Abstract
Hypoxia-inducible factors (HIFs) are transcriptional regulators that mediate the cellular response to low oxygen levels. By stimulating the expression of angiogenic growth factors such as vascular endothelial growth factor (VEGF), they trigger the neovascularization of tissues under physiologic and pathologic conditions. Here, we have investigated the endothelial cell–autonomous HIF function in blood vessel growth and development by expressing a dominant-negative HIF mutant (HIFdn) that inhibits the transcriptional responses mediated by both HIF-1 and HIF-2, specifically in endothelial cells of transgenic mice. HIFdn transgenic embryos were growth retarded and died around E11.5. Primitive vascular networks were established, but vascular remodeling in the yolk sac and in the embryo proper was defective, and vascular sprouts failed to invade the neuroepithelium. In addition, heart looping was incomplete, and the ventricles of the heart were thin-walled and lacked trabeculation. Similar cardiovascular defects have been observed in Tie2–deficient mouse embryos. Consistently, HIFdn transgenic embryos expressed reduced levels of the endothelial angiopoietin receptor, Tie-2, whereas other endothelial markers, such as PECAM-1, Tie-1, and VE-cadherin were not affected. These results show that HIFs in endothelial cells are essential for embryonic heart and blood vessel development and control angiogenesis and vascular remodeling.
Introduction
Growing tissues have a continuous demand for oxygen and nutrients. Tissue hypoxia efficiently triggers the growth of new blood vessels in both physiologic and pathologic processes. For example, hypoxia stimulates the compensatory formation of blood vessels in coronary heart disease, stroke, or in tumors by up-regulating the expression of angiogenic factors such as vascular endothelial growth factor (VEGF).1,2 It has also been suggested that during embryonic vessel development, regions of relative hypoxia attract growing blood vessels.3 The cellular responses to low oxygen levels are mediated by transcription factors called hypoxia-inducible factors (HIFs). At present, 3 different HIF family members are known: HIF-1, HIF-2, and HIF-3. HIFs are heterodimers composed of 1 of 3 different HIF-α subunits, HIF-1α,4 HIF-2α,5-7 or HIF-3α,8,9 and the common HIF-1β (ARNT) subunit. Under normoxic conditions, the HIF-α subunits are rapidly degraded. This process involves the posttranslational modification of HIF-α by a recently discovered family of prolyl hydroxylases (PHDs).10 Hydroxylated prolyl residues of the HIF-α subunits are recognized by the von Hippel-Lindau protein, a part of the E3 ubiquitin ligase complex that marks the HIF-α subunits for degradation by the proteasome.11-13 . Under hypoxic conditions, PHDs are inactive and HIF-α subunits accumulate in the nucleus, where they transactivate their target genes. In addition, growth factor signaling modulates HIF activity and stability in a hypoxia-independent fashion.10,14 Whereas HIF-1α is expressed ubiquitously, HIF-2α and HIF-3α show a spatially restricted expression pattern, HIF-2 being predominantly expressed in endothelial cells during ontogeny.5-7
HIF-1 and HIF-2 stimulate the expression of essential endothelial receptor tyrosine kinases, such as Tie-2, Flt-1, and Flk-1.7,15-17 A recent study characterized the transcriptional regulation of vascular endothelial cell responses to hypoxia by HIF-1 in vitro.18 Collectively, these data suggest that HIF-1 and HIF-2 exert important functions in endothelial cells and might not only stimulate the expression of angiogenic factors by nonendothelial cells adjacent to blood vessels but also mediate cell-autonomous activation of endothelial cells.
However, gene-targeting experiments in mice have largely failed to demonstrate an essential function for HIFs in the developing endothelium. Hif1a–deficient mouse embryos showed impaired extraembryonic and intraembryonic vessel formation, defective neural tube closure, and increased mesenchymal cell death, leading to embryonic death around embryonic day 10 (E10).19-21 However, the endothelial cell selective loss of Hif1a did not affect vascular development but, rather, pathologic angiogenesis in the adult mouse.22 This suggested that HIF-1 exerts its function in embryonic angiogenesis primarily by regulating paracrine factors such as VEGF rather than by stimulating endothelial cell intrinsic processes. Unexpectedly, several independent studies reported that the targeted inactivation of Hif2a did not affect mouse embryonic vascular development: Hif2a–deficient embryos died from defective catecholamine metabolism and bradycardia,23 suffered from defects in neonatal lung maturation,24 or developed a syndrome of multiorgan pathology and biochemical abnormalities.25 Only one group reported vascular defects in the yolk sac vasculature, which were rescued by HIF-2α.26,27 Thus, it remained unclear whether HIFs have an essential function in endothelial cells during vascular development or whether functional redundancy within the HIF family obscured such a function.
Here, we report that HIF activity in endothelial cells is essential for embryonic blood vessel development. HIF-mediated gene transcription was blocked by the expression of a dominant-negative HIF mutant (HIFdn) that inhibited the activity of both HIF-1-and HIF-2. HIFdn expression was directed to endothelial cells of transgenic mice from promoter/enhancer regions of the Flk1 gene (Flk1 p/e). The Flk1 p/e–HIFdn transgenic embryos were severely growth retarded, had multiple cardiovascular defects, and died around E11.5. The expression of the angiopoietin receptor, Tie-2, was greatly reduced in these mice. These results establish an essential role for endothelial HIFs in the formation of the heart, in angiogenic sprouting, and in the remodeling of the embryonic vascular system.
Materials and methods
Cell culture and firefly luciferase reporter gene assays
HEK293 cells were cultured in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal calf serum (FCS; PAA Laboratories, Linz, Austria). Human umbilical endothelial (HUE) cells were cultured in MCDB131 medium (CC Pro, Neustadt, Germany) containing 8% FCS and 4 mM l-glutamine. The murine VEGFR-2 (Flk1) promoter (nt –640 to +299)28 or the human VEGFR-2 (KDR) promoter (nt –763 to +296) fused to the luciferase gene were used as HIF-2–responsive reporters, and the human VEGF promoter (nt –2848 to –984)-luciferase29 as HIF-1–responsive reporter. In a typical experiment, 1 μg reporter plasmid, 0.3 μg CMV promoter–β-galactosidase, and 1 μg pcDNA3-HIF-1α or pcDNA3-HIF-2α,16 and/or 1 μg pcDNA3-HIFdn,17 respectively, were transfected using Superfect reagent (Qiagen, Hilden, Germany). Cells were harvested in lysis buffer (Dual light kit; Applied Biosystems, Bedford, MA) and assayed for luciferase and β-galactosidase activity in a luminometer (Berthold, Nashua, NH) using the respective substrates (Dual light kit; Applied Biosystems). To correct for variable transfection efficiency, the ratio of luciferase and β-galactosidase activities was determined. At least 3 independent transfection assays were performed for each experiment.
Construction of the Flk1–HIFdn transgene and generation of transgenic mice
A 955-bp cDNA fragment (with C-terminal FLAG tag) for a dominant-negative HIF-2α mutant lacking the C-terminal transactivation domains and the N-terminal DNA binding domain (HIFdn, amino acids 24 to 325)17 was fused to a 939-bp promoter fragment and a 2.3-kb intron fragment of mouse Flk1 gene.16 Transgenic mice were generated by pronuclear microinjection of DNA (5 to 10 ng/μL) into fertilized mouse oocytes from superovulated C57BL/6 × C3H mice. Oocytes were reimplanted into pseudopregnant CD1 fosters. Embryos were genotyped by polymerase chain reaction (PCR) using primers HIFdn forward (forw) 5′-CCACAGCATG GACATGAAGTT-3′ and FLAG reverse (rev) 5′-CTTATCGTCG TCGTCTTTGTA-3′: 94°C for 5 minutes, 35 cycles of 94°C for 1 minute, 56°C for 1 minute, 72°C for 1 minute, and 72°C for 5 minutes.
RNA preparation and RT-PCR analysis
Total RNA was isolated from the yolk sacs or distal trunks of E10.5 to E13.5 embryos using RNeasy Mini Kit (Qiagen). Aliquots of 1.5 μg RNA were reverse transcribed using Superscript II (Invitrogen) and random hexameric primers (Roche, Mannheim, Germany). To exclude contamination of genomic DNA, a control reverse transcriptase (RT) reaction without reverse transcriptase was performed. Primers for PECAM-1, Flk-1, Tie-2, VE-cadherin, Ang-1, and Tie-1 were described previously.30,31 Primers for VEGF were VEGF forw 5′-CTGCTCTCTT GGGTGCACTGG-3′ and VEGF rev 5′-CACCGCCTTG GCTTGTCACAT-3′. Expression of the HIFdn transgene was detected by using the HIFdn forw and FLAG rev primers (see “Construction of the flk-1–HIFdn transgene and generation of transgenic mice.”).
Histology and immunohistochemistry
Embryos were dissected, embedded in Tissue-Tek (Sakura, Giessen, Germany), and frozen on dry ice–isopentane. Frozen sections (8 to 12 μm) were stained using the Vectastain-Peroxidase kit (Vector Laboratories, Burlingame, CA). Peroxidase activity was visualized using the AEC substrate kit as suggested by the supplier (Vector Laboratories). Sections were counterstained with hematoxylin solution and covered with Aquatex (Merck, Darmstadt, Germany). Primary antibodies used were rat monoclonal anti–Flk-1 antibody,32 rat monoclonal anti–PECAM-1 (Mec13.3),33 and rat monoclonal anti–Tie-2 (4G8; Silenus Laboratories, Melbourne, Australia). A light microscope (Axiophot; Zeiss, Oberkochen, Germany) with digital camera HC-300Z (Fuji, Düsseldorf, Germany) was used for documentation. Objectives used, as well as image acquisition and processing, were as described for immunofluorescence staining. HIFdn-FLAG was detected by immunohistochemistry using the Mouse-on-Mouse Kit (Vector Laboratories). The mouse monoclonal anti-FLAG M2 antibody (Sigma, St Louis, MO) was used as primary antibody (1:500 dilution). Peroxidase reaction was performed using the AEC kit.
Whole-mount immunohistochemistry
Embryos were fixed in 4% paraformaldehyde/PBS at 4°C. The embryos were dehydrated in methanol, bleached in 5% H2O2, and then stepwise rehydrated to PBS. After blocking with 3% nonfat dry milk powder/0.1% Triton X-100 in PBS (PBSMT), embryos were incubated with rat monoclonal anti–PECAM-1 antibody (Mec13.3, diluted 1:10 in PBSMT). Following washing in PBSMT, the embryos were incubated with the secondary antibody (biotinylated goat anti–rat IgG, [Vector Laboratories] diluted 1:200). Embryos were then incubated with the ABC complex (Vector Laboratories) and, for color development, incubated in AEC solution according to the instructions of the manufacturer (Vector Laboratories). Embryos were postfixed in 2% paraformaldehyde/0.1% glutaraldehyde at 4°C. Then they were cleared in 50% and 70% glycerol before photographic documentation using a dissecting microscope (Stemi SV11; Zeiss) with a digital camera (DSC-S75; Sony, Cologne, Germany). A Zeiss Plan Achromat S 1.0 ×/0.45 numeric aperture (NA) objective was used. Images were acquired and processed using Adobe Photoshop 5 software (Adobe Systems, Munich, Germany).
Immunofluorescence staining
HUE cells were seeded on glass coverslips and transiently transfected with pcDNA3/HIF-2α-FLAG or pcDNA3/HIFdn-FLAG using Superfect transfection reagent (Qiagen). Forty-eight hours after transfection, the cells were fixed and stained using the mouse monoclonal anti-FLAG M2 antibody (diluted 1:500; Sigma). As secondary antibody the goat anti–mouse Alexa594 antibody (Molecular Probes, Leiden, Netherlands) was used in a 1:2000 dilution. The cells were mounted in Mowiol (Merck) and analyzed with an epifluorescence microscope (Axiophot; Zeiss) equipped with the following Zeiss Epiplan Neofluar objectives: 10 ×/0.25 NA, 20 ×/0.5 NA, 50 × oil/1.0 NA, and 100 × oil/1.3 NA. For image acquisition and processing, a Zeiss Axiocam digital camera and Axiovision (Zeiss) or Adobe Photoshop 5 software were used.
Results
A deletion mutant of HIF-2α was generated (Figure 1A) that lacked the C-terminal transactivation domains, the oxygen-dependent destruction domain (ODD), and part of the N-terminal DNA binding domain.17 To test this mutant functionally in vitro, HIF-1– or HIF-2–responsive promoters fused to the luciferase reporter gene were stimulated by transient overexpression of the respective HIF-α factors in HEK293 cells. The basal activity of the VEGF promoter and of the Flk1 promoter was significantly reduced by HIFdn expression. Expression of HIF-1α resulted in a 2-fold stimulation of the VEGF promoter, and coexpression of HIFdn completely inhibited this effect (Figure 1B). The Flk1 promoter was stimulated by HIF-2α as previously shown.16,17 Coexpression of the HIFdn mutant led to a 56% reduction of promoter stimulation (Figure 1C). Transfection of larger amounts of HIFdn plasmid did not decrease Flk1 promoter activity any further (data not shown). The same effects were observed in transfected human endothelial cells (Figure 1D). These experiments showed that HIFdn blocks both HIF-1– and HIF-2–dependent gene expression in a dominant-negative manner. Because the PAS domain is preserved in the HIFdn mutant, it is likely that HIFdn, like HIF-2α, forms heterodimers with ARNT. A similar dominant-negative HIF-1α construct was shown previously to dimerize with ARNT and to block normal HIF-1 DNA binding.34 Sequestering ARNT in the cytoplasm would inhibit the formation of functional HIF-1 and HIF-2 heterodimers and consequently block HIF-1 and HIF-2 function. To determine the cellular localization of HIF-2α and HIFdn, HUE cells were transiently transfected with HIF-2α or HIFdn expression vectors. Immunofluorescence for the FLAG epitope revealed that HIF-2α was exclusively located in the nuclei of transfected cells, whereas HIFdn was distributed in the cytoplasm, most likely because of the deletion of a nuclear localization signal (Figure 1E).
Structure and function of HIFdn in vitro. (A) Domain structure of HIF-2α and HIFdn. (B-D) Luciferase reporter gene assays. HEK293 cells were transfected with VEGF promoter-luc (B) or Flk1 promoter-luc (C) reporter constructs. HUE cells were transfected with KDR promoter-luc reporter gene (D). Expression of HIFdn reduced the basal activity of these promoters and inhibited the stimulatory effects of HIF-1α or HIF-2α, respectively. (E) Overexpression of HIF-2α or HIFdn in HUE cells and immunofluorescence staining against the FLAG epitope revealed nuclear localization of HIF-2α, whereas HIFdn was detected in the cytoplasm. Bar indicates 25 μm. #P < .05; *P < .03; **P < .005, unpaired 2-tailed t test.
Structure and function of HIFdn in vitro. (A) Domain structure of HIF-2α and HIFdn. (B-D) Luciferase reporter gene assays. HEK293 cells were transfected with VEGF promoter-luc (B) or Flk1 promoter-luc (C) reporter constructs. HUE cells were transfected with KDR promoter-luc reporter gene (D). Expression of HIFdn reduced the basal activity of these promoters and inhibited the stimulatory effects of HIF-1α or HIF-2α, respectively. (E) Overexpression of HIF-2α or HIFdn in HUE cells and immunofluorescence staining against the FLAG epitope revealed nuclear localization of HIF-2α, whereas HIFdn was detected in the cytoplasm. Bar indicates 25 μm. #P < .05; *P < .03; **P < .005, unpaired 2-tailed t test.
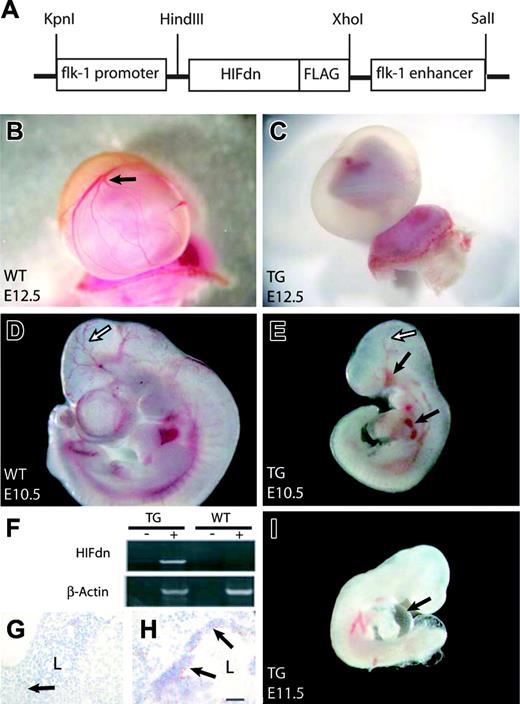
To investigate the potential effects of blocking HIF activity in the developing vascular system, we generated transgenic mice that expressed HIFdn in an endothelial cell–specific manner. A cDNA for HIFdn was fused to promoter/enhancer elements of the murine Flk1 gene (Flk1 p/e; Figure 2A). The Flk1 p/e directs the expression of transgenes specifically to the endothelium in mouse embryos starting at around E8.0.16,35,36 Due to embryonic lethality, transgenic mouse lines could not be generated. We therefore analyzed embryos that developed from microinjected oocytes. In total, 63 transgenic embryos were dissected between E10.5 and E14.5. About 30% (19 of 63) of the Flk1 p/e–HIFdn transgenic embryos were growth retarded and showed severe vascular disorganization. Most transgenic embryos were alive and had beating hearts at E10.5. We focused our analysis on embryos at E10.5 and E11.5, because embryonic lethality increased at later developmental stages. Transgene expression was confirmed by RT-PCR using a specific primer for the FLAG tag that allowed us to distinguish between transgenic HIFdn and endogenous HIF-2α expression (Figure 2F). Immunohistochemistry for the FLAG epitope revealed transgene expression in blood vessels (Figure 2G-H). As expected, not all transgenics expressed HIFdn, most likely because the transgene was integrated in transcriptionally inactive regions of the genome. Transgene expression correlated with the observed phenotype, whereas nonexpressers showed no morphologic abnormalities (data not shown). Henceforth, the term “HIFdn transgenic embryos” is used to refer to the Flk1 p/e–HIFdn transgenic embryos that displayed the morphologic abnormalities and expressed the transgene.
Generation of Flk1 p/e–HIFdn transgenic embryos. (A) Expression vector for the generation of HIFdn transgenic mouse embryos. (B) Well-vascularized yolk sac of a wild-type embryo at E12.5 (arrow indicates branching vessels). (C) The yolk sac of a transgenic littermate was not perfused and lacked large vessels. (D) Wild-type embryo at E10.5 with well-organized cephalic vascular tree (arrow). (E) HIFdn transgenic embryos at E10.5 were growth retarded, had a disorganized vasculature (open arrow), and showed hemorrhages (filled arrows). (F) In transgenic embryos, HIFdn was detected by RT-PCR in the RT reaction (+) but not in control reaction without reverse transcriptase (–). (G-H) FLAG-tagged HIFdn was detected by immunohistochemistry in blood vessels of transgenics (H, arrows) but not in vessels of wild-type embryos (G, arrow). L indicates vessel lumen; bar, 50 μm. (I) Transgenic embryos at E11.5 displayed pericardial edema (arrow). Magnification: 8 × (B,C), 12 × (D,E,I).
Generation of Flk1 p/e–HIFdn transgenic embryos. (A) Expression vector for the generation of HIFdn transgenic mouse embryos. (B) Well-vascularized yolk sac of a wild-type embryo at E12.5 (arrow indicates branching vessels). (C) The yolk sac of a transgenic littermate was not perfused and lacked large vessels. (D) Wild-type embryo at E10.5 with well-organized cephalic vascular tree (arrow). (E) HIFdn transgenic embryos at E10.5 were growth retarded, had a disorganized vasculature (open arrow), and showed hemorrhages (filled arrows). (F) In transgenic embryos, HIFdn was detected by RT-PCR in the RT reaction (+) but not in control reaction without reverse transcriptase (–). (G-H) FLAG-tagged HIFdn was detected by immunohistochemistry in blood vessels of transgenics (H, arrows) but not in vessels of wild-type embryos (G, arrow). L indicates vessel lumen; bar, 50 μm. (I) Transgenic embryos at E11.5 displayed pericardial edema (arrow). Magnification: 8 × (B,C), 12 × (D,E,I).
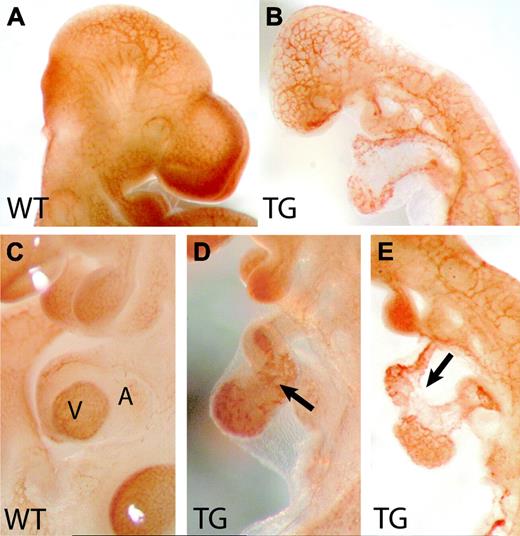
Vascular disorganization in Flk1 p/e–HIFdn transgenic mice. Whole-mount immunohistochemistry for PECAM-1. (A) Wild-type embryo at E10.5 with well-developed vessel system. (B) The cephalic vasculature in transgenic littermates remained a primitive plexus. (C) Higher magnification of a wild-type heart. V indicates ventricle; A, atrium. (D-E) Transgenic hearts were not properly looped but remained linear tubes. The endocardium was patchy and irregularly shaped (arrows). Magnification: 32 × (A,B), 50 × (C,D,E).
Vascular disorganization in Flk1 p/e–HIFdn transgenic mice. Whole-mount immunohistochemistry for PECAM-1. (A) Wild-type embryo at E10.5 with well-developed vessel system. (B) The cephalic vasculature in transgenic littermates remained a primitive plexus. (C) Higher magnification of a wild-type heart. V indicates ventricle; A, atrium. (D-E) Transgenic hearts were not properly looped but remained linear tubes. The endocardium was patchy and irregularly shaped (arrows). Magnification: 32 × (A,B), 50 × (C,D,E).
Whereas wild-type embryos had a well-perfused and hierarchically structured yolk sac vasculature with branching vitelline vessels (Figure 2B), the yolk sac vasculature of HIFdn transgenic embryos was disarranged without large-caliber vessels and lacked perfusion (Figure 2C). The HIFdn transgenic embryos appeared pale and showed focal hemorrhage (Figure 2E). As compared with wild-type littermates (Figure 2D), they were severely growth retarded (Figure 2E,I). The intraembryonic vasculature appeared overall disarranged, especially in the cephalic region where the branches of the internal carotid artery were not visible (Figure 2E). Moreover, in most transgenic embryos, pericardial edema was detectable (Figure 2I, arrow), indicating defects in the circulatory system. Whole-mount immunohistochemistry for PECAM-1 was performed to detect vascular structures at E10.5 (Figure 3). Whereas wild-type embryos displayed a hierarchically structured vascular tree in the head (Figure 3A), HIFdn transgenic embryos had a primitive, meshwork-like vasculature (Figure 3B), indicating a failure in the remodeling of the primitive vascular plexus of the head. The major embryonic vessels, like the dorsal aorta and the cardinal vein, were formed. Also, the intersomitic vessels were established in wild-type as well as in HIFdn transgenic embryos at this stage (data not shown). Heart development in HIFdn transgenic mouse embryos was retarded. Only a primitive linear heart tube was observed (Figure 3D-E), whereas wild-type littermates had an S-shaped heart tube, bringing atria and ventricle into close proximity (Figure 3C). Moreover, PECAM-1 staining of the endocardial layer revealed a patchy and irregular pattern as compared with wild-type embryos (Figure 3D).
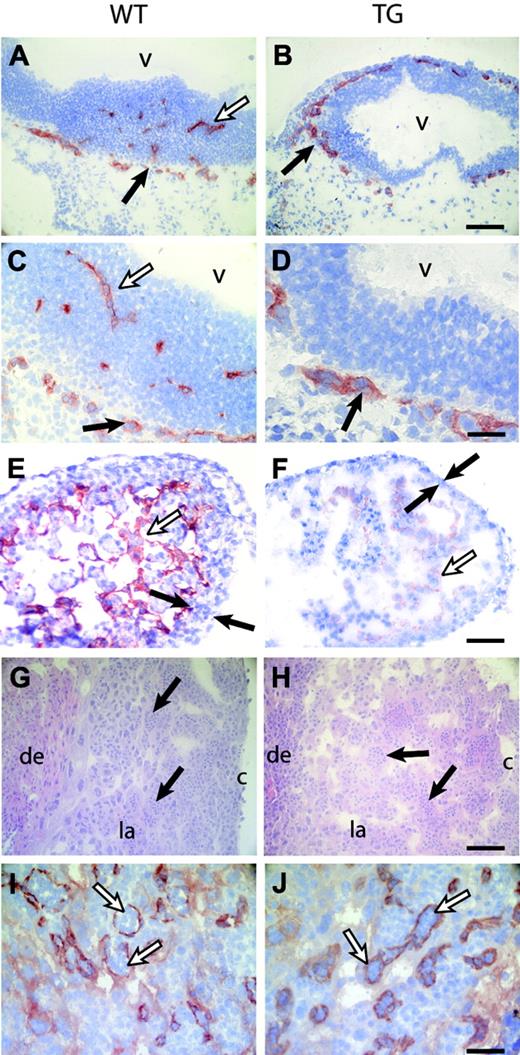
The vessels in the head were visualized by PECAM-1 or Flk-1 staining in wild-type animals (Figure 4A,C and data not shown). In HIFdn transgenics, the perineural vascular plexus (PNV) formed normally but failed to invade the neuropithelium (Figure 4B,D). Analysis of serial sections of whole embryos confirmed that the neuroepithelium of the brain and spinal cord remained avascular, demonstrating that sprouting angiogenesis did not occur (data not shown). The hearts of wild-type embryos developed a solid myocardium with multiple layers of cardiomyocytes. The inner surface of the ventricles showed muscular trabeculae covered with endocardial cells (Figure 4E). In contrast, hearts of HIFdn transgenic embryos had a very thin myocardium and poorly established trabeculation (Figure 4F). Whereas the endocardial layer of wild-type embryos was regularly shaped, HIFdn transgenic embryos showed only disorganized islands of PECAM-1–positive cells (Figure 4E,F).
Defects in angiogenesis and heart development in Flk1 p/e–HIFdn transgenic embryos. Immunohistochemistry for PECAM-1 (except for panels G-H). (A-D) Sections of the head region at E10.5. (A,C) The PNV was formed in wild-type embryos (filled arrows), and vessels invaded the neuroepithelium (open arrows). (B,D) In transgenic embryos, the PNV was formed (arrows) but failed to sprout into the neural tissue. (E) Ventricles of the heart consisted of multiple layers of myocytes (filled arrows) and trabeculae lined with endothelial cells (open arrow) in wild-type embryos at E11.5. (F) HIFdn transgenic embryos had a thin myocard (filled arrows), poorly established trabeculation, and disorganized endocardium (open arrow). (G-H) Hematoxylin and eosin (H+E) staining of placentae at E10.5. Embryonic vessels were filled with primitive nucleated erythrocytes (arrows). (I-J) PECAM-1 staining revealed endothelial cells of embryonic vessels in the labyrinthine layer (arrows). The degree of vascularization was equal in wild-type and transgenic placentae. v indicates brain ventricle; de, maternal deciduas; la, labyrinthine layer; c, chorionic plate. Bars in panels B,F,H, 100 μm; panels D,J, 50 μm. Magnification in panels A, C, E, G, and I is the same as in panels B, D, F, H, and J, respectively.
Defects in angiogenesis and heart development in Flk1 p/e–HIFdn transgenic embryos. Immunohistochemistry for PECAM-1 (except for panels G-H). (A-D) Sections of the head region at E10.5. (A,C) The PNV was formed in wild-type embryos (filled arrows), and vessels invaded the neuroepithelium (open arrows). (B,D) In transgenic embryos, the PNV was formed (arrows) but failed to sprout into the neural tissue. (E) Ventricles of the heart consisted of multiple layers of myocytes (filled arrows) and trabeculae lined with endothelial cells (open arrow) in wild-type embryos at E11.5. (F) HIFdn transgenic embryos had a thin myocard (filled arrows), poorly established trabeculation, and disorganized endocardium (open arrow). (G-H) Hematoxylin and eosin (H+E) staining of placentae at E10.5. Embryonic vessels were filled with primitive nucleated erythrocytes (arrows). (I-J) PECAM-1 staining revealed endothelial cells of embryonic vessels in the labyrinthine layer (arrows). The degree of vascularization was equal in wild-type and transgenic placentae. v indicates brain ventricle; de, maternal deciduas; la, labyrinthine layer; c, chorionic plate. Bars in panels B,F,H, 100 μm; panels D,J, 50 μm. Magnification in panels A, C, E, G, and I is the same as in panels B, D, F, H, and J, respectively.
Because defective placental vascularization in Arnt–/– embryos has been reported,37,38 we analyzed the placenta of HIFdn embryos. Embryonic vessels that contained nucleated primitive erythrocytes invaded the maternal tissue (Figure 4G,I). The transgenic placentas were well vascularized and showed no obvious defect in the connection of embryonic and maternal vessels in the labyrinthine layer (Figure 4H,J).
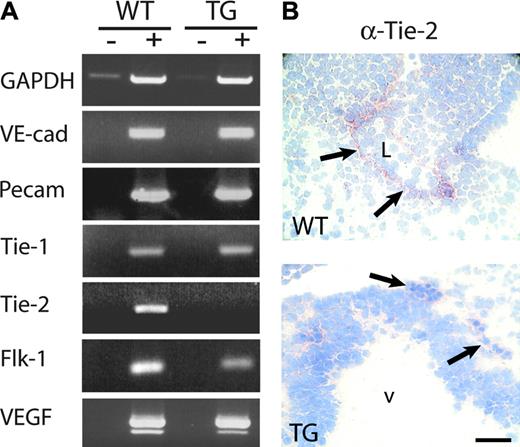
To identify the causes of the observed phenotypic abnormalities and the endothelial target genes affected by HIF inhibition, we prepared RNA from wild-type and HIFdn transgenic embryos and performed semiquantitative RT-PCR for endothelial marker genes. RT-PCR for GAPDH (25 cycles) confirmed that equal amounts of wild-type and transgenic cDNA were used (Figure 5A). No difference in the expression levels of VE-cadherin, Tie-1, and PECAM-1 mRNA were observed between wild-type and transgenic mouse embryos. This indicates that the number of endothelial cells was not reduced in HIFdn transgenic mouse embryos. In contrast, Tie-2 mRNA was greatly reduced in transgenic mouse embryos (Figure 5A) (n = 4). The expression of Flk-1 mRNA was also down-regulated in 4 of 7 transgenic embryos investigated. This indicates that the endothelial cell–specific expression of HIFdn had specific effects on gene expression and did not impair the formation of the primitive endothelium. Immunohistochemistry confirmed the results of the RT-PCR analysis (Figure 5B). While wild-type embryos expressed Tie-2 in endothelial cells, no staining was found in blood vessels of transgenic embryos (Figure 5B). The mRNA levels for the respective receptor ligands (which are expressed predominantly by nonendothelial cells) were also examined. As expected, VEGF expression levels were unaltered in HIFdn transgenic embryos (Figure 5A), because VEGF is mainly expressed by nonendothelial cells in vivo.39,40 Angiopoietin-1 transcript levels were also similar in wild-type and transgenic embryos (data not shown).
Reduced Tie-2 expression in Flk1 p/e–HIFdn transgenic embryos. (A) RNA was isolated from wild-type (WT) and transgenic (TG) embryos at E10.5. RT-PCR (+) and a control reaction without RT (–) were performed. No difference in the transcription levels of VE-cadherin, PECAM-1, Tie-1, or VEGF was observed. Flk-1 mRNA was reduced in transgenic embryos, and Tie-2 was below detection level after 35 PCR cycles (n = 4). (B) Immunohistochemistry of embryo sections. Tie-2 was detectable in endothelial cells of wild-type embryos (arrows, top panel) but was absent from vessels in transgenic littermates (arrows, bottom panel). v indicates brain ventricle; L, lumen. Bar in panel B, 50 μm.
Reduced Tie-2 expression in Flk1 p/e–HIFdn transgenic embryos. (A) RNA was isolated from wild-type (WT) and transgenic (TG) embryos at E10.5. RT-PCR (+) and a control reaction without RT (–) were performed. No difference in the transcription levels of VE-cadherin, PECAM-1, Tie-1, or VEGF was observed. Flk-1 mRNA was reduced in transgenic embryos, and Tie-2 was below detection level after 35 PCR cycles (n = 4). (B) Immunohistochemistry of embryo sections. Tie-2 was detectable in endothelial cells of wild-type embryos (arrows, top panel) but was absent from vessels in transgenic littermates (arrows, bottom panel). v indicates brain ventricle; L, lumen. Bar in panel B, 50 μm.
Discussion
Blood vessel growth in response to hypoxia is mediated by HIFs. In addition to hypoxia, growth factor signaling via MAP kinases leads to the stabilization and activation of HIF. HIF-1 stimulates the expression of VEGF and other potent angiogenic factors.41,42 Endothelial cells respond to hypoxia by up-regulating Tie-2 and VEGF receptors.43-46 Prime candidates for intrinsic regulators of angiogenesis are HIF-1, which regulates endothelial genes in response to hypoxia,18 and HIF-2, which stimulates Tie2 and Flk1 promoter activity in vitro.7,15-17 However, the endothelial cell–specific ablation of HIF-1α did not interfere with normal vascular development, whereas it impaired tumor angiogenesis in adult mice.22 Similarly, because HIF-2α null embryos did not show consistent vascular abnormalities,23-26 an endothelial cell intrinsic function of HIF-2 during development appeared unlikely. We suspected that the loss of HIF-1 function can be partially compensated by HIF-2 and vice versa. To circumvent the problem of functional redundancy, we designed a dominant-negative approach that interferes with the endothelial activity of both HIF-1 and HIF-2. The HIFdn mutant used in our study was highly specific because it interfered with HIF target genes known from in vitro studies but did not with other endothelial markers, such as PECAM-1, Tie-1, and VE-cadherin.
HIFdn transgenic embryos showed multiple cardiovascular defects. Endothelial cells and primitive vessels were formed, but the vascular network remained immature and disorganized, indicating that the initial phase of vessel formation (vasculogenesis) was not disturbed. The absence of a vasculogenesis phenotype was expected because the Flk1 p/e elements used in our study direct HIFdn transgene expression to committed endothelial cells but are not active in progenitor cells of the hemangioblastic lineage.16,35 Whether HIFs have a cell-autonomous function in endothelial progenitor commitment/proliferation during vasculogenesis remains to be determined.
The observed phenotypic abnormalities in transgenic HIFdn mouse embryos included defects in yolk sac circulation, sprouting angiogenesis, and vascular remodeling, especially in the head region. Also, heart development was impaired because the endocardium was disarranged, the ventricle walls were thin, and trabeculae were poorly formed. Because placental vascularization was not impaired by the inhibition of endothelial HIF activity, it can be excluded that the observed embryonic phenotype is secondary to defective placentation, as it was observed in Arnt-null mice.37,38 The inhibition of HIF activity in endothelial cells had a less severe effect on vascular development and embryonic survival than the loss of HIF-1α expression alone in all cells, which results in embryonic lethality by E10.5,20 confirming that there is also a requirement for HIF-1 in nonendothelial cells for normal vascular development to occur.
The defects observed in HIFdn transgenic embryos showed remarkable similarities to the phenotypes of Tie2 knock-out mice. Tie-2 is an essential receptor tyrosine kinase expressed by endothelial cells during vascular morphogenesis. Tie2 null mice display defects in vascular remodeling, sprouting angiogenesis, and ventricular trabeculation. These malformations might result from defective perivascular cell recruitment47,48 and/or endothelial cell survival.47,49,50 Tie-2 serves as receptor for angiopoietin-1 (Ang-1) and angiopoietin-2. The observation that Tie2 and Ang1 null mice display similar phenotypes supports the hypothesis that Ang-1, via binding to and activating Tie-2, regulates reciprocal interactions between perivascular cells and endothelial cells.51 Thus, it appears that the large reduction of Tie-2 expression is a primary cause of the phenotype in HIFdn transgenic mouse embryos. Our results show that endothelial HIF activity is required for Tie2 expression in vivo. This was not directly shown in either of the Hif1a, Hif2a, or Arnt knock-out mice because gene inactivation in these mice was not tissue specific and the embryos died because of nonvascular defects like mesenchymal cell death or defective placenta formation.
Flk-1 mRNA expression was reduced but not completely down-regulated in approximately half of the HIFdn transgenic embryos examined. Flk-1 expression is induced by hypoxia.43 We have shown that the Flk1 promoter is stimulated by HIF-2α and, to a lesser extent, by HIF-1α16,17 and that HIF-2α cooperates with the transcription factor Ets-1 to transactivate Flk1. Other transcriptional regulators of Flk1 expression include SCL/Tal1, GATA factors,16,52 and HoxB5.53 The absence of HIF-1 or HIF-2 in such a transcriptional complex causes only a moderate reduction of Flk1 promoter transactivation (Figure 1). The observation that Tie-2 is completely down-regulated in transgenic HIFdn embryos argues against the possibility that the expression levels of the HIFdn transgene were not high enough to block Flk-1 transcription completely.
In conclusion, we provide direct functional evidence that HIF activity in the developing endothelium is essential for embryonic cardiovascular development. Although endothelial HIFs control a broad spectrum of genes in endothelial cells,18 a particularly critical target is the essential endothelial receptor tyrosine kinase, Tie-2.
Prepublished online as Blood First Edition Paper, September 27, 2005; DOI 10.1182/blood-2005-07-3033.
Supported by grants of the Deutsche Forschungsgemeinschaft (DFG-Br 1336 [G.B.] and DFG-Fl223 [I.F.]) and of the Bundesministerium für Bildung und Forschung (G.B.).
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Dr Anke Nicolaus for helpful discussions.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal