Abstract
The Sleeping Beauty (SB) transposon system is a nonviral DNA delivery system in which a transposase directs integration of an SB transposon into TA-dinucleotide sites in the genome. To determine whether the SB transposon system can mediate stable gene expression in human T cells, primary peripheral blood lymphocytes (PBLs) were nucleofected with SB vectors carrying a DsRed reporter gene. Plasmids containing the SB transposase on the same molecule as (cis) or on a molecule separate from (trans) the SB transposon mediated long-term and stable reporter gene expression in human primary T cells. Sequencing of transposon:chromosome junctions confirmed that stable gene expression was due to SB-mediated transposition. In other studies, PBLs were successfully transfected using the SB transposon system and shown to stably express a fusion protein consisting of (1) a surface receptor useful for positive T-cell selection and (2) a “suicide” gene useful for elimination of transfected T cells after chemotherapy. This study is the first report demonstrating that the SB transposon system can mediate stable gene transfer in human primary PBLs, which may be advantageous for T-cell–based gene therapies.
Introduction
Genetic modification of peripheral blood lymphocytes (PBLs) or hematopoietic stem cells (HSCs) has been shown to be promising in the treatment of cancer,1,2 transplant complications,3 viral infections,2,4 and immunodeficiencies.5 It also holds great promise in the study of T-cell biology.6 In addition, redirecting T-cell specificity via chimeric receptors for tumor antigens or T-cell receptor (TCR) gene transfer has offered great potential for immunotherapy of cancer.1,2,7,8
Most protocols involving gene transfer into T cells use oncoretroviral vectors that require cellular replication for vector integration into host chromosomes. Although T cells can be induced to proliferate ex vivo, it has been shown that extended ex vivo culture may alter biologic properties of T cells, including CD4+/CD8+ T-cell ratio, TCR repertoire, and cytokine secretion.9-12 In addition, methylation of transcriptional regulatory sequences can occur in oncoretroviral proviruses, resulting in reduced or silenced transgene expression.13
Recently, attention has been focused on HIV-1–derived lentiviruses, which have been shown to transduce a variety of slowly or nondividing cells, including unstimulated T cells.14-17 However, the procedure required to produce high-titer lentiviral vectors remains complex and costly. Furthermore, both oncoretroviral and lentiviral vector integration show a preference for transcriptionally active genes, including proto-oncogenes and signaling genes.18-22 Insertional mutagenesis has become a serious concern after 3 of 20 patients with X-linked severe combined immunodeficiency developed an acute leukemia-like syndrome following infusion of genetically modified HSCs using oncoretroviral vectors.19,23
Genetic engineering of human T cells with plasmid DNA has been investigated for the treatment of B-cell leukemia and lymphoma.24-26 However, there were several limitations of this approach: (1) the T cells required activation prior to gene transfer; (2) random integration after electroporation was of low efficiency; (3) coexpressed drug resistance genes in T cells are immunogenic and thus their efficacy for in vivo applications is unknown; and (4) this procedure requires at least 1.5 months and is labor intensive.
The Sleeping Beauty (SB) transposon system recently has been used as a promising tool for high-level, persistent transgene expression that can be delivered in a nonviral plasmid form.27-29 SB is a synthetic DNA transposon of the Tc1/mariner superfamily that was “reawakened” by Hackett and colleagues through molecular reconstruction and mediates DNA transfer via a “cut-and-paste” mechanism.27 SB transposase mediates transposition by recognition of the short inverted/direct repeat (IR/DR) sequences of the transposon, resulting in the excision of the transposon and insertion into a TA-dinucleotide sequence in chromosomal DNA. The SB transposase has an N-terminal DNA-binding domain, a nuclear localization signal, and a C-terminal catalytic domain that is characterized by the DDE motif (containing 2 invariable aspartic acid residues and a glutamic acid residue) and is responsible for excision and subsequent integration of transposon DNA.27-29 The SB transposase can be introduced by expression of a transposase-encoding DNA molecule either on the same DNA molecule as the one containing the SB transposon (cis delivery) or on a separate DNA molecule (trans delivery). SB transposons have been shown to mediate transposition and long-term expression in a wide range of vertebrate cells and tissues, including cultured mammalian cells,27,30,31 mouse liver and lung tissues,32,33 mouse embryonic stem cells,34 and in mammalian germ-line transgenesis and insertional mutagenesis.35
Because of SB's capability to bring about stable DNA-mediated gene transfer into a variety of cells and tissues, there has been considerable interest in its potential use in gene therapy applications. However, up to this point, the use of SB for stable gene transfer and expression in primary human cell populations has not been demonstrated. Here we demonstrate that the SB transposon system can effectively mediate stable gene transfer in primary human T cells. These results demonstrate that the SB transposon system represents a viable approach to accomplishing nonviral, DNA-mediated gene transfer in primary human T cells and have implications for experimental and therapeutic application of this approach in these and other primary human cellular gene transfer targets.
Materials and methods
SB transposons and transposase-encoding plasmids
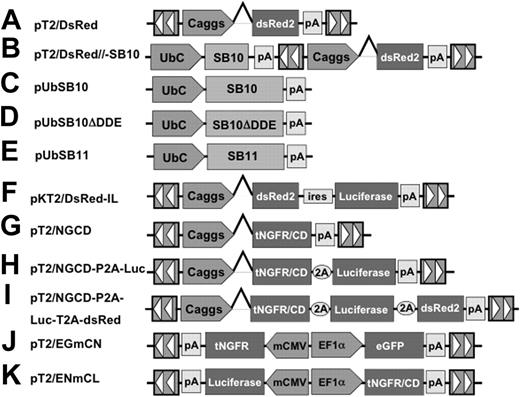
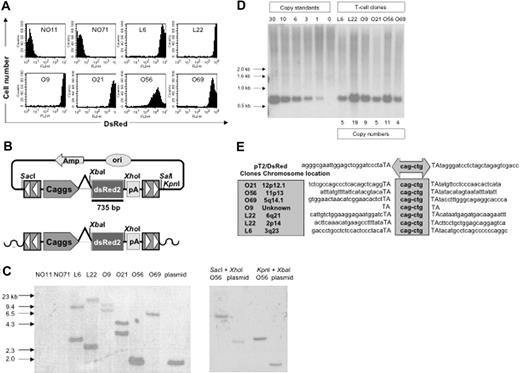
SB transposons (Figures 1A-K) were constructed using standard molecular cloning techniques and the IR/DR sequences previously described.36 pT2/DsRed (Figure 1A) is an SB transposon trans vector in which the Discosoma sp DsRed reporter gene (BD Biosciences, Mountain View, CA) is transcriptionally regulated by the cytomegalovirus (CMV) enhancer/chicken β-actin promoter/β-globin intron fusion sequence (Caggs).37 pT2/DsRed//-SB10 (Figure 1B) is an SB cis vector in which both the DsRed reporter gene and SB10 transposase are contained on the same plasmid. pUbSB10 (Figure 1C) contains the human ubiquitin (Ub) promoter (Invitrogen, Carlsbad, CA) regulating transposase expression. pUbSB10ΔDDE (Figure 1D) encodes an inactive SB transposase due to deletion of the DDE catalytic domain.27 pUbSB11 (Figure 1E) encodes an improved transposase in which a few transposase amino acids were modified to enhance transposition efficiency.30 pKT2/DsRed-IL (Figure 1F) is a bicistronic SB (pT2) trans vector containing the DsRed coding sequence, the encephalomyocarditis virus (ECMV) internal ribosome entry site (IRES), and the firefly luciferase (Luc) (Promega, Madison, WI) coding sequence.55 pT2/NGCD (Figure 1G) is an SB trans vector encoding a truncated human nerve growth factor receptor (tNGFR)/yeast cytosine deaminase (CD) fusion gene termed NGCD.38 pT2/NGCD-P2A-Luc (Figure 1H) and pT2/NGCD-P2A-Luc-T2A-DsRed (Figure 1I) were bicistronic and tricistronic vectors using the 2A peptide.39,40 P2A and T2A are “self-cleaving” 2A peptides derived from porcine teschovirus-1 and Thosea asigna virus that have been shown to express multiple genes.41 The SB transposon vectors pT2/EGmCN (Figure 1J) and pT2/ENmCL (Figure 1K) contain synthetic bidirectional promoters regulating dual gene expression. The minimal CMV promoter was obtained from an HIV-1–derived lentiviral vector42 (generously provided by Dr Luigi Naldini, San Raffaele Telethon Institute for Gene Therapy, Milan, Italy), and the human elongation factor 1α promoter (EF1α) was obtained from lentiviral vector pEF1α GFP.17
The Sleeping Beauty (SB) transposon vectors used in this study. The SB transposon system consists of 2 components: the inverted repeat/direct repeats (IR/DR, indicated by arrowheads) flanking the gene of interest, and the expression cassette encoding the transposase. Caggs is a chimeric promoter derived from chicken β-actin and cytomegalovirus immediate-early promoter sequences. UbC indicates human ubiquitin C promoter; SB10, transposase; SB10ΔDDE, inactive transposase due to deletion of the catalytic domain; tNGFR/CD, truncated human nerve growth factor receptor and cytosine deaminase fusion gene; P2A and T2A, “self-cleaving” 2A peptides derived from porcine teschovirus-1 and Thosea asigna virus have been shown to express multiple genes. pT2/EGmCN and pT2/ENmCL were bidirectional SB vectors in which synthetic bidirectional promoters were used. mCMV indicates minimal CMV promoter; EF1α, human elongation factor 1α promoter; IRES, internal ribosomal entry sites; DsRed2, red fluorescent protein; Luciferase, bioluminescent reporter gene; and pA, polyadenylation signal.
The Sleeping Beauty (SB) transposon vectors used in this study. The SB transposon system consists of 2 components: the inverted repeat/direct repeats (IR/DR, indicated by arrowheads) flanking the gene of interest, and the expression cassette encoding the transposase. Caggs is a chimeric promoter derived from chicken β-actin and cytomegalovirus immediate-early promoter sequences. UbC indicates human ubiquitin C promoter; SB10, transposase; SB10ΔDDE, inactive transposase due to deletion of the catalytic domain; tNGFR/CD, truncated human nerve growth factor receptor and cytosine deaminase fusion gene; P2A and T2A, “self-cleaving” 2A peptides derived from porcine teschovirus-1 and Thosea asigna virus have been shown to express multiple genes. pT2/EGmCN and pT2/ENmCL were bidirectional SB vectors in which synthetic bidirectional promoters were used. mCMV indicates minimal CMV promoter; EF1α, human elongation factor 1α promoter; IRES, internal ribosomal entry sites; DsRed2, red fluorescent protein; Luciferase, bioluminescent reporter gene; and pA, polyadenylation signal.
Human T-cell gene transfer
PBLs were obtained through Ficoll-Hypaque (Mediatech Cellgro, Herndon, VA) from buffy coat purchased from Memorial Blood Centers (Minneapolis, MN) or blood from healthy donors after informed consent. Peripheral blood mononuclear cells (PBMCs) were washed 2 times with phosphate-buffered saline (PBS) (Invitrogen) containing 0.5% bovine serum albumin (BSA) (Sigma, St Louis, MO). PBLs were transfected using an Amaxa Nucleofector device in combination with the human T-cell nucleofector kit according to the manufacturer's instructions (Amaxa, Gaithersburg, MD). Briefly, 100 μL of 5 × 106 PBLs mixed with plasmid was transferred to the cuvette provided with the kit and nucleofected. The cells were immediately transferred into 24-well plates containing 37°C prewarmed human T-cell medium consisting of RPMI-1640 (Invitrogen), 10% human serum, 10 mM HEPES, 2 mM l-glutamine, 50 U/mL penicillin, and 50 μg/mL streptomycin. Human serum was purchased from SeraCare Life Sciences (Oceanside, CA) or prepared in our laboratory from healthy HIV-1 seronegative donors' blood collected by the Blood Donor Center, University of Minnesota Medical Center, Fairview. All donors provided informed consent according to the Institutional Review Board–approved study.
After nucleofection, the cells were analyzed for transgene expression by fluorescence-activated cell sorting (FACS) Calibur (FACSCalibur; BD Biosciences) on day 1 or day 2 or activated by exposure to anti-CD3 and anti-CD28 beads (anti-CD3/28 beads)43 at 1:3 target-to-bead ratio in the presence of human interleukin-2 (IL-2) (hIL-2, 50 IU/mL, Chiron, Emeryville, CA). After activation for 3 to 5 days, the beads were removed. The activated T cells were maintained in human T-cell medium plus IL-2 (50 IU/mL) and IL-7 (10 ng/mL, R&D Systems, Minneapolis, MN) and restimulated once every 10 to 14 days with anti-CD3/28 beads or OKT3 (Ortho Biotech, Raritan, NJ) as previously described.43,44 Cell samples were periodically harvested for FACS analysis of transgene expression.
T-cell cloning
Southern hybridization analysis
Southern blotting was performed essentially as previously described.45 Briefly, 10 μg of genomic DNA extracted from each T-cell clone or 6 pg of the plasmid pT2/DsRed supplemented with 10 μg of the genomic DNA from DsRed– T-cell clone NO71 was digested with XbaI and SalI overnight and electrophoresed through 0.9% agarose gel. A 735-bp EcoRI fragment encoding the DsRed sequence was isolated from pT2/DsRed and radiolabeled by random priming for use as the DNA probe.
Transgene copy number analysis
DsRed transgene copies per genome equivalent were determined by Southern hybridization as described in “Southern hybridization analysis.” One copy number of the plasmid pT2/DsRed was calculated as described.46 Band intensities were quantified using a PhosphorImager (Molecular Dynamics, Sunnyvale, CA).
Hirt DNA isolation and analysis
Unintegrated low-molecular-weight DNA (Hirt DNA) was prepared as previously described.47 High-molecular-weight genomic DNA also was isolated from the same T cells using the Puregene DNA purification kit (Gentra Systems, Minneapolis, MN). Both Hirt DNA (500 μg) and genomic DNA (500 μg) were used to transform 25 μL of ElectroMAX DH10B cells (Invitrogen) by using a Bio-Rad electroporator. Colonies resulting from each bacterial transformation were quantified, and plasmid DNA was extracted from 20 transformants of each group, if any, for further analysis.
PCR cloning of insertion sites
The splinkerette polymerase chain reaction (PCR) technique and linker sequences used to recover sequences flanking transposon inserts on the 3′ side were described previously.33,35 Genomic DNA (1 μg) from DsRed+ and DsRed– T-cell clones was isolated using the Puregene DNA purification kit, digested with BfaI, and ligated to the linker. Primary PCR was performed with primers 5′-gtaatacgactcactatagggc-3′ and 5′-ctggaattttccaagctgtttaaaggcacagtcaac-3′ under the following conditions: 94°C for 2 minutes, then 25 cycles of 94°C for 15 seconds, 60°C for 30 seconds, and 72°C for 90 seconds. The PCR products were diluted, and nested PCR was carried out under the same conditions using primers 5′-agggctccgcttaagggac-3′) and 5′-gacttgtgtcatgcacaaagtagatgtcc-3′. Nested products were separated by electrophoresis through 2% agarose gel. Specific bands were excised, and DNA was purified using the Qiaquick gel extraction kit (Qiagen, Valencia, CA), cloned into pGEM-T vector (Promega), transformed into ElectroMax DH10B (Invitrogen), and sequenced by the Advanced Genetics Analysis Center at the University of Minnesota. The resulting sequences were subjected to BlastN analysis against the human genome using the ENSEMBL database.
Flow cytometric analysis
A hybridoma secreting monoclonal antibody (mAb) against NGFR (clone 20.4, murine IgG1) was purchased from the American Type Culture Collection (Manassas, VA). Purified anti-NGFR mAb was produced by Taconic (Germantown, NY). mAbs including murine IgG1 isotype control and fluorescein isothiocyanate (FITC)–conjugated mouse IgG1, anti–human CD4, CD8, and CD19 were purchased from BD Biosciences. Goat anti–mouse IgG conjugated with FITC was purchased from Caltag (Burlingame, CA). Flow cytometric analysis was carried out on a FACSCalibur using CellQuest software (BD Biosciences).
Cytotoxicity assays
Cytosine deaminase function was determined in colorimetric cytotoxicity assays as previously described.38
Statistical analyses
Statistical analyses were conducted on the SAS 9.1 software (Cary, NC). The gene expression data (percent DsRed+ cells) were skewed, and therefore all analyses were completed on a log scale. The data were analyzed using a general linear mixed model to adjust for subject correlation.48 General linear mixed models were used for hypothesis testing and to produce covariate adjusted geometric means and 95% confidence intervals (CI) at each time point and dose level. All hypothesis tests were 2-sided and adjusted for multiple comparisons using the Bonferroni multiple comparisons adjustment (Tables 1-2). The Wilcoxon rank sum test was used to compare the difference between control and experimental groups in Table 3.49 P values less than .05 were considered statistically significant.
Sequential analysis of SB cis vector-mediated gene expression in human T cells as related to DNA amounts used for nucleofection
. | pT2/DsRed . | . | pT2/DsRed/-SB10 . | . | . | ||
|---|---|---|---|---|---|---|---|
| Dose, by day . | Geometric mean (n = 10) . | 95% Cl . | Geometric mean (n = 10) . | 95% Cl . | P . | ||
| Day 21 | |||||||
| 10 μg | 0.58 | 0.28, 1.20 | 3.33 | 1.63, 6.80 | < .001 | ||
| 5 μg | 0.22 | 0.11, 0.44 | 1.62 | 0.79, 3.30 | < .001 | ||
| Day 28 | |||||||
| 10 μg | 0.20 | 0.11, 0.36 | 3.17 | 1.74, 5.80 | < .001 | ||
| 5 μg | 0.06 | 0.03, 0.11 | 1.26 | 0.69, 2.30 | < .001 | ||
. | pT2/DsRed . | . | pT2/DsRed/-SB10 . | . | . | ||
|---|---|---|---|---|---|---|---|
| Dose, by day . | Geometric mean (n = 10) . | 95% Cl . | Geometric mean (n = 10) . | 95% Cl . | P . | ||
| Day 21 | |||||||
| 10 μg | 0.58 | 0.28, 1.20 | 3.33 | 1.63, 6.80 | < .001 | ||
| 5 μg | 0.22 | 0.11, 0.44 | 1.62 | 0.79, 3.30 | < .001 | ||
| Day 28 | |||||||
| 10 μg | 0.20 | 0.11, 0.36 | 3.17 | 1.74, 5.80 | < .001 | ||
| 5 μg | 0.06 | 0.03, 0.11 | 1.26 | 0.69, 2.30 | < .001 | ||
Cl indicates confidence intervals.
Sequential analysis of SB trans vector-mediated gene expression in human T cells as related to DNA amounts used for nucleofection
pT2/DsRed +/- pUbSB10 . | . | . | . | |||
|---|---|---|---|---|---|---|
| Dose, by day . | Geometric mean (n = 8) . | 95% Cl . | P(vs 0 μg) . | |||
| Day 21 | ||||||
| 20 μg | 7.07 | 3.80, 13.16 | < .001 | |||
| 10 μg | 11.04 | 5.93, 20.56 | < .001 | |||
| 5 μg | 7.03 | 3.78, 13.09 | < .001 | |||
| 2.5 μg | 3.63 | 1.95, 6.76 | < .001 | |||
| 0 μg | 0.07 | 0.04, 0.13 | ||||
| Day 28 | ||||||
| 20 μg | 6.64 | 3.99, 11.03 | < .001 | |||
| 10 μg | 10.78 | 6.49, 17.92 | < .001 | |||
| 5 μg | 7.00 | 4.21, 11.63 | < .001 | |||
| 2.5 μg | 3.80 | 2.29, 6.32 | < .001 | |||
| 0 μg | 0.12 | 0.07, 0.20 | ||||
pT2/DsRed +/- pUbSB10 . | . | . | . | |||
|---|---|---|---|---|---|---|
| Dose, by day . | Geometric mean (n = 8) . | 95% Cl . | P(vs 0 μg) . | |||
| Day 21 | ||||||
| 20 μg | 7.07 | 3.80, 13.16 | < .001 | |||
| 10 μg | 11.04 | 5.93, 20.56 | < .001 | |||
| 5 μg | 7.03 | 3.78, 13.09 | < .001 | |||
| 2.5 μg | 3.63 | 1.95, 6.76 | < .001 | |||
| 0 μg | 0.07 | 0.04, 0.13 | ||||
| Day 28 | ||||||
| 20 μg | 6.64 | 3.99, 11.03 | < .001 | |||
| 10 μg | 10.78 | 6.49, 17.92 | < .001 | |||
| 5 μg | 7.00 | 4.21, 11.63 | < .001 | |||
| 2.5 μg | 3.80 | 2.29, 6.32 | < .001 | |||
| 0 μg | 0.12 | 0.07, 0.20 | ||||
Analysis of Hirt and genomic DNA isolated from SB transposon-nucleofected human primary T cells and clones
. | Total colonies . | No. of colonies analyzed . | pT2/DsRed . | pUbSB10 . | Unknown . |
|---|---|---|---|---|---|
| Hirt DNA | |||||
| Donor 1: pT2/DsRed, day 1 | 3.7 × 107* | 20 | 19 | 0 | 1 |
| Donor 1: pT2/DsRed + pUbSB10, day 1 | 6.7 × 107* | 20 | 8 | 12 | 0 |
| Donor 1: pT2/DsRed, day 7 | 2 × 105* | 20 | 18 | 0 | 2 |
| Donor 1: pT2/DsRed + pUbSB10, day 7 | 1 × 106* | 20 | 9 | 11 | 0 |
| Donor 1: pT2/DsRed + pUbSB10, sorted, more than 50 days | 0† | — | — | — | — |
| Donor 2: pT2/DsRed + pUbSB10, sorted, more than 50 days | 0† | — | — | — | — |
| T-cell clones | |||||
| NO71 | 0† | — | — | — | — |
| L6 | 2† | 2 | 0 | 1 | 1 |
| L22 | 2† | 0 | 0 | 0 | 2 |
| O9 | 4† | 0 | 0 | 0 | 4 |
| O21 | 19† | 0 | 0 | 0 | 19 |
| O56 | 1† | 0 | 0 | 0 | 1 |
| O69 | 4† | 0 | 0 | 1 | 3 |
| pUC19 control | 7.5 × 1010 | — | — | — | — |
| Genomic DNA | |||||
| Donor 1: pT2/DsRed, day 1 | 3.8 × 108‡ | 20 | 20 | 0 | 0 |
| Donor 1: pT2/DsRed + pUbSB10, day 1 | 3.4 × 108‡ | 20 | 11 | 9 | 0 |
| Donor 1: pT2/DsRed, day 7 | 2.5 × 105‡ | 20 | 20 | 0 | 0 |
| Donor 1: pT2/DsRed + pUbSB10, day 7 | 1.2 × 106‡ | 20 | 5 | 15 | 0 |
| Donor 1: pT2/DsRed + pUbSB10, sorted, more than 50 days | 0§ | — | — | — | — |
| Donor 2: pT2/DsRed + pUbSB10, sorted, more than 50 days | 0§ | — | — | — | — |
| T-cell clones | |||||
| NO71 | 1§ | 1 | 1 | 0 | 0 |
| L6 | 0§ | — | — | — | — |
| L22 | 0§ | — | — | — | — |
| O9 | 0§ | — | — | — | — |
| O21 | 0§ | — | — | — | — |
| O56 | 0§ | — | — | — | — |
| O69 | 0§ | — | — | — | — |
| pUC19 control | 1 × 1011 | — | — | — | — |
. | Total colonies . | No. of colonies analyzed . | pT2/DsRed . | pUbSB10 . | Unknown . |
|---|---|---|---|---|---|
| Hirt DNA | |||||
| Donor 1: pT2/DsRed, day 1 | 3.7 × 107* | 20 | 19 | 0 | 1 |
| Donor 1: pT2/DsRed + pUbSB10, day 1 | 6.7 × 107* | 20 | 8 | 12 | 0 |
| Donor 1: pT2/DsRed, day 7 | 2 × 105* | 20 | 18 | 0 | 2 |
| Donor 1: pT2/DsRed + pUbSB10, day 7 | 1 × 106* | 20 | 9 | 11 | 0 |
| Donor 1: pT2/DsRed + pUbSB10, sorted, more than 50 days | 0† | — | — | — | — |
| Donor 2: pT2/DsRed + pUbSB10, sorted, more than 50 days | 0† | — | — | — | — |
| T-cell clones | |||||
| NO71 | 0† | — | — | — | — |
| L6 | 2† | 2 | 0 | 1 | 1 |
| L22 | 2† | 0 | 0 | 0 | 2 |
| O9 | 4† | 0 | 0 | 0 | 4 |
| O21 | 19† | 0 | 0 | 0 | 19 |
| O56 | 1† | 0 | 0 | 0 | 1 |
| O69 | 4† | 0 | 0 | 1 | 3 |
| pUC19 control | 7.5 × 1010 | — | — | — | — |
| Genomic DNA | |||||
| Donor 1: pT2/DsRed, day 1 | 3.8 × 108‡ | 20 | 20 | 0 | 0 |
| Donor 1: pT2/DsRed + pUbSB10, day 1 | 3.4 × 108‡ | 20 | 11 | 9 | 0 |
| Donor 1: pT2/DsRed, day 7 | 2.5 × 105‡ | 20 | 20 | 0 | 0 |
| Donor 1: pT2/DsRed + pUbSB10, day 7 | 1.2 × 106‡ | 20 | 5 | 15 | 0 |
| Donor 1: pT2/DsRed + pUbSB10, sorted, more than 50 days | 0§ | — | — | — | — |
| Donor 2: pT2/DsRed + pUbSB10, sorted, more than 50 days | 0§ | — | — | — | — |
| T-cell clones | |||||
| NO71 | 1§ | 1 | 1 | 0 | 0 |
| L6 | 0§ | — | — | — | — |
| L22 | 0§ | — | — | — | — |
| O9 | 0§ | — | — | — | — |
| O21 | 0§ | — | — | — | — |
| O56 | 0§ | — | — | — | — |
| O69 | 0§ | — | — | — | — |
| pUC19 control | 1 × 1011 | — | — | — | — |
Hirt and genomic DNA were isolated as described in “Materials and methods” from transiently nucleofected PBLs on days 1 and 7, and from stably transposed bulk T cells as well as T-cell clones. Freshly isolated PBLs (120 × 106 cells) were nucleofected with pT2/DsRed (5 × 106 cells per 5 μg) or pT2/DsRed plus pUbSB10 (5 μg each) and incubated with human T-cell medium for 24 hours at 37°C. After FACS analysis to confirm DsRed expression, half of the transfected PBLs were washed once with PBS and used to isolate Hirt and genomic DNA, and the other half of the cells were cocultured with anti-CD3/28 beads for an additional 3 days at 37°C. Stable DsRed+ (> 95%) bulk T cells also were established using FACS sorting from 2 donors' PBLs 4 weeks after nucleofection with pT2/DsRed plus pUBSB10. The Wilcoxon rank sum test was used to analyze the difference between control (* or †) and experimental groups (‡ or §) (all P < .01).
— indicates not done.
Results
SB transposon–mediated gene transfer and long-term expression in human primary T cells
To test our hypothesis that the SB transposon can mediate integration and long-term transgene expression in human primary T cells, freshly isolated PBLs from healthy adult blood donors were first nucleofected with the SB transposon plasmid pT2/DsRed (Figure 1A) encoding a red fluorescent protein (DsRed) without the SB10 encoded transposase, or with pT2/DsRed//-SB10 (Figure 1B) containing both a DsRed transposon and an SB10 transposase coding sequence on the same molecule. We conducted a dose-response experiment in which different amounts (10, 5, 2.5, 1.2, and 0.6 μg) of pT2/DsRed or pT2/DsRed//-SB10 were nucleofected per 5 × 106 PBLs. A greater proportion of DsRed-positive cells (47.0%, 49.0%, 15.7%, 2.4%, and 1.0%, respectively) were observed on day 9 after transfection with pT2/DsRed than with pT2/DsRed//-SB10. However, this DsRed expression in the pT2/DsRed-transfected cells declined and became undetectable after 50 days in culture (data not shown), most likely due to dilution or loss of episomal plasmids during cellular division. Importantly, under the same culture conditions, DsRed expression (27.8%-45.1%) was maintained in cells transfected with pT2/DsRed//-SB10 at 50 days after transfection. DsRed expression also was observed at day 22 in T cells nucleofected with pT2/DsRed//-SB10, but not in those that were nucleofected with pT2/DsRed (data not shown). Under these conditions, the highest level of long-term gene expression (45.1%) was observed when T cells were nucleofected with 10 μg of pT2/DsRed//-SB10. Furthermore, when 3 independent PBL populations were nucleofected with 5, 10, or 20 μg of cis transposase-containing plasmid (pT2/DsRed//-SB10), the highest level (5.8%, 4.6%, and 27.4% for PBL2, PBL3, and PBL4, respectively) of stable gene transfer 38 days after nucleofection was observed for those cultures transfected with 10 μg rather than 20 μg (3.2%, 4.0%, and 1.8%) or 5 μg (0.6%, 2.7%, and 1.9%) of plasmid. Subsequent nucleofection experiments described in Tables 1, 2, 3 and Figures 2, 3, 4, 5, 6, 7 were carried out with 10 μg and 5 μg of plasmid to more thoroughly evaluate the efficiency of stable gene transfer and expression in human T cells.
Transposition in PBLs requires a functional transposase that can be delivered by cis or trans vectors
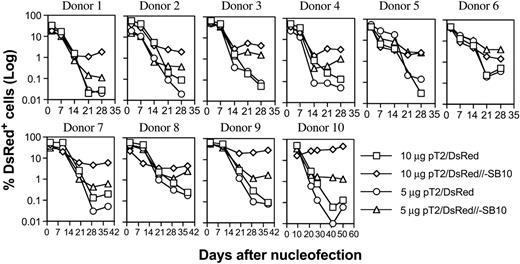
PBLs from 10 different donors were nucleofected with pT2/DsRed or pT2/DsRed//-SB10 (10 μg and 5 μg), and DsRed expression was monitored periodically by flow cytometric analysis. As shown in Figure 2, the percentages of DsRed+ cells mediated by both plasmids were similar on days 1 and 2, 7, and 14. However, the percentage of DsRed+ cells remained significantly higher for the pT2/DsRed//-SB10 transfected cells than for the pT2/DsRed transfected cells in all 10 donor cell populations at 3 weeks after transfection. We conclude that 10 μg and 5 μg of the SB cis plasmid mediated a geometric average of 3.33% and 1.62% of stable gene expression in human primary PBLs, compared to 0.58% and 0.22% mediated by SB transposon in the absence of transposase, respectively (Table 1, day 21).
Stable transgene expression in human primary PBLs by the SB transposon cis construct. Freshly isolated PBLs (5 × 106) were nucleofected at concentrations of 10 or 5 μg trans vector pT2/DsRed or cis vector pT2/DsRed//-SB10 using the T-cell Nucleofector kit (Amaxa, Gaithersburg, MD). On day 1 or 2 after nucleofection, a fraction of cells was analyzed for DsRed expression by flow cytometry, and the remaining cells were stimulated with anti-CD3/28 beads for 3 to 5 days. After removal of beads, the cells were cultured in human T-cell medium with IL-2 (50 IU/mL) and IL-7 (10 ng/mL). DsRed transgene expression was analyzed at the indicated time points by flow cytometry. The cells were restimulated once every 10 to 14 days. The data from 10 individual PBLs are shown.
Stable transgene expression in human primary PBLs by the SB transposon cis construct. Freshly isolated PBLs (5 × 106) were nucleofected at concentrations of 10 or 5 μg trans vector pT2/DsRed or cis vector pT2/DsRed//-SB10 using the T-cell Nucleofector kit (Amaxa, Gaithersburg, MD). On day 1 or 2 after nucleofection, a fraction of cells was analyzed for DsRed expression by flow cytometry, and the remaining cells were stimulated with anti-CD3/28 beads for 3 to 5 days. After removal of beads, the cells were cultured in human T-cell medium with IL-2 (50 IU/mL) and IL-7 (10 ng/mL). DsRed transgene expression was analyzed at the indicated time points by flow cytometry. The cells were restimulated once every 10 to 14 days. The data from 10 individual PBLs are shown.
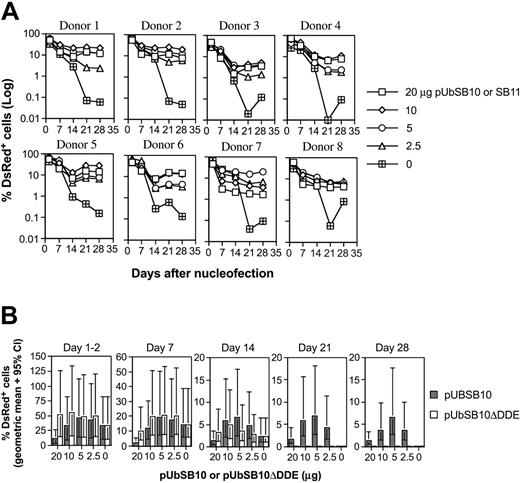
Because the efficiency of the transposition process in cultured cells30,50 and mice34,51 has been shown to be affected by the amount of transposase present in a given cell, we decided to evaluate the effect of trans delivery on stable gene expression in human PBLs. Co-nucleofection of 5 μg pT2/DsRed along with varying amounts (20, 10, 5, 2.5, and 0 μg per 5 × 106 PBLs) of transposase expression vectors pUbSB10 (donors 1-6 in Figure 3A) or pUbSB1130 (donors 7-8 in Figure 3A) into 8 independent PBLs was performed. On day 1 and 2 after transfection, 40% to 60% of the cells were DsRed+ in all the plasmid groups. However, upon extended culture, those cells transfected with pT2/DsRed alone gradually lost transgene expression, whereas a significant percentage of the cells co-transfected with pUbSB10 or pUbSB11 remained DsRed+. This difference was statistically significant on days 21 and 28 after transfection (Table 2). The most effective amount of transposase-encoding plasmid provided in trans in these experiments was 10 μg, which resulted in 11% of stably transfected cells (Table 2). Co-delivery of higher amounts of transposase-encoded plasmid (20 μg) did not increase the level of stable DsRed-expressing cells, in agreement with previous observations that overproduction of the SB transposase can inhibit transposition.30 Alternatively, it is possible that more cell death could result from a higher amount of the transposase plasmid used for transfection. In addition, we reproducibly observed that PBLs transfected with a higher amount of the transposase-expressing plasmid responded poorly to anti-CD3/28 bead activation, suggesting that high amounts of transposase or plasmid in general were detrimental to cellular function (data not shown). We thus observed that delivery of SB transposase–encoding plasmid in trans effectively mediated stable gene expression in primary T cells, exhibiting about 3-fold (11% vs 3% with 10 μg plasmid on day 21) in potency in comparison with the cis vector (Tables 1-2).
Stable transgene expression in human primary PBLs by the SB transposon trans construct and functional transposase. (A) SB trans delivery. Freshly isolated PBLs (5 × 106) were co-nucleofected with 5 μg of pT2/DsRed and different amounts of pUbSB10 or pUbSB11 (donors 7 and 8 only) (20, 10, 5, 2.5, and 0 μg). DsRed expression was analyzed on days 1, 2, 7, 14, 21, and 28. The data from 8 individual PBLs are shown. (B) Comparison of wild-type and mutant SB10 in trans delivery. Columns represent geometric means with 95% confidence intervals of 6 independent experiments. P values to compare SB10 and SB10ΔDDE on days 21 and 28 were all less than .001.
Stable transgene expression in human primary PBLs by the SB transposon trans construct and functional transposase. (A) SB trans delivery. Freshly isolated PBLs (5 × 106) were co-nucleofected with 5 μg of pT2/DsRed and different amounts of pUbSB10 or pUbSB11 (donors 7 and 8 only) (20, 10, 5, 2.5, and 0 μg). DsRed expression was analyzed on days 1, 2, 7, 14, 21, and 28. The data from 8 individual PBLs are shown. (B) Comparison of wild-type and mutant SB10 in trans delivery. Columns represent geometric means with 95% confidence intervals of 6 independent experiments. P values to compare SB10 and SB10ΔDDE on days 21 and 28 were all less than .001.
The DDE catalytic domain of transposase is required for transposition.27 To verify the necessity of catalytically active SB transposase for transposition in T cells, we tested for transposition in 6 human primary PBL cultures, comparing wild-type and mutant forms of SB transposase. The construct, pUbSB10ΔDDE, expresses a truncated transposase polypeptide that contains specific DNA-binding and nuclear localization signal domains but lacks the DDE catalytic domain.27 As shown in Figure 3B, the mutant construct was incapable of mediating stable gene expression in human PBLs nucleofected at all concentrations (20, 10, 5, and 2.5 μg) of plasmids tested, confirming that the catalytic DDE domain is necessary for transposition in human primary T cells.
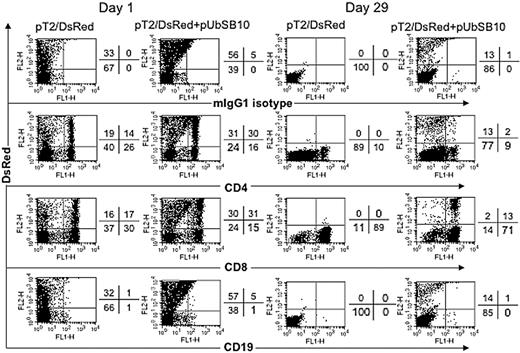
To define the mixed PBL populations that were effectively transfected and stably expressing the DsRed transgene, PBLs from 2 donors were nucleofected with pT2/DsRed alone or with pT2/DsRed plus pUbSB10, and then the cells were immunophenotyped by flow cytometric analysis at day 1 and day 29 after transfection. As shown in Figure 4, both CD4+ and CD8+ T cells in pT2/DsRed or pT2/DsRed+pUbSB10 nucleofected PBLs expressed DsRed at day 1. Upon extended culture until day 29 with anti-CD3/28 bead stimulation, loss of DsRed gene expression was observed in both CD4+ and CD8+ T cells transfected with pT2/DsRed alone, whereas a proportion of CD4+ and CD8+ T cells in pT2/DsRed+pUbSB10 transfected PBLs remained DsRed+. These results demonstrate that SB can mediate stable transgene expression in both CD4 and CD8 T cells.
Immunophenotyping of transfected T cells. PBLs from 2 donors were nucleofected with pT2/DsRed or with pT2/DsRed plus pUbSB10, or without DNA (mock, data not shown), and then the cells were stained with anti-CD4, CD8, CD19, and isotype mouse IgG1 mAbs on days 1, 15 (not shown), and 29 for analysis by flow cytometry. Representative data from one donor is shown. Similar data from at least 3 other donors were obtained 3 to 4 weeks after transfection (not shown).
Immunophenotyping of transfected T cells. PBLs from 2 donors were nucleofected with pT2/DsRed or with pT2/DsRed plus pUbSB10, or without DNA (mock, data not shown), and then the cells were stained with anti-CD4, CD8, CD19, and isotype mouse IgG1 mAbs on days 1, 15 (not shown), and 29 for analysis by flow cytometry. Representative data from one donor is shown. Similar data from at least 3 other donors were obtained 3 to 4 weeks after transfection (not shown).
Molecular analysis of transposition in isolated human T-cell clones
PBLs from 2 donors (PBL31 and PBL32) were nucleofected with 5 μg of pT2/DsRed in combination with 20, 10, 5, 2.5, and 0 μg of pUbSB10. Three weeks after transfection, when stable gene expression was detected in PBLs, a limiting dilution assay was applied to generate single T-cell clones. Eighty and 56 clones were generated from PBL31 and PBL32, respectively, and flow cytometric analysis identified 2 DsRed+ clones (L6 and L22) from PBL31 and 4 clones (O9, O21, O56, and O69) from PBL32. DsRed+ clones were maintained in culture for up to 4 months without losing observable transgene expression, compared to DsRed– clones (NO11 and NO71) (Figure 5A). NO71, O56, and O69 clones were CD4+ and NO11, L6, L22, O9, and O21 clones were CD8+ (data not shown). Transgene expression in the majority (5 of 6) of cloned T cells was very high (> 1000 mean fluorescence intensity) (Figure 5A). Southern hybridization analysis was conducted on genomic DNA isolated from these clones, using enzyme combinations that cut once both inside and outside of the transposon sequence, thus generating DsRed-hybridizing bands of a different size for each transposon integrant (Figure 5B). The results showed 1 to 2 integrants among the DsRed+ clones, but not in the DsRed– clones (Figure 5C). Surprisingly, all the clones had 4 to 19 copy numbers, suggesting that a high level of transgene expression mediated by SB transposons may be due to high copy numbers of the integrated transgene (Figure 5D). Notably, one band in clone O56 showed a similar size as the plasmid control, indicating the possibility that random plasmid integration may have occurred in this clone. However, this possibility was eliminated using a combination of different restriction enzymes for digestion of the genomic DNA from T-cell clone O56 and transposon plasmid pT2/DsRed. As shown in Figure 5C, different sizes of genomic DNA bands derived from O56 T-cell clone were generated using different restriction enzymes in comparison with the plasmid control.
Molecular analyses of transposition in human T cells. (A) DsRed transgene expression in transposed T-cell clones 4 months after gene transfer. (B) Schematic representation of a DsRed probe used in this study. Top row shows the circular transposon-encoded plasmid nucleofected into T cells and the location of the 735-bp DsRed probe. Bottom row shows the loss of flanking restriction enzyme recognition sequences after transposase-mediated integration into genomic DNA (wavy line). (C) Integration assay by Southern blotting. Southern blot of genomic DNA digested with SalI and XbaI and hybridized with a DsRed probe. The SalI site located outside the IR/DR is lost, resulting in the absence of a detectable 1.7-kb plasmid-specific band. Southern blot of genomic DNA from clone O56 digested with SacI and XhoI as well as KpnI and XbaI and hybridized with the same probe. The SacI and KpnI sites flanking IR/DRs are lost, resulting in the absence of detectable 2.8- and 1.7-kb plasmid-specific bands, respectively. (D) Determination of per-cell copy numbers by Southern blot. A 1-copy standard of pT2/DsRed was equal to 11 pg. Copy standards were mixed with 10 μg genomic DNA isolated from bulk activated T cells. Transgene copy numbers for each T-cell clone were calculated from the intensities of the bands spanning the DsRed gene in comparison to a copy number standard after linear regression analysis. Band intensities were quantified using a PhosphorImager. (E) Transposition assay. Transposon insertion site sequences were determined using a linker-mediated PCR technique described in “Materials and methods.” Plasmid backbone sequences of pT2/DsRed are shown. Duplicated TA dinucleotide target site is in capital letters. Transposon-specific sequences are in the center box. Amplified transposon:chromosome junction sequences were subjected to BlastN analysis against the human genome using the ENSEMBL database (chromosome locations indicated in shaded box on the left). Genome-specific primers were designed in accordance with identified chromosome positions to amplify the opposing flanking sequence and confirm insertion into a single TA dinucleotide.
Molecular analyses of transposition in human T cells. (A) DsRed transgene expression in transposed T-cell clones 4 months after gene transfer. (B) Schematic representation of a DsRed probe used in this study. Top row shows the circular transposon-encoded plasmid nucleofected into T cells and the location of the 735-bp DsRed probe. Bottom row shows the loss of flanking restriction enzyme recognition sequences after transposase-mediated integration into genomic DNA (wavy line). (C) Integration assay by Southern blotting. Southern blot of genomic DNA digested with SalI and XbaI and hybridized with a DsRed probe. The SalI site located outside the IR/DR is lost, resulting in the absence of a detectable 1.7-kb plasmid-specific band. Southern blot of genomic DNA from clone O56 digested with SacI and XhoI as well as KpnI and XbaI and hybridized with the same probe. The SacI and KpnI sites flanking IR/DRs are lost, resulting in the absence of detectable 2.8- and 1.7-kb plasmid-specific bands, respectively. (D) Determination of per-cell copy numbers by Southern blot. A 1-copy standard of pT2/DsRed was equal to 11 pg. Copy standards were mixed with 10 μg genomic DNA isolated from bulk activated T cells. Transgene copy numbers for each T-cell clone were calculated from the intensities of the bands spanning the DsRed gene in comparison to a copy number standard after linear regression analysis. Band intensities were quantified using a PhosphorImager. (E) Transposition assay. Transposon insertion site sequences were determined using a linker-mediated PCR technique described in “Materials and methods.” Plasmid backbone sequences of pT2/DsRed are shown. Duplicated TA dinucleotide target site is in capital letters. Transposon-specific sequences are in the center box. Amplified transposon:chromosome junction sequences were subjected to BlastN analysis against the human genome using the ENSEMBL database (chromosome locations indicated in shaded box on the left). Genome-specific primers were designed in accordance with identified chromosome positions to amplify the opposing flanking sequence and confirm insertion into a single TA dinucleotide.
To confirm that the stable DsRed expression in the DsRed+ T-cell clones was the result of transposition rather than unintegrated episomal DNA, a splinkerette PCR technique was used to recover sequences flanking transposon inserts on both the 5′ and the 3′ sides. SB-mediated transposition requires a TA-dinucleotide for integration. As shown in Figure 5E, 5 of the 6 clones analyzed contained novel chromosomal sequences at TA sites, the benchmark for transposition. These junction sequences were mapped to determine their locations in human chromosomes. Moreover, both Hirt and genomic DNA isolated from stably transfected DsRed+ bulk PBLs and T-cell clones were incapable of transforming E coli. However, DNA isolated from transiently nucleofected PBLs at both day 1 and day 7 efficiently transformed the bacteria (Table 3, P < .01). Collectively, these results provide molecular evidence for SB-mediated transposition in human T cells.
Dual gene expression in human T cells. PBLs from 2 donors were nucleofected with the SB dual gene plasmid pT2/EGmCN (5 μg) ± pUbSB10 (10, 5, 2.5, and 0 μg). The cells were activated by anti-CD3/28 beads on day 2 and assayed for eGFP and NGFR expression on days 7 (data not shown) and 21. Similar data were also obtained from another donor.
Dual gene expression in human T cells. PBLs from 2 donors were nucleofected with the SB dual gene plasmid pT2/EGmCN (5 μg) ± pUbSB10 (10, 5, 2.5, and 0 μg). The cells were activated by anti-CD3/28 beads on day 2 and assayed for eGFP and NGFR expression on days 7 (data not shown) and 21. Similar data were also obtained from another donor.
Development of SB transposon vectors expressing multiple genes
Some gene therapy applications may require coexpression of several genes coordinately. Currently, the use of an internal ribosome entry site (IRES) is one strategy commonly chosen for translation of 2 coding sequences by the same message. In preliminary studies, we tested whether the encephalomyocarditis virus (ECMV) IRES could be used to regulate expression of 2 genes from the same messenger RNA in the context of an SB transposon. PBLs from 2 donors were co-nucleofected with the SB transposon pKT2/DsRed-IL (Figure 1F) expressing both DsRed and the IRES-luciferase fusion sequence along with the transposase expression plasmid pUbSB10. Following continuous culture for up to 32 days, there were no significant DsRed+ cells (< 0.2%), although a significant fraction of DsRed+ cells could be detected on days 1 and 2 (ranging from 12% to 40%). However, over the same time period, the positive mono-cistronic control plasmid pT2/DsRed co-transfected with pUbSB10 gave rise to 2% to 3% DsRed+ cells (data not shown). Alternatively, an 18 amino acid 2A self-cleaving oligopeptide from foot-and-mouth disease virus can be used for expression of multiple, discrete proteins from a polyprotein generated from a single open reading frame.39,40 Recently, Vignali's group has shown that 2A peptides can be used to co-express up to 4 proteins from a single retroviral vector.41 To evaluate the potential of the 2A peptide system in the context of an SB transposon, we constructed SB plasmids (Figure 1G-I) designed to express NGCD (a fusion gene of truncated nerve growth factor receptor and cytosine deaminase),38 DsRed, and luciferase. Nerve growth factor receptor (NGFR) was used as a marker for selection of the transfected cells. Cytosine deaminase (CD) converts 5-fluorocytosine (5-FC) to the toxic metabolite 5-fluorouracil (5-FU) and has been investigated as a potential tool for selective cellular eradication in the case of graft-versus-host disease (GVHD) caused by donor T cells after bone marrow transplantation. Our results demonstrated that downstream gene expression with use of the 2A in SB transposons (Figure 1H-I) was not improved (data not shown). Moreover, expression of the upstream gene was significantly reduced by the 2A sequence compared to the vector without the 2A (pT2/NGCD, Figure 1G) (data not shown).
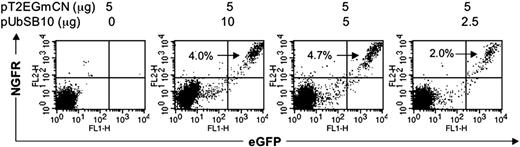
It recently has been reported that lentiviral vectors containing synthetic bidirectional promoters can mediate coordinate dual-gene expression.42 We cloned bidirectional promoters into a modified SB transposon vector (Figure 1J). PBLs from 2 donors were nucleofected with the bidirectional SB vector pT2/EGmCN carrying EGFP and NGFR genes in combination with different amounts of pUbSB10. As shown in Figure 6, 2% to 4.7% of PBLs co-transfected with pUbSB10 were positive for both GFP and NGFR 3 weeks after transfection, while neither gene product was expressed in cells nucleofected with pT2/EGmCN alone after only 7 days in culture (data not shown). We conclude that synthetic bidirectional promoters can be used for stable expression of 2 gene products in human primary T cells after nucleofection of SB transposon vectors.
Application of SB-mediated gene transfer in human T cells
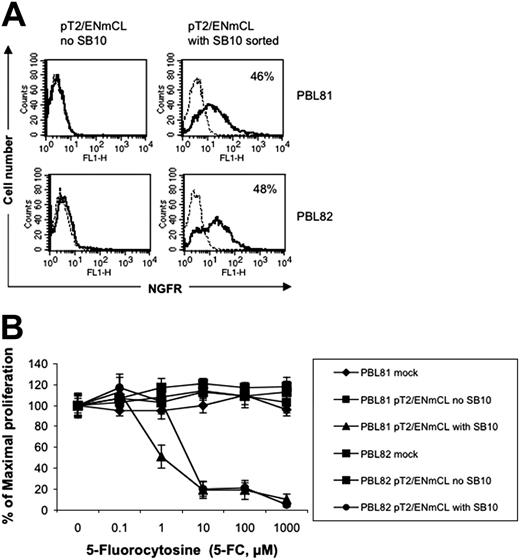
To test if the SB transposon system can be used to express a therapeutically relevant gene product, a NGCD38 “suicide” gene was coexpressed with luciferase in the SB vector pT2/ENmCL (Figure 1K). PBLs from 2 independent donors nucleofected with pT2/ENmCL alone were minimally positive (0.1% and 0.4%, respectively) for NGFR expression 7 days after transfection, whereas 8.5% and 6.3% of T cells nucleofected with both pT2/ENmCL and pUbSB10 (10 μg) expressed NGFR (data not shown). Single cell sorting of nucleofected T cells for NGFR 9 days after transfection generated a population of cells that was 46% to 48% positive for NGFR (Figure 7A). In vitro cytotoxicity assays performed with this sorted cell population demonstrated that NGCD+ cells could be killed in a dose-dependent manner by 5-FC, while NGCD– cells were resistant to treatment with 5-FC and remained viable (Figure 7B). It is likely that NGCD– cells in the NGCD sorted population were killed through a bystander effect.52 Luciferase activity in the NGCD sorted cells was only marginally detectable, suggesting that successfully nucleofected T cells were not as receptive to firefly luciferase expression as for DsRed or NGCD, or that the relatively larger size of the firefly luciferase gene (1.6 kb) was a limiting factor. Nevertheless, these results demonstrate that SB transposons can effectively mediate integration and stable expression of therapeutically relevant transgenes in human primary T cells.
Functional therapeutic gene expression in human T cells by SB-mediated transposition. (A) NGFR expression in the SB-nucleofected T cells (solid line). A murine IgG1 isotype mAb was used (broken line). (B) Cell killing by 5-FC. Human T cells nucleofected with SB transposon DNA (2 × 105 cells/well) or CEM cells (104 cells/well) transduced with retrovirus expressing NGCD were exposed to different concentrations of 5-fluorocytosine (5-FC, Sigma) in 96-well flat-bottom plates for 5 days. A colorimetric assay kit (CellTiter 96, Promega) based on the conversion of a tetrazolium dye to formazan was used to quantitate the number of viable cells on day 5. Absorbance was measured at 565 nm using a microplate spectrophotometer (Bio-Tek Instruments, Winooski, VT). CEM cells alone or transduced with retrovirus expressing NGCD were used as negative and positive controls (data not shown). The data represent mean and standard deviation of 10 replicates. This experiment was repeated twice with similar results. Similar results also were obtained with the plasmid pT2/NGCD (data not shown).
Functional therapeutic gene expression in human T cells by SB-mediated transposition. (A) NGFR expression in the SB-nucleofected T cells (solid line). A murine IgG1 isotype mAb was used (broken line). (B) Cell killing by 5-FC. Human T cells nucleofected with SB transposon DNA (2 × 105 cells/well) or CEM cells (104 cells/well) transduced with retrovirus expressing NGCD were exposed to different concentrations of 5-fluorocytosine (5-FC, Sigma) in 96-well flat-bottom plates for 5 days. A colorimetric assay kit (CellTiter 96, Promega) based on the conversion of a tetrazolium dye to formazan was used to quantitate the number of viable cells on day 5. Absorbance was measured at 565 nm using a microplate spectrophotometer (Bio-Tek Instruments, Winooski, VT). CEM cells alone or transduced with retrovirus expressing NGCD were used as negative and positive controls (data not shown). The data represent mean and standard deviation of 10 replicates. This experiment was repeated twice with similar results. Similar results also were obtained with the plasmid pT2/NGCD (data not shown).
Discussion
Three important observations concerning SB transposon–mediated human T-cell gene transfer have emerged in this study. First, both SB cis and trans vectors can mediate stable gene transfer and expression in human primary CD4 and CD8 T cells without prior T-cell activation for transfection. This suggests that transfected PBLs could be infused into a recipient immediately after transfection, although we do not have evidence that stable transgene-expressing cells can be maintained in vivo. Second, under the conditions tested, the SB trans vector appeared to be more efficient than the SB cis vector in mediating stable gene transfer. Third, PBLs expressing a “suicide” gene as a result of SB-mediated transposition can be killed by a specific drug, demonstrating one potential application of SB transposon–mediated gene transfer in human T cells. We thus demonstrate for the first time that SB transposons can mediate transposition in a primary human T-cell population, with implications for the application of SB in human gene therapy.
Our results showed that an SB trans vector mediated at least 3-fold higher gene transfer in human primary T cells than a cis vector, even though transposase expression was regulated by the same promoter (Tables 1-2). We favor the explanation that the trans vectors (pT2/DsRed, 6551 bp; pUbSB10, 5348 bp) are smaller in size than the cis vector (pT2/DsRed//-SB10, 9289 bp) and consequently may display higher transfection efficiency. Our data also demonstrated that the amount of SB transposase–encoded vector is critical in mediating a high level of transgene expression in human T cells. As shown in Figures 2 and 3, 5 to 10 μg of either the cis vector or the SB10 transposase–expressing vector gave the highest level of gene transfer, whereas overexpression or underexpression of transposase resulted in poor long-term expression, suggesting that overexpression of the transposase has an inhibitory effect on transposition in human T cells.29,30
We found transposition efficiency in human primary T cells by the SB transposon to be about 3% for the cis vector and 11% for the trans vector (Tables 1-2, day 21 and 10 μg), while lentiviral vectors typically can achieve about 50% transduction efficiency in human unstimulated primary T cells.17 However, we believe that this relatively low transposition efficiency in human T cells can be enhanced with improved transposon vectors50,53 and new DNA delivery methods, including nanoparticle DNA delivery systems.54 Two groups have reported that hyperactive versions of the SB transposase can improve transposition efficiency by increasing DNA-binding affinity of the transposase.50,53 It has been reported that these hyperactive transposases alone or in combination with transiently expressed high-mobility group box 1 (HMGB1), a cellular co-factor of the SB transposase,50 or a third-generation transposon (pT3)53 can increase transposition by 4- to 10-fold compared to the SB10 transposase used in this study. Thus, it is of great interest in future studies to test whether newer generations of transposon vectors can improve T-cell gene transfer and therapy. It also is of importance to compare the SB gene delivery system with the current gold standard of lentiviral vectors in human primary T-cell gene transfer.
We demonstrate that high-level transgene expression mediated by SB transposons in human T cells could be maintained at least 4 months in vitro (Figure 5A). It has been shown that viral vector-transduced PBLs often lose transgene expression rapidly upon transplantation in vivo because they are not in an environment of constant cytokine stimulation.2 Therefore, it is essential to test the survival and expression kinetics of the SB-transfected PBLs in vivo to obtain a complete assessment of this novel gene delivery system.
An important feature of the SB transposon system in mediating stable gene transfer in human primary T cells is the directed chromosomal integration at target TA dinucleotide sites. As shown in Figure 5E, of 6 clones analyzed, 5 clones contained novel chromosomal sequences, and these junction sequences were mapped to different human chromosomes. Yant and colleagues have mapped 1336 insertion sites in primary and cultured mammalian cells and found that SB-mediated integration is associated with less regional preference than retroviruses and is not significantly influenced by transcriptional activity.31 These results are in contrast to most integrating oncoretrovirus- or lentivirus-based vectors that preferentially integrate into transcriptionally active sites, including proto-oncogenes or signaling genes,18-23 which theoretically may result in a higher predisposition toward uncontrolled proliferation and tumorigenesis. Therefore, SB transposon vectors might prove safer for genetic therapy. Further studies are still required to analyze SB insertion sites in T cells and more thoroughly evaluate the potential therapeutic value of SB-mediated gene transfer in human T cells.
The SB transposon system offers several advantages for T-cell gene transfer and therapy over widely used virus-based or conventional mammalian DNA vectors.28,29 First, plasmid DNA is simpler, more stable, less immunogenic, and more straightforward to manufacture. Second, integration of exogenous DNA sequence by transposition suggests the potential for persistent transgene expression, thus limiting the need for repeated administration of the therapy and reduced cost. Third, there may be improved safety since integration is restricted to target TA sites of the human genome with no preference for active genes.31 Fourth, there is no apparent need for T-cell activation before transfection. Thus, the duration of ex vivo culture is reduced, and alterations in T-cell phenotypes and functions are minimal.
Prepublished online as Blood First Edition Paper, September 27, 2005; DOI 10.1182/blood-2005-05-2133.
Supported in part by Children's Cancer Research Fund, Howard Hughes Medical Institute, grants from the Minnesota Medical Foundation, Grant-in-Aid of Research, Artistry and Scholarship of the Graduate School, University of Minnesota (X.Z.), National Institute of General Medical Sciences (NIGMS) training grant (T32 GM08347) (A.C.W.), Fanconi Anemia Research Fund (J.T.), the Arnold and Mabel Beckman Foundation (R.S.M.) and National Institutes of Health (NIH) grant R01 52952 (B.R.B.). X.Z. is a recipient of the American Society of Hematology Junior Faculty Scholar Award. This investigation was conducted in a facility constructed with support from Research Facilities Improvement Program Grant (CO6 CA062526-01) from the National Center for Research Resources, NIH (X.Z.).
R.S.M. has declared a financial interest in Discovery Genomics, Inc.
X.H. designed and performed research and analyzed data. A.C.W. designed and performed research, analyzed the data, and wrote the paper. L.B., D.T., and J.T. designed and performed research. P.J.O., B.L.L., and C.H.J. provided critical reagents. R.S.M. provided critical reagents, designed research, analyzed data, and wrote the paper. B.R.B. designed research, analyzed data, and wrote the paper. X.Z. initiated, designed and performed research, analyzed data, and wrote the paper.
R.S.M. and B.R.B. contributed equally to this work.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Dr Luigi Naldini and Mario Amendola for providing us with the HIV-1–derived lentiviral vector containing bidirectional promoters; Scott Bell and Zhimei Wang for helpful assistance in luciferase and immunophenotyping assays, respectively; Joseph S. Koopmeiners and Dr Baolin Wu for statistical analysis of the data; and the University of Minnesota Cancer Center Flow Cytometry Core Facility for assistance with the flow cytometric analyses. We are grateful to Drs Perry Hackett, John Wagner, and Stephen Jameson for critical reading of the manuscript; Drs Thom Saunders and David Largaespada for helpful discussions in copy number analysis; and Ms Arlys Clements for secretarial assistance.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal