) from parental cells and their antifolate-resistant subline before and after treatment with Bt2-cAMP, IBMX, or pCPT-cGMP (A), pCPT-cGMP, pCPT-cGMP plus okadaic acid, or okadaic acid alone (B), were first incubated with [32P]-labeled GC-oligonucleotide, resolved by electrophoresis on polyacrylamide gels, and analyzed by a Phosphorimager.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/107/2/10.1182_blood-2005-07-2743/4/m_zh80020689610003.jpeg?Expires=1768415713&Signature=JuHbwE4weHSV3f1mJtYjm~GRPuj776vIgcljngzgTGC2fkNR~x4yMV4jad65jpMgvacnWYkc1UYkRcq6GCEf5eJ8ods5MnztM3-ZhmB0sBgXAEv~cIdY33FIwz8Pn~MZ28DY0kIuHsgO3II44FzyYpkhh05xb1zODtUc~9YgHVLZPpou83Ho-87cJ0zYD9c5L0rDnK9a1Mf04IOiG5zeaOSJO5zEuKg8wHSkSao6Agmhyv8N6A5oXCua~5BwGVBIwxypufFrbcsOW7b25jNhxgv8C9ttvEZiYya2hl8F-XdMkd63HHLXpIViiG~2e2Ymo9OtjTiwpZ6pcThp8KRylg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal