Abstract
The Bcr-Abl fusion kinase drives oncogenesis in chronic myeloid leukemia (CML). CML patients are currently treated with the Abl tyrosine kinase inhibitor imatinib, which is effective in early stages of the disease. However, resistance to imatinib arises in later disease stages primarily because of a Bcr-Abl mutation. To gain deeper insight into Bcr-Abl signaling pathways, we generated phosphotyrosine profiles for 6 cell lines that represent 3 Bcr-Abl fusion types by using immunoaffinity purification of tyrosine phosphopeptides followed by tandem mass spectrometry. We identified 188 nonredundant tyrosine-phosphorylated sites, 77 of which are novel. By comparing the profiles, we found a number of phosphotyrosine sites common to the 6 cell lines regardless of cellular background and fusion type, several of which are decreased by imatinib treatment. Comparison of this Bcr-Abl signature with the profile of cells expressing an alternative imatinib-sensitive fusion kinase, FIP1L1-PDGFRα, revealed that these kinases signal through different pathways. This phosphoproteomic study of the Bcr-Abl fusion kinase highlights novel disease markers and potential drug-responsive biomarkers and adds novel insight into the oncogenic signals driven by the Bcr-Abl kinase.
Introduction
Chronic myelogenous leukemia (CML) is a stem cell disease characterized by the overproliferation of myeloid cells. CML afflicts 1 in 100 000 people and constitutes 20% of all adult forms of leukemia.1,2 This disease is characterized by the presence of the t(9;22)(q34;q11) Philadelphia chromosome (Ph) translocation3 arising from fusion of a portion of the breakpoint cluster region (BCR) gene with the Abl tyrosine kinase, leading to constitutive tyrosine kinase activity and increased phosphorylation of downstream targets. These phosphorylation events are critical for the continued proliferation and survival of the leukemic cells (reviewed in Deininger and Druker4 ).
CML is unique in that the expression of the Bcr-Abl fusion is strongly implicated as the only oncogenic abnormality in early stages of the disease.5 In fact, inhibition of Abl tyrosine kinase activity by treatment with imatinib (Gleevec, STI571; Novartis, Basel, Switzerland) during the chronic phase of the disease can lead to complete remission.6 Unfortunately, patients treated with imatinib during the accelerated or blast crisis phases often have relapses and die of the disease.7,8 The mechanism of resistance to imatinib is predominantly through mutations occurring in the Abl kinase domain that affect binding of the drug to the kinase or gene amplification of Bcr-Abl.9,10
Studies are ongoing to develop the use of drug combinations or alternative therapies for patients who are resistant to imatinib. In CML models, the efficacy of combining imatinib with rapamycin is demonstrated in cell lines expressing drug-resistant kinases.11 Additionally, combination therapies using PI3K inhibitors with imatinib are beneficial in models of late-stage CML.12 The ability to pinpoint the alternative signal transduction pathways that are activated is necessary for further research into combination therapy. Studying downstream mediators of Bcr-Abl provides insight into the molecular mechanisms of CML and may lead to further understanding of disease progression and resistance to imatinib. In addition, these studies could lead to the identification of alternative targets in the event of resistance.
Tyrosine phosphorylation is less common than serine and threonine phosphorylation,13 making the study of phosphorylated tyrosine residues challenging. To enhance the detection of the phosphotyrosine content of the cellular proteome, we digested cell lysates and enriched for phosphotyrosine containing peptides by immunoprecipitating these peptides with antiphosphotyrosine antibody followed by liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS).14 In this study, we profiled tyrosine phosphorylation in 6 Bcr-Abl cell lines and identified a series of phosphorylated peptides common to Bcr-Abl–containing cells, regardless of cellular background or fusion type. We refer to this group of phosphotyrosine sites, consistently associated with activated Bcr-Abl, as a Bcr-Abl kinase phosphotyrosine signature. We demonstrated that some of these phosphorylated peptides were responsive to treatment with imatinib. In addition, we found differences in phosphorylation sites identified within different Bcr-Abl fusion proteins that were confirmed by targeted tandem mass spectrometry. Finally, using data-dependent mass spectrometry and stable isotope labeling by amino acids in cell culture (SILAC) after imatinib treatment, we compared the phosphotyrosine signature of Bcr-Abl–containing cell lines with a cell line containing another imatinib-sensitive fusion kinase, FIP1L1-PDGFRα, which is implicated in hypereosinophilic syndrome.15,16 Our results demonstrate that the Bcr-Abl signature is distinct from the phosphotyrosine profile found downstream of PDGFRα; thus, it does not simply represent imatinib-responsive phosphosites.
Materials and methods
Experimental procedures
Antibody to CD2AP was obtained from Santa Cruz Biotechnology (Santa Cruz, CA). All other antibodies for immunoblotting, protein, and phosphopeptide immunoprecipitation were from Cell Signaling Technology (Danvers, MA).
Cell culture, treatment, and lysis
All cell culture reagents were purchased from Invitrogen (Carlsbad, CA). K562, KU812, and SUP-B15 cells were obtained from American Type Culture Collection (Manassas, VA). BV173, KCL22, and SD1 cells were obtained from the German National Resource Centre for Biological Material (DSMZ Braunschweig, Germany). EoL-1 cells were a kind gift from Donald Small (Department of Oncology and Comparative Medicine, Johns Hopkins University School of Medicine, Baltimore, MD). Cells were grown in a 5% CO2 incubator at 37°C. BV173, KCL22, KU812, SD1, SUP-B15, and EoL-1 were grown in RPMI 1640 with 10% fetal bovine serum and penicillin/streptomycin, whereas K562 was cultured in DMEM containing 10% fetal bovine serum and penicillin/streptomycin. Cells were treated for 2 hours (immunoblotting) and 3 hours (SILAC) with 10 μM imatinib. Cells were lysed in 1 × cell lysis buffer (20 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1 mM Na2 EDTA, 1 mM EGTA, 1% Triton, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 1 mM Na3 VO4, 1 μg/mL leupeptin) supplemented with Complete, Mini, EDTA-free protease inhibitor cocktail (Roche, Indianapolis, IN) and sonicated. Cell lysates were cleared by centrifugation at 10 000g for 5 minutes.
Protein immunoprecipitation
Cell lysates were normalized based on protein concentration. Cells were immunoprecipitated using phosphotyrosine antibody (pY100) followed by Western blots to total CD2AP and VASP. Immunoprecipitation efficiency was measured using the p-cdc2 (Y15) antibody.
Phosphopeptide immunoprecipitation
Cells (2 × 108) were lysed in urea lysis buffer (20 mM HEPES, pH 8.0, 9 M urea, 1 mM sodium vanadate, 2.5 mM sodium pyrophosphate, and 1 mM β-glycerophosphate) at 1.25 × 108 cells/mL and sonicated. Sonicated lysates were cleared by centrifugation at 8000g, and proteins were reduced and alkylated as described previously.14 Samples were diluted with 20 mM HEPES, pH 8.0, to a final urea concentration of 2 M. Trypsin (1 mg/mL in 0.001 M HCl) was added to the clarified lysate at 1:100 vol/vol. Samples were digested overnight at room temperature. After digestion, lysates were acidified to a final concentration of 1% TFA. Peptide purification was carried out using Sep-Pak C18 columns (Waters, Milford, MA), as described previously.14 After purification, all elutions (8%, 12%, 15%, 18%, 22%, 25%, 30%, 35%, and 40% acetonitrile in 0.1% TFA) were combined into one fraction. For SILAC analysis, the elutions were combined into 2 (K562) or 3 (EoL-1) fractions and lyophilized. Dried peptides were resuspended in 1.4 mL (1 fraction) or 1.0 mL (2 and 3 fractions) of MOPS IP buffer (50 mM MOPS/NaOH, pH 7.2, 10 mM Na2HPO4, 50 mM NaCl), and insoluble material was removed by centrifugation at 500g for 10 minutes. The phosphotyrosine monoclonal antibody P-Tyr-100 from ascites fluid was coupled noncovalently to protein G agarose (Roche, Indianapolis, IN) at 4 mg/mL beads overnight at 4°C. After coupling, antibody resin was washed twice with PBS and 3 times with MOPS IP buffer (5- to 10-bead vol buffer for each wash). Immobilized antibody (40 μL, 160 μg) was added as a 1:1 slurry in MOPS IP buffer to the solubilized peptide fraction, and the mixture was incubated overnight at 4°C. The immobilized antibody beads were washed 3 times with 1 mL MOPS IP buffer and twice with 1 mL water with ice-cold solutions. Peptides were eluted from beads by incubation with 60 μL of 0.1% TFA followed by a second elution with 40 μL of 0.1% TFA, and the fractions were combined.
SILAC analysis of imatinib-treated cells
K562 and EoL-1 cells were grown in DMEM or RPMI, respectively, lacking arginine and lysine supplemented with 10% dialyzed fetal bovine serum, penicillin/streptomycin, and l-lysine/HCl and l-arginine/HCl (Sigma, St Louis, MO) for light cultures or l-arginine/HCl (U-13C6, 98%) and l-lysine:2 HCl (U-13C6, 98%; U-15N2, 98%) (Cambridge Isotope Laboratories, Andover, MA) for heavy cultures, as previously described.17,18 Cells were grown to a density of approximately 1 × 106 cells/mL for a total of 1 × 108 cells per each cell culture type (2 × 108 cells total). After lysis, heavy and light cultures were combined and carried through the phosphopeptide immunoprecipitation protocol.
Analysis by LC-MS/MS
Peptides in the IP eluate (53 μL) were concentrated and separated from eluted antibody using ZipTip μC18 columns (Millipore, Billerica, MA). Peptides were eluted from the microcolumns with 1 μL of 60% MeCN and 0.1% TFA into 7.6 μL of 0.4% acetic acid/0.005% heptafluorobutyric acid (HFBA). The sample was loaded onto a 10 cm × 75 μm PicoFrit capillary column (New Objective, Ringoes, NJ) packed with Magic C18 AQ reversed-phase resin (Michrom Bioresources, Auburn, CA) using a Famos autosampler with an inert sample injection valve (Dionex, Sunnyvale, CA). The column was developed with a 45-minute linear gradient of acetonitrile in 0.4% acetic acid/0.005% HFBA delivered at 280 nL/min (Ultimate; Dionex, Sunnyvale, CA). Tandem mass spectra were collected in a data-dependent manner with an LCQ Deca XP Plus ion trap mass spectrometer (ThermoFinnigan, San Jose, CA), using a top-4 method, a dynamic exclusion repeat count of 1, and a repeat duration of 0.5 minute. TurboSequest (ThermoFinnigan) searches were performed against the National Center for Biotechnology Information (NCBI) human database released on February 23, 2004 containing 27 175 proteins allowing oxidized methionine (M+16) and phosphorylation (Y+80) as dynamic modifications. SILAC samples were analyzed using a top-1 method, with all other analysis conditions identical to those used for data-dependent analysis. After reviewing the spectra assigned to phosphotyrosine sequences and normalizing for protein levels, ratios of the integrated peak areas for phosphopeptide quantification were obtained using Xpress (ThermoFinnigan).
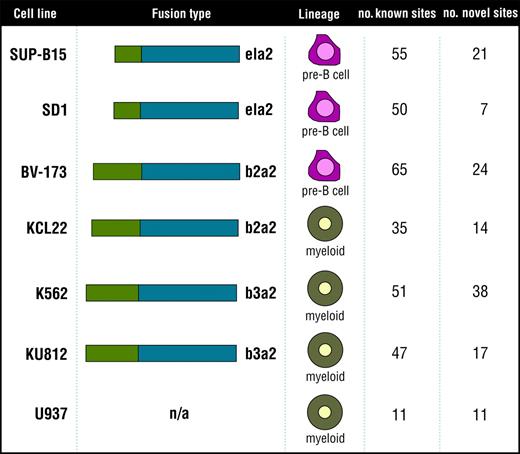
Cell lines analyzed to identify the Bcr-Abl signature. The 7 cell lines analyzed to identify the Bcr-Abl signature (SUP-B15, SD1, BV173, KCL22, K562, KU812, and the control cell line U937) are listed along with fusion type, cell lineage, and number of known and novel tyrosine phosphorylation sites identified in each cell type.
Cell lines analyzed to identify the Bcr-Abl signature. The 7 cell lines analyzed to identify the Bcr-Abl signature (SUP-B15, SD1, BV173, KCL22, K562, KU812, and the control cell line U937) are listed along with fusion type, cell lineage, and number of known and novel tyrosine phosphorylation sites identified in each cell type.
Targeted analysis of Bcr-Abl phosphopeptides was performed by placing the m/z values for the target peptides on the parent mass list. MS/MS spectra of the targeted mass values were collected using a top-2 method, a dynamic exclusion repeat count of 3, and a repeat duration of 0.5 minute. MS/MS spectra from data-dependent, SILAC, and targeted analyses were evaluated and reviewed as previously described.14 For data-dependent analysis, samples were analyzed in duplicate, and the data sets were combined after review.
Results
Phosphorylation sites common to 6 Bcr-Abl–containing cell lines
Two predominant Bcr-Abl fusion proteins are involved in CML. The b2a2 fusion contains the first 13 exons of BCR fused to exon 2 of ABL (e13a2). The b3a2 contains BCR through exon 14 fused to exon 2 of ABL (e14a2).2 In addition, a p190 form of the Bcr-Abl protein occurs from a fusion of exon 1 of BCR to exon 2 of ABL (e1a2). The p190 Bcr-Abl oncogene is rarely seen in CML but is more often found in acute lymphoblastic leukemia (ALL).19 In this study, we compared 6 cell lines—K562, KU812, KCL22, BV173, SD1, SUP-B15—expressing 3 fusions from 2 distinct cellular backgrounds, myeloid and pre-B cells. K562 and KU812 are myeloid cells that contain the b3a2 Bcr-Abl fusion, and the myeloid cell line KCL22 contains the b2a2 fusion protein. BV173, SD-1, and SUP-B15 are pre-B cells. As is the case for KCL22, BV173 express the b2a2 fusion protein. SD1 and SUP-B15 are ALL cell lines containing the p190 (e1a2) Bcr-Abl fusion.20 The phosphotyrosine profiles of these 6 cell lines were compared with U937, a myeloid leukemia cell line lacking the Bcr-Abl fusion protein (Figure 1).
To gain a global understanding of Bcr-Abl signaling, we used phosphotyrosine peptide immunoprecipitation, as described earlier,14 and identified 188 tyrosine phosphorylation sites within 110 proteins using the 6 Bcr-Abl and the control U937 cell lines (a complete list is available in Table S1 on the Blood website; see the Supplemental Tables link at the top of the online article), many of which were kinases (Table 1). One hundred eighty-three of the identified sites were found in the Bcr-Abl kinase–containing cells (a subset of these sites was also found in the U937 cell line). Of these 183 sites, 72 are novel, unpublished sites according to searches against published in vivo phosphorylation sites as referenced in the PhosphoSite database (available at www.phosphosite.org).21 Several of the sites were within proteins involved in Bcr-Abl transformation of cells, such as Gab2,22 Dok,23,24 Bcr, and Abl.25 Many phosphorylation sites identified are implicated in Bcr-Abl signaling, such as STAT5A (Y694), several tyrosines in the adaptor protein Dok (Y296, Y315, Y362, Y377, Y398, Y409), the ERK activation loop tyrosines, Bcr (Y177, Y246, Y436, Y591), and all 6 tyrosine sites found in Abl. In addition, we identified novel phosphorylation sites in several of these proteins, including Dok (Y337, Y341, Y449) and Bcr sites (Y598, Y644). Of special interest, Gab1 is involved in other activated tyrosine kinase pathways but is thought not to be related to Bcr-Abl signaling.26 We identified several known Gab1 phosphorylation sites, including Y259, Y317, Y373, Y406, and Y659. In addition, we identified Y242 as a novel Gab1 phosphorylation site. Bcr-Abl is known to phosphorylate cytoskeletal regulating proteins such as Dok1.27 In addition, Drosophila Abl kinase phosphorylates the Drosophila Ena/VASP protein, which is also involved in cytoskeletal regulation.28 In our analysis, we showed the phosphorylation of several cytoskeleton proteins, including known sites in β-actin (Y294), VASP (Y39), and vimentin (Y53 and Y61). We also purified phosphopeptides containing novel phosphorylation sites on actinin α1 (Y246), cofilin 1 (Y68), and plakophilin 3 (Y84).
Kinases identified in the Bcr-Abl–containing and control cell lines
PSDB ID . | pTyr Site . | Peptide . | BV173 . | K562 . | KCL22 . | KU812 . | SD1 . | SUP-B15 . | U937 . | Identified in K562 IMAC* . | Novel site . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Lipid kinase | |||||||||||
| PIK3R1 | 580 | DQyLMWLTQK | + | - | - | - | - | + | - | - | - |
| PIK3R2 | 467 | EYDQLYEEyTR | + | - | - | - | - | - | - | + | |
| Protein kinase | |||||||||||
| Abl | 115 | NGQGWVPSNyITPVNSLEK | + | + | + | + | - | + | - | - | - |
| Abl | 185 | INTASDGKLyVSSESR | + | + | + | + | + | + | - | + | - |
| Abl | 226 | NKPTVyGVSPNYDK | + | + | + | + | + | + | - | - | - |
| Abl | 253 | LGGGQyGEVYEGVWK | - | + | - | - | - | - | - | + | - |
| Abl | 257 | LGGGQYGEVyEGVWK | - | + | + | - | - | + | - | + | - |
| Abl | 393 | LMTGDTyTAHAGAK | + | + | + | + | + | + | - | + | - |
| Ack | 596 | KPTyDPVSEDQDPLSSDFKR | - | - | - | - | + | - | - | - | - |
| Bcr | 177 | GHGQPGADAEKPFyVNVEFHHER | + | + | + | + | + | + | - | + | - |
| Bcr | 246 | SSESSCGVDGDyEDAELNPR | - | + | - | - | - | - | - | - | - |
| Bcr | 436 | KTGQIWPNDGEGAFHGDADGSFGTPPGyGCAADR | - | + | - | - | - | - | - | - | + |
| Bcr | 591 | LASQLGVyR | - | + | - | - | - | - | - | + | - |
| Bcr | 598 | AFVDNyGVAMEMAEK | - | + | - | - | - | - | - | - | + |
| Bcr | 644 | NSLETLLyKPVDR, NSLETLLyKPVDRVTR | + | + | + | + | - | - | - | + | - |
| Btk | 223 | KVVALyDYMPMNANDLQLR | + | + | - | - | - | + | - | - | - |
| Btk | 225 | KVVALYDyMPMNANDLQLR | + | - | - | - | - | - | - | - | + |
| Btk | 361 | HLFSTIPELINyHQHNSAGLISR | + | - | - | - | - | - | - | - | + |
| Cdc2 | 15 | IEKIGEGTyGVVYK | + | + | + | + | + | + | + | - | - |
| Cdc2 | 19 | IGEGTYGVVyKGR | - | + | - | - | + | - | - | - | - |
| Cdk2 | 15 | VEKIGEGTyGVVYK | + | + | + | + | + | + | + | - | + |
| Cdk2 | 15 | IGEGTyGTVFK | + | - | - | - | - | - | - | - | - |
| DYRK1A | 145 | KVYNDGYDDDNyDYIVK | - | + | + | - | - | + | - | - | - |
| DYRK1A | 319 | IyQYIQSR | - | - | - | - | - | - | + | - | + |
| DYRK1A | 321 | IYQyIQSR | + | + | + | + | + | + | + | - | - |
| DYRK2 | 309 | VYTyIQSR | + | - | - | - | - | - | + | - | + |
| ERK1 | 204 | IADPEHDHTGFLTEyVATR | + | + | + | + | + | + | + | - | - |
| ERK2 | 187 | VADPDHDHTGFLTEyVATR | + | + | + | + | + | + | + | - | - |
| Fyn | 214 | KLDNGGYyITTR | - | + | - | - | - | - | - | - | + |
| Fyn | 420 | LIEDNEyTAR | + | - | - | - | + | - | - | - | - |
| GSK3-β | 216 | GEPNVSyICSR | + | + | + | + | + | + | + | - | - |
| Hck | 209 | TLDNGGFyISPR | - | - | - | - | - | - | + | - | + |
| Hck | 411 | VIEDNEyTAR | + | - | - | - | - | - | - | - | - |
| HIPK1 | 352 | AVCSTyLQSR | + | + | + | + | + | + | + | - | - |
| HIPK3 | 359 | TVCSTyLQSR | + | - | + | - | - | + | - | - | + |
| IGF1R | 1189 | DIYETDyYRK | - | - | - | - | + | - | - | - | - |
| Lck | 192 | NLDNGGFyISPR | - | - | - | - | + | - | - | - | - |
| Lck | 505 | SVLEDFFTATEGQyQPQP | - | + | - | - | + | + | - | - | - |
| Lyn | 193 | SLDNGGyYISPR | - | + | - | - | - | - | - | - | + |
| Lyn | 508 | AEERPTFDYLQSVLDDFYTATEGQyQQQP | + | - | - | - | - | + | + | - | - |
| P38-α | 182 | HTDDEMTGyVATR | - | + | + | + | + | + | + | - | - |
| PKCD | 313 | SDSASSEPVGIyQGFEK | + | - | - | - | - | - | - | - | - |
| PRP4 | 849 | LCDFGSASHVADNDITPyLVSR | + | + | + | + | + | + | + | - | - |
| SgK223 | 390 | CLGLTGEPQPPAHPQEATQPEPIyAESTK | + | - | - | - | - | + | - | - | - |
| Syk | 323 | QESTVSFNPyEPELAPWAADKGPQR | - | - | - | - | + | - | - | - | - |
| Syk | 348 | EALPMDTEVyESPyADPEEIRPK | - | - | - | - | + | - | - | + | - |
| Syk | 352 | EALPMDTEVYESPyADPEEIRPK | - | - | - | - | + | - | - | + | - |
PSDB ID . | pTyr Site . | Peptide . | BV173 . | K562 . | KCL22 . | KU812 . | SD1 . | SUP-B15 . | U937 . | Identified in K562 IMAC* . | Novel site . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Lipid kinase | |||||||||||
| PIK3R1 | 580 | DQyLMWLTQK | + | - | - | - | - | + | - | - | - |
| PIK3R2 | 467 | EYDQLYEEyTR | + | - | - | - | - | - | - | + | |
| Protein kinase | |||||||||||
| Abl | 115 | NGQGWVPSNyITPVNSLEK | + | + | + | + | - | + | - | - | - |
| Abl | 185 | INTASDGKLyVSSESR | + | + | + | + | + | + | - | + | - |
| Abl | 226 | NKPTVyGVSPNYDK | + | + | + | + | + | + | - | - | - |
| Abl | 253 | LGGGQyGEVYEGVWK | - | + | - | - | - | - | - | + | - |
| Abl | 257 | LGGGQYGEVyEGVWK | - | + | + | - | - | + | - | + | - |
| Abl | 393 | LMTGDTyTAHAGAK | + | + | + | + | + | + | - | + | - |
| Ack | 596 | KPTyDPVSEDQDPLSSDFKR | - | - | - | - | + | - | - | - | - |
| Bcr | 177 | GHGQPGADAEKPFyVNVEFHHER | + | + | + | + | + | + | - | + | - |
| Bcr | 246 | SSESSCGVDGDyEDAELNPR | - | + | - | - | - | - | - | - | - |
| Bcr | 436 | KTGQIWPNDGEGAFHGDADGSFGTPPGyGCAADR | - | + | - | - | - | - | - | - | + |
| Bcr | 591 | LASQLGVyR | - | + | - | - | - | - | - | + | - |
| Bcr | 598 | AFVDNyGVAMEMAEK | - | + | - | - | - | - | - | - | + |
| Bcr | 644 | NSLETLLyKPVDR, NSLETLLyKPVDRVTR | + | + | + | + | - | - | - | + | - |
| Btk | 223 | KVVALyDYMPMNANDLQLR | + | + | - | - | - | + | - | - | - |
| Btk | 225 | KVVALYDyMPMNANDLQLR | + | - | - | - | - | - | - | - | + |
| Btk | 361 | HLFSTIPELINyHQHNSAGLISR | + | - | - | - | - | - | - | - | + |
| Cdc2 | 15 | IEKIGEGTyGVVYK | + | + | + | + | + | + | + | - | - |
| Cdc2 | 19 | IGEGTYGVVyKGR | - | + | - | - | + | - | - | - | - |
| Cdk2 | 15 | VEKIGEGTyGVVYK | + | + | + | + | + | + | + | - | + |
| Cdk2 | 15 | IGEGTyGTVFK | + | - | - | - | - | - | - | - | - |
| DYRK1A | 145 | KVYNDGYDDDNyDYIVK | - | + | + | - | - | + | - | - | - |
| DYRK1A | 319 | IyQYIQSR | - | - | - | - | - | - | + | - | + |
| DYRK1A | 321 | IYQyIQSR | + | + | + | + | + | + | + | - | - |
| DYRK2 | 309 | VYTyIQSR | + | - | - | - | - | - | + | - | + |
| ERK1 | 204 | IADPEHDHTGFLTEyVATR | + | + | + | + | + | + | + | - | - |
| ERK2 | 187 | VADPDHDHTGFLTEyVATR | + | + | + | + | + | + | + | - | - |
| Fyn | 214 | KLDNGGYyITTR | - | + | - | - | - | - | - | - | + |
| Fyn | 420 | LIEDNEyTAR | + | - | - | - | + | - | - | - | - |
| GSK3-β | 216 | GEPNVSyICSR | + | + | + | + | + | + | + | - | - |
| Hck | 209 | TLDNGGFyISPR | - | - | - | - | - | - | + | - | + |
| Hck | 411 | VIEDNEyTAR | + | - | - | - | - | - | - | - | - |
| HIPK1 | 352 | AVCSTyLQSR | + | + | + | + | + | + | + | - | - |
| HIPK3 | 359 | TVCSTyLQSR | + | - | + | - | - | + | - | - | + |
| IGF1R | 1189 | DIYETDyYRK | - | - | - | - | + | - | - | - | - |
| Lck | 192 | NLDNGGFyISPR | - | - | - | - | + | - | - | - | - |
| Lck | 505 | SVLEDFFTATEGQyQPQP | - | + | - | - | + | + | - | - | - |
| Lyn | 193 | SLDNGGyYISPR | - | + | - | - | - | - | - | - | + |
| Lyn | 508 | AEERPTFDYLQSVLDDFYTATEGQyQQQP | + | - | - | - | - | + | + | - | - |
| P38-α | 182 | HTDDEMTGyVATR | - | + | + | + | + | + | + | - | - |
| PKCD | 313 | SDSASSEPVGIyQGFEK | + | - | - | - | - | - | - | - | - |
| PRP4 | 849 | LCDFGSASHVADNDITPyLVSR | + | + | + | + | + | + | + | - | - |
| SgK223 | 390 | CLGLTGEPQPPAHPQEATQPEPIyAESTK | + | - | - | - | - | + | - | - | - |
| Syk | 323 | QESTVSFNPyEPELAPWAADKGPQR | - | - | - | - | + | - | - | - | - |
| Syk | 348 | EALPMDTEVyESPyADPEEIRPK | - | - | - | - | + | - | - | + | - |
| Syk | 352 | EALPMDTEVYESPyADPEEIRPK | - | - | - | - | + | - | - | + | - |
For a complete listing of phosphotyrosine peptides found in these samples, see Table S1. Bcr-Abl—containing cell lines were BV173, K562, KCL22, KU812, SD1, and SUP-B15. Control cell line was U937. PSB ID is a protein identifier referenced from PhosphoSite.21
Sites identified in K562 IMAC appear in Salomon et al.40
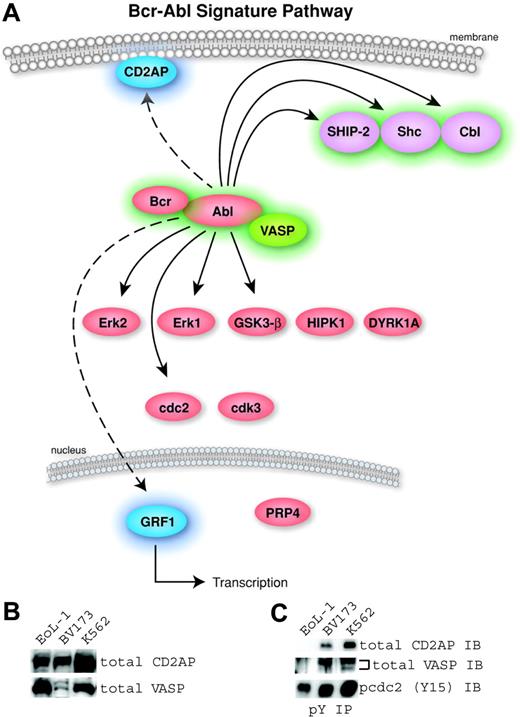
Identification of a Bcr-Abl kinase signature. (A) A pathway diagram of the 16 common proteins from Table 2 with cellular localization is shown. The proteins without highlighting were found in the Bcr-Abl–containing cells and U937 cells and were therefore not considered part of a signature for Bcr-Abl. The 6 signature proteins with a previous link to Bcr-Abl were highlighted in green (Abl, Bcr, Cbl, SHIP-2, Shc, and VASP). The signature proteins not previously identified in Bcr-Abl signaling are highlighted in blue (CD2AP and GRF1). (B) Expression of 2 novel human signature proteins in Bcr-Abl–containing (BV173 and K562) and control (EoL-1) cells was confirmed by Western blotting using antibodies to total CD2AP and VASP. (C) Phosphorylation in Bcr-Abl–containing cells was validated using antiphosphotyrosine immunoprecipitation followed by Western blotting using total CD2AP or VASP antibodies. Immunoblotting for p-cdc2 (Y15) was used to control for the efficiency of the phosphotyrosine immunoprecipitation because this site is ubiquitously phosphorylated in cell lines.
Identification of a Bcr-Abl kinase signature. (A) A pathway diagram of the 16 common proteins from Table 2 with cellular localization is shown. The proteins without highlighting were found in the Bcr-Abl–containing cells and U937 cells and were therefore not considered part of a signature for Bcr-Abl. The 6 signature proteins with a previous link to Bcr-Abl were highlighted in green (Abl, Bcr, Cbl, SHIP-2, Shc, and VASP). The signature proteins not previously identified in Bcr-Abl signaling are highlighted in blue (CD2AP and GRF1). (B) Expression of 2 novel human signature proteins in Bcr-Abl–containing (BV173 and K562) and control (EoL-1) cells was confirmed by Western blotting using antibodies to total CD2AP and VASP. (C) Phosphorylation in Bcr-Abl–containing cells was validated using antiphosphotyrosine immunoprecipitation followed by Western blotting using total CD2AP or VASP antibodies. Immunoblotting for p-cdc2 (Y15) was used to control for the efficiency of the phosphotyrosine immunoprecipitation because this site is ubiquitously phosphorylated in cell lines.
Our analysis identified 19 phosphorylation sites from 16 proteins common to all 6 cell lines (Table 2). Of these 19 sites, 8 are not specific to Bcr-Abl signaling. Six sites—Cdc2 (Y15), Cdk3 (Y15), DYRK1A (Y321), GSK3-β (Y216), HIPK1 (Y352), and PRP4 (Y849)—have been identified in many cell lines including the U937 cell line, which does not contain Bcr-Abl.14 The remaining 2 sites are phosphorylation sites in ERK1 (Y204) and ERK2 (Y187), indicating the activation of these kinases. These phosphosites were inhibited by imatinib in Bcr-Abl–containing cells and are known to be involved in cell growth through the activation of many tyrosine kinases. Although these sites are relevant to Bcr-Abl signaling, we did not consider them part of a Bcr-Abl signature since they were also found in the U937 cells. A pathway diagram containing the 16 common proteins found in this study is shown (Figure 2A). The proteins with a previous link to Bcr-Abl signaling (Bcr, Abl, SHIP-2, Shc, and Cbl) are highlighted in green. Two Bcr-Abl signature proteins, CD2AP (CD2 associated protein) and the transcription factor GRF1 (glucocorticoid receptor DNA-binding factor 1), have no known involvement with Bcr-Abl signaling and are represented in blue in the common pathway.
Phosphotyrosine peptides common to Bcr-Abl–containing cells
PSDB ID . | NCBI access no. . | pTyr site . | Peptide . | Bcr-Abl link . | Present in U937 . | Present in EoL-1 . |
|---|---|---|---|---|---|---|
| Abl | NP_005148 | 185 | INTASDGKLyVSSESR | + | - | - |
| Abl | NP_005148 | 226 | NKPTVyGVSPNYDK | + | - | - |
| Abl | NP_005148 | 393 | LMTGDTyTAHAGAK | + | - | - |
| Bcr | NP_004318 | 177 | GHGQPGADAEKPFyVNVEFHHER | + | - | - |
| Cbl | NP_005179 | 674 | IKPSSSANAlySLAAR | + | - | + |
| CD2AP | NP_036252 | 548 | DTCYSPKPSVyLSTPSSASK | - | - | - |
| cdc2 | NP_001777 | 15 | IEKIGEGTyGWYK | - | + | + |
| cdk3 | NP_001249 | 15 | VEKIGEGTyGWYK | - | + | - |
| DYRK1A | NP_001387 | 321 | IYQylQSR | - | + | + |
| GRF1 | NP_004482 | 1106 | NEEENlySVPHDSTQGK | - | - | - |
| GSK3-β | NP_002084 | 216 | GEPNVSylCSR | - | + | + |
| HIPK1 | NP_689909 | 352 | AVCSTyLQSR | - | + | - |
| SHIP-2 | NP_001558 | 986 | NSFNNPAyYVLEGVPHQLLPPEPPSPAR | + | - | + |
| SHIP-2 | NP_001558 | 1135 | TLSEVDyAPAGPAR | + | - | - |
| Erk1 | NP_002737 | 204 | IADPEHDHTGFLTEyVATR | + | + | + |
| Erk2 | NP_002736 | 187 | VADPDHDHTGFLTEyVATR | + | + | + |
| PRP4 | NP_003904 | 849 | LCDFGSASHVADNDITPyLVSR | - | + | + |
| Shc | NP_892113 | 427 | ELFDDPSyVNVQNLDKAR | + | - | + |
| VASP | NP_001008736 | 39 | VQlyHNPTANSFR | + | - | - |
PSDB ID . | NCBI access no. . | pTyr site . | Peptide . | Bcr-Abl link . | Present in U937 . | Present in EoL-1 . |
|---|---|---|---|---|---|---|
| Abl | NP_005148 | 185 | INTASDGKLyVSSESR | + | - | - |
| Abl | NP_005148 | 226 | NKPTVyGVSPNYDK | + | - | - |
| Abl | NP_005148 | 393 | LMTGDTyTAHAGAK | + | - | - |
| Bcr | NP_004318 | 177 | GHGQPGADAEKPFyVNVEFHHER | + | - | - |
| Cbl | NP_005179 | 674 | IKPSSSANAlySLAAR | + | - | + |
| CD2AP | NP_036252 | 548 | DTCYSPKPSVyLSTPSSASK | - | - | - |
| cdc2 | NP_001777 | 15 | IEKIGEGTyGWYK | - | + | + |
| cdk3 | NP_001249 | 15 | VEKIGEGTyGWYK | - | + | - |
| DYRK1A | NP_001387 | 321 | IYQylQSR | - | + | + |
| GRF1 | NP_004482 | 1106 | NEEENlySVPHDSTQGK | - | - | - |
| GSK3-β | NP_002084 | 216 | GEPNVSylCSR | - | + | + |
| HIPK1 | NP_689909 | 352 | AVCSTyLQSR | - | + | - |
| SHIP-2 | NP_001558 | 986 | NSFNNPAyYVLEGVPHQLLPPEPPSPAR | + | - | + |
| SHIP-2 | NP_001558 | 1135 | TLSEVDyAPAGPAR | + | - | - |
| Erk1 | NP_002737 | 204 | IADPEHDHTGFLTEyVATR | + | + | + |
| Erk2 | NP_002736 | 187 | VADPDHDHTGFLTEyVATR | + | + | + |
| PRP4 | NP_003904 | 849 | LCDFGSASHVADNDITPyLVSR | - | + | + |
| Shc | NP_892113 | 427 | ELFDDPSyVNVQNLDKAR | + | - | + |
| VASP | NP_001008736 | 39 | VQlyHNPTANSFR | + | - | - |
Bcr-Abl-containing cell lines were BV173, K562, KCL22, KU812, SD1, and SUP-B15. Control cell line was U937. Peptides found in the U937 and EoL-1 cells are indicated.
In this study, tyrosine phosphorylation of the novel Bcr-Abl signature proteins CD2AP and VASP (VASP is associated with Bcr-Abl signaling in Drosophila but not in human cells) was not detected in cells that do not express the Bcr-Abl kinase. It is possible that cells lacking Bcr-Abl have little or no expression of these proteins. To confirm expression of novel signature proteins in the cell lines analyzed, we performed immunoblots of total CD2AP and VASP using whole cell lysates of control (EoL-1) and Bcr-Abl–containing cells (BV173 and K562). The expression of CD2AP and VASP proteins was detectable in the 3 cell lines (Figure 2B). To confirm tyrosine phosphorylation of these proteins, we immunoprecipitated with phosphotyrosine antibody followed by Western blots for total CD2AP and VASP (Figure 2C). The Bcr-Abl–containing, but not the control, cells showed tyrosine phosphorylation of CD2AP and VASP. Abl (Y185, Y226, and 393), Bcr (Y177), Cbl (Y674), CD2AP (Y548), GRF1 (Y1106), SHIP-2 (Y986 and Y1135), Shc (Y427), and VASP (Y39) can be defined as a phosphotyrosine signature for the Bcr-Abl kinase regardless of cell lineage or fusion type.
Differential phosphorylation of the Bcr-Abl protein in alternative fusions
Initial data-dependent phosphopeptide analysis suggested differential phosphorylation of Bcr-Abl among the 3 fusion types. Given that all identified Bcr-Abl phosphorylation sites were present in K562 cells, we compared a representative cell line for each Bcr-Abl fusion type to K562. During analysis, the mass spectrometer chooses the 4 most intense peptides eluting from the LC column for MS/MS fragmentation. Because of sample complexity, peptides may not be identified during this initial analysis. To overcome this issue, phosphotyrosine peptides from the 4 representative cell lines were subjected to a targeted tandem mass spectrometric analysis of all observed Bcr-Abl phosphopeptides. This targeted strategy expands the effective dynamic range of the mass spectrometer by focusing analysis time exclusively on the phosphopeptides of interest, resulting in a greater chance of identification (Table 3).
Targeted analysis of phosphopeptides found in the Bcr and Abl proteins
PSDB ID and pTyr site . | Peptide . | K562 (b3a2) . | KU812 (b3a2) . | BV-173 (b2a2) . | SUP-B15 (e1a2) . |
|---|---|---|---|---|---|
| Abl | |||||
| 115 | NGQGWVPSNyITPVNSLEK | + | + | + | + |
| 185 | INTASDGKLyVSSESR | + | + | + | + |
| 226 | NKPTVyGVSPNYDKWEMER | + | + | + | + |
| 253 | LGGGQyGEVYEGVWK | + | - | - | - |
| 257 | LGGGQYGEVyEGVWK | + | + | + | + |
| 393 | LMTGDTyTAHAGAK | + | + | + | + |
| Bcr | |||||
| 177 | KGHGQPGADAEKPFyVNVEFHHER | + | + | + | + |
| 246 | SSESSCGVDGDyEDAELNPR | + | - | + | + |
| 436 | KTGQIWPNDGEGAFHGDADGSFGTPPGyGCAADR | + | - | - | - |
| 591 | LASQLGVyR | + | + | - | - |
| 598 | AFVDNyGVAMEMAEK | + | - | - | - |
| 644 | NSLETLLyKPVDR | + | + | + | - |
PSDB ID and pTyr site . | Peptide . | K562 (b3a2) . | KU812 (b3a2) . | BV-173 (b2a2) . | SUP-B15 (e1a2) . |
|---|---|---|---|---|---|
| Abl | |||||
| 115 | NGQGWVPSNyITPVNSLEK | + | + | + | + |
| 185 | INTASDGKLyVSSESR | + | + | + | + |
| 226 | NKPTVyGVSPNYDKWEMER | + | + | + | + |
| 253 | LGGGQyGEVYEGVWK | + | - | - | - |
| 257 | LGGGQYGEVyEGVWK | + | + | + | + |
| 393 | LMTGDTyTAHAGAK | + | + | + | + |
| Bcr | |||||
| 177 | KGHGQPGADAEKPFyVNVEFHHER | + | + | + | + |
| 246 | SSESSCGVDGDyEDAELNPR | + | - | + | + |
| 436 | KTGQIWPNDGEGAFHGDADGSFGTPPGyGCAADR | + | - | - | - |
| 591 | LASQLGVyR | + | + | - | - |
| 598 | AFVDNyGVAMEMAEK | + | - | - | - |
| 644 | NSLETLLyKPVDR | + | + | + | - |
Five of the 6 Abl sites were phosphorylated in all cell lines analyzed; Y253 was identified only in K562 cells. As expected, Bcr (Y177), a known phosphorylation site implicated in Gab2 binding and Bcr-Abl–mediated transformation,22,29 was phosphorylated in all 4 cell lines. Five other Bcr sites were differentially phosphorylated among the 4 cell lines analyzed. Some differences resulted from the sequence of the fusion. Y591-, Y598-, and Y644-containing peptides were not present in the e1a2 fusion form because it only contains Bcr residues 1-426. Other notable unpredicted differences in phosphorylation pattern included the detection of Bcr (Y591) phosphorylation in the b3a2 fusion but not in the b2a2 fusion protein. Phosphorylation of this residue could be relevant to reported phenotypic differences between the b3a2 and the b2a2 fusion types.30
Imatinib-responsive phosphorylation sites
Identifying phosphorylation changes after kinase inhibition is necessary for identifying drug targets within disease models. To assess changes in phosphorylation after drug treatment, we used SILAC17,18 to compare untreated and imatinib-treated K562 cells. We arbitrarily required at least a 2-fold change in phosphopeptide intensity as a threshold to define an imatinib-sensitive site. For most drug-responsive phosphorylation sites in our quantitative study, drug treatment resulted in phosphorylation diminished to the extent that the treated phosphopeptide was undetectable beneath the mass spectrometer's baseline background signal. In these cases, numeric quantitation is based on the intensity of the untreated peptide compared with the noise level to provide a minimum decrease in phosphorylation level in response to drug treatment. Therefore, peptides observed at high signal-to-noise ratios are reported to have decreased in abundance to a higher minimum extent than peptides observed at lower signal-to-noise ratios. Through SILAC analysis, we confirmed the known imatinib sensitivity of several phosphorylation sites, including signature sites such as the Bcr-Abl protein itself, and other downstream effectors, including STAT5A, Shc, and ERK. In addition, we observed drug sensitivity in novel phosphorylation sites such as CD2AP (Y548), the tight junction protein ZO2 (Y1118), and LIM (Y142) (Table 4). Phosphorylation sites that were found in most cell lines, including U937 (Cdc2, DYRK1A, GSK3-β, and PRP4 [at the sites previously indicated]) were not sensitive to imatinib treatment (Table 4).
SILAC analysis of K562 cells to quantify phosphorylation site responsiveness to imatinib
PSDB ID . | pTyr site . | Peptide . | Control/treated . |
|---|---|---|---|
| Peptides that are less abundant following imatinib treatment | |||
| Adaptor/scaffold | |||
| Abi-1 | 213 | TLEPVKPPTVPNDyMTSPAR | > 2 |
| CD2AP | 548 | DTCYSPKPSVyLSTPSSASK | > 2 |
| Gab1 | 406 | DASSQDCyDIPR | > 4 |
| Shc | 427 | ELFDDPSyVNVQNLDK | > 10 |
| Protein kinase | |||
| Abl | 115 | NGQGWVPSNyITPVNSLEK | > 10 |
| Abl | 393 | LMTGDTyTAHAGAK | > 20 |
| Abl | 393 | VADFGLSRLMTGDTyTAHAGAK | > 10 |
| Bcr | 177 | GHGQPGADAEKPFyVNVEFHHER | > 2 |
| Bcr | 177 | KGHGQPGADAEKPFyVNVEFHHER | > 10 |
| Bcr | 246 | SSESSCGVDGDyEDAELNPR | > 2 |
| Bcr | 591 | LASQLGVyR | > 5 |
| Bcr | 644 | NSLETLLyKPVDR | > 5 |
| ERK2 | 187 | VADPDHDHTGFLTEyVATR | > 2 |
| Lipid phosphatase | |||
| SHIP-2 | 986 | NSFNNPAYyVLEGVPHQLLPPEPPSPAR | > 20 |
| SHIP-2 | 1135 | TLSEVDyAPAGPAR | > 10 |
| Transcription factor | |||
| STAT5A | 694 | AVDGyVKPQIK | > 20 |
| Other | |||
| LIM | 142 | YTEFyHVPTHSDASK | > 2 |
| ZO2 | 1118 | IEIAQKHPDIyAVPIK | > 2 |
| Peptide intensities remaining unchanged following imatinib treatment | |||
| Protein kinase | |||
| Cdc2 | 15 | IEKIGEGTyGVVYK | 1.10* |
| Cdc2 | 15 | IGEGTyGVVYK | 1.04* |
| Cdc2 | 15 | IGEGTyGVVYKGR | 1.04* |
| DYRK1A | 321 | IYQyIQSR | 1.04* |
| GSK3-β | 216 | GEPNVSyICSR | 1.22* |
| HIPK1 | 352 | AVCSTyLQSR | 1.03* |
| p-38-α | 182 | HTDDEMTGyVATR | 1.61 |
| PRP4 | 849 | LCDFGSASHVADNDITPyLVSR | 1.14* |
| Other | |||
| eEF1A-1 | 29 | STTTGHLIyK | 1.33 |
| Peptides without definitive quantification | |||
| Adaptor/scaffold | |||
| Cbl | 674 | IKPSSSANAIySLAARPLPVPK | ND |
| Other | |||
| Calmodulin | 100 | VFDKDGNGyISAAELR | ND |
PSDB ID . | pTyr site . | Peptide . | Control/treated . |
|---|---|---|---|
| Peptides that are less abundant following imatinib treatment | |||
| Adaptor/scaffold | |||
| Abi-1 | 213 | TLEPVKPPTVPNDyMTSPAR | > 2 |
| CD2AP | 548 | DTCYSPKPSVyLSTPSSASK | > 2 |
| Gab1 | 406 | DASSQDCyDIPR | > 4 |
| Shc | 427 | ELFDDPSyVNVQNLDK | > 10 |
| Protein kinase | |||
| Abl | 115 | NGQGWVPSNyITPVNSLEK | > 10 |
| Abl | 393 | LMTGDTyTAHAGAK | > 20 |
| Abl | 393 | VADFGLSRLMTGDTyTAHAGAK | > 10 |
| Bcr | 177 | GHGQPGADAEKPFyVNVEFHHER | > 2 |
| Bcr | 177 | KGHGQPGADAEKPFyVNVEFHHER | > 10 |
| Bcr | 246 | SSESSCGVDGDyEDAELNPR | > 2 |
| Bcr | 591 | LASQLGVyR | > 5 |
| Bcr | 644 | NSLETLLyKPVDR | > 5 |
| ERK2 | 187 | VADPDHDHTGFLTEyVATR | > 2 |
| Lipid phosphatase | |||
| SHIP-2 | 986 | NSFNNPAYyVLEGVPHQLLPPEPPSPAR | > 20 |
| SHIP-2 | 1135 | TLSEVDyAPAGPAR | > 10 |
| Transcription factor | |||
| STAT5A | 694 | AVDGyVKPQIK | > 20 |
| Other | |||
| LIM | 142 | YTEFyHVPTHSDASK | > 2 |
| ZO2 | 1118 | IEIAQKHPDIyAVPIK | > 2 |
| Peptide intensities remaining unchanged following imatinib treatment | |||
| Protein kinase | |||
| Cdc2 | 15 | IEKIGEGTyGVVYK | 1.10* |
| Cdc2 | 15 | IGEGTyGVVYK | 1.04* |
| Cdc2 | 15 | IGEGTyGVVYKGR | 1.04* |
| DYRK1A | 321 | IYQyIQSR | 1.04* |
| GSK3-β | 216 | GEPNVSyICSR | 1.22* |
| HIPK1 | 352 | AVCSTyLQSR | 1.03* |
| p-38-α | 182 | HTDDEMTGyVATR | 1.61 |
| PRP4 | 849 | LCDFGSASHVADNDITPyLVSR | 1.14* |
| Other | |||
| eEF1A-1 | 29 | STTTGHLIyK | 1.33 |
| Peptides without definitive quantification | |||
| Adaptor/scaffold | |||
| Cbl | 674 | IKPSSSANAIySLAARPLPVPK | ND |
| Other | |||
| Calmodulin | 100 | VFDKDGNGyISAAELR | ND |
ND indicates not determined.
Cdc2, DYRK1A, GSK3-beta, HIPK1 and PRP4 were not expected to be drug sensitive from the signature data, and can be viewed as a representative ratio of control/treated.
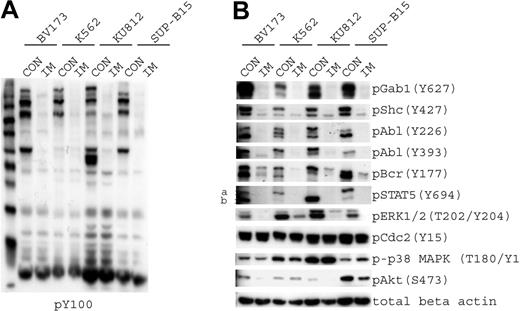
We chose a subset of phosphorylation sites that we could examine using phosphospecific antibodies to confirm the targeted and SILAC experimental results. The 4 cell lines showed a marked reduction in phosphotyrosine signaling after imatinib treatment when measured by phosphotyrosine immunoblots (Figure 3A). We confirmed that Gab1, Shc, Bcr, Abl, ERK, and STAT5A tyrosine phosphorylation decreased after imatinib treatment. The tyrosine phosphorylation of Cdc2 and p38 MAPK remained unchanged after treatment, as was measured by SILAC (Figure 3B).
Imatinib-responsive phosphoproteins. (A) The 4 cell lines that were used for targeted analysis (BV173, K562, KU812, and SUP-B15) were treated with diluent (CON) or imatinib (IM) to inhibit Bcr-Abl kinase activity. Samples were immunoblotted using antiphosphotyrosine antibody. (B) Cells treated as described in panel A were immunoblotted for phosphoproteins that exhibited a change in phosphorylation level, as determined by SILAC analysis in K562 cells. Phospho-Gab1 (Y406) was identified by mass spectrometry, but a phosphospecific antibody that recognizes this site was not available; therefore, we used a p-Gab1 (Y627) antibody. Protein loading was normalized with the use of β-actin.
Imatinib-responsive phosphoproteins. (A) The 4 cell lines that were used for targeted analysis (BV173, K562, KU812, and SUP-B15) were treated with diluent (CON) or imatinib (IM) to inhibit Bcr-Abl kinase activity. Samples were immunoblotted using antiphosphotyrosine antibody. (B) Cells treated as described in panel A were immunoblotted for phosphoproteins that exhibited a change in phosphorylation level, as determined by SILAC analysis in K562 cells. Phospho-Gab1 (Y406) was identified by mass spectrometry, but a phosphospecific antibody that recognizes this site was not available; therefore, we used a p-Gab1 (Y627) antibody. Protein loading was normalized with the use of β-actin.
Comparison of phosphotyrosine profiles of an alternative imatinib-sensitive fusion kinase
Imatinib is also used clinically to inhibit kinases other than Bcr-Abl, such as PDGFR and c-Kit.31,32 These kinases may use different oncogenic signaling pathways, but they may also share important components required to transform cells. To evaluate the similarities and differences in the imatinib response of 2 of these kinases, we compared our profile of tyrosine-phosphorylated peptides in Bcr-Abl–containing cells to that of the EoL-1 cell line, a hypereosinophilic (HES) leukemic cell line that expresses a fusion between the FIP1L1 protein with PDGFRα.15,16 Through phosphotyrosine profiling, we identified 51 nonredundant phosphopeptides representing 49 phosphorylation sites (Table S2), many of which are kinases, including both portions of the fusion kinase FIP1L1 and PDGFRα (Table 5). The phosphorylation sites found in the PDGFRα kinase (Y742, Y762, Y768, and Y1018) were all previously reported33,34 (reviewed in Ikuno et al35 ). Interestingly, we found 2 novel sites, Y95 and Y98, within the PDGFRα fusion partner FIP1L1. Unlike the Bcr-Abl cell lines, we identified many phosphorylated peptides from the regulatory and catalytic subunits of PI3K and the phosphorylated PI3K adaptor protein BCAP (Y392) (Tables 4 and S2) implicated in B cell receptor signaling to PI3K and Akt.36 Three of the Bcr-Abl signature peptides—Cbl (Y674), Shc (Y427), and SHIP-2 (Y986)—were also identified in the EoL-1 cell line.
Kinases identified in the FIP1L1-PDGFRα cell line EoL-1
PSDB ID . | pTyr sites . | Peptide . | Novel site . |
|---|---|---|---|
| Lipid kinase | |||
| PIK3CB | 772 | EALSDLQSPLNPCVILSELyVEK | Yes |
| PIK3CD | 440 | CLYyMWPSVPDEKGELLNPTGTVR | Yes |
| PIK3CD | 485 | SNPNTDSAAALLICLPEVAPHPVYyPALEK | Yes |
| PIK3CD | 524 | GSGELyEHEKDLVWK | Yes |
| PIK3R1 | 580 | DQyLMWLTQK | No |
| Protein kinase | |||
| Ack | 518 | KPTyDPVSEDQDPLSSDFKR | No |
| Cdc2 | 15 | IEKIGEGTyGVVYK | No |
| Cdc2 | 19 | IEKIGEGTYGVVyK | No |
| DYRK1A | 321 | IYQyIQSR | No |
| ERK1 | 204 | IADPEHDHTGFLTEyVATR | No |
| ERK2 | 187 | VADPDHDHTGFLTEyVATR | No |
| FIP1L1* | 95 | TGAPQyGSYGTAPVNLNIK | Yes |
| FIP1L1* | 98 | TGAPQYGSyGTAPVNLNIK | Yes |
| GSK3-β | 216 | GEPNVSyICSR | No |
| Hck | 411 | VIEDNEyTAR | No |
| Hck | 522 | NRPEERPTFEYIQSVLDDFYTATESQyQQQP | No |
| Lyn | 507 | EKAEERPTFDYLQSVLDDFYTATEGQyQQQP | No |
| PDGFRα | 742 | QADTTQyVPMLER | No |
| PDGFRα | 762, 768 | SLyDRPASyK | No |
| PDGFRα | 1018 | LSADSGyIIPLPDIDPVPEEEDLGKR | No |
| PRP4 | 849 | LCDFGSASHVADNDITPyLVSR | No |
| Syk | 323 | QESTVSFNPyEPELAPWAADKGPQR | No |
PSDB ID . | pTyr sites . | Peptide . | Novel site . |
|---|---|---|---|
| Lipid kinase | |||
| PIK3CB | 772 | EALSDLQSPLNPCVILSELyVEK | Yes |
| PIK3CD | 440 | CLYyMWPSVPDEKGELLNPTGTVR | Yes |
| PIK3CD | 485 | SNPNTDSAAALLICLPEVAPHPVYyPALEK | Yes |
| PIK3CD | 524 | GSGELyEHEKDLVWK | Yes |
| PIK3R1 | 580 | DQyLMWLTQK | No |
| Protein kinase | |||
| Ack | 518 | KPTyDPVSEDQDPLSSDFKR | No |
| Cdc2 | 15 | IEKIGEGTyGVVYK | No |
| Cdc2 | 19 | IEKIGEGTYGVVyK | No |
| DYRK1A | 321 | IYQyIQSR | No |
| ERK1 | 204 | IADPEHDHTGFLTEyVATR | No |
| ERK2 | 187 | VADPDHDHTGFLTEyVATR | No |
| FIP1L1* | 95 | TGAPQyGSYGTAPVNLNIK | Yes |
| FIP1L1* | 98 | TGAPQYGSyGTAPVNLNIK | Yes |
| GSK3-β | 216 | GEPNVSyICSR | No |
| Hck | 411 | VIEDNEyTAR | No |
| Hck | 522 | NRPEERPTFEYIQSVLDDFYTATESQyQQQP | No |
| Lyn | 507 | EKAEERPTFDYLQSVLDDFYTATEGQyQQQP | No |
| PDGFRα | 742 | QADTTQyVPMLER | No |
| PDGFRα | 762, 768 | SLyDRPASyK | No |
| PDGFRα | 1018 | LSADSGyIIPLPDIDPVPEEEDLGKR | No |
| PRP4 | 849 | LCDFGSASHVADNDITPyLVSR | No |
| Syk | 323 | QESTVSFNPyEPELAPWAADKGPQR | No |
For a complete listing of phosphotyrosine peptides found in this sample, see Table S2.
Fusion partner with PDGFRα.
We analyzed EoL-1 cells using SILAC to identify the imatinib-responsive phosphorylation sites in EoL-1 cells. In this analysis, 2 of the proteins common to the Bcr-Abl signature (Shc and SHIP-2) were also responsive to imatinib in the PDGFRα-containing cell line (Table 6). In addition, PDGFRα and FIP1L1 are dephosphorylated after imatinib treatment, as predicted.15,16 Three of the phosphorylation sites, PIK3CB (Y504 and Y505) and PIK3R1 (Y467), from PI3K were also responsive to imatinib treatment. These data suggest that the phosphorylation of PI3K subunits may play an important role in FIP1L1-PDGFRα–mediated signaling, perhaps mediating the activation of Akt survival signals.
SILAC analysis of imatinib-treated EoL-1 cells
PSDB ID . | pTyr sites . | Peptide . | Fold reduction, control/treated . | Found in Bcr-Abl signature . |
|---|---|---|---|---|
| Peptides that are less abundant after imatinib treatment | ||||
| Adaptor/scaffold | ||||
| Shc | 427 | ELFDDPSyVNVQNLDK | > 10 | Yes* |
| Protein kinase | ||||
| ERK2 | 187 | VADPDHDHTGFLTEyVATR | > 10 | No |
| FIP1L1 | 98 | TGAPQYGSyGTAPVNLNIK | > 2 | No |
| PDGFR-α | 742 | QADTTQyVPMLER | > 2 | No |
| PDGFR-α | 762 | SLyDRPASYK | > 2 | No |
| PDGFR-α | 762, 768 | SLyDRPASyK | > 10 | No |
| Lipid kinase | ||||
| PIK3CB | 504, 505 | KQPYyyPPFDK | > 10 | No |
| PIK3R1 | 467 | SREYDRLyEEYTR | > 2 | No |
| Lipid phosphatase | ||||
| SHIP | 914 | APPCSGSSITEIINPNyMGVGPFGPPMPLHVK | > 2 | No |
| SHIP-2 | 986 | NSFNNPAyYVLEGVPHQLLPPEPPSPAR | > 5 | Yes* |
| Transcription factor | ||||
| STAT5A | 694 | AVDGyVKPQIK | > 5 | No |
| Other | No | |||
| Calmodulin | 100 | VFDKDGNGyISAAELR | > 2 | No |
| hnRNP-I | 127 | GQPIyIQFSNHK | > 2 | No |
| Peptide intensities remaining unchanged after imatinib treatment | ||||
| Protein kinase | ||||
| Cdc2 | 15 | IGEGTyGVVYK | 1.89 | No |
| DYRK1A | 321 | IYQyIQSR | 1.23 | No |
| GSK3-β | 216 | GEPNVSyICSR | 0.81 | No |
| PRP4 | 849 | LCDFGSASHVADNDITPyLVSR | 0.96 | No |
| Syk | 323 | QESTVSFNPyEPELAPWAADKGPQR | 0.70 | No |
| Lipid kinase | ||||
| PIK3CD | 485 | SNPNTDSAAALLICLPEVAPHPVYyPALEK | 1.39 | No |
| PIK3CD | 524 | GSGELyEHEKDLVWK | 1.20 | No |
| PIK3R1 | 580 | DQyLMWLTQK | 1.12 | No |
| PIK3R2 | 453 | VYHQQyQD | 1.72 | No |
| Protein phosphatase | ||||
| SHP-2 | 62 | IQNTGDyYDLYGGEK | 1.33 | No |
| SHP-2 | 580 | EDSARVyENVGLMQQQK | 1.33 | No |
| Other | ||||
| PAG | 317 | SREEDPTLTEEEISAMySSVNKPGQLVNK | 0.92 | No |
| PAG | 417 | ENDyESISDLQQGR | 0.61 | No |
| PSMB6 | 59 | TTTGSyIANR | 1.25 | No |
| THTP | 30 | LQELGGTLEyR | 0.68 | No |
PSDB ID . | pTyr sites . | Peptide . | Fold reduction, control/treated . | Found in Bcr-Abl signature . |
|---|---|---|---|---|
| Peptides that are less abundant after imatinib treatment | ||||
| Adaptor/scaffold | ||||
| Shc | 427 | ELFDDPSyVNVQNLDK | > 10 | Yes* |
| Protein kinase | ||||
| ERK2 | 187 | VADPDHDHTGFLTEyVATR | > 10 | No |
| FIP1L1 | 98 | TGAPQYGSyGTAPVNLNIK | > 2 | No |
| PDGFR-α | 742 | QADTTQyVPMLER | > 2 | No |
| PDGFR-α | 762 | SLyDRPASYK | > 2 | No |
| PDGFR-α | 762, 768 | SLyDRPASyK | > 10 | No |
| Lipid kinase | ||||
| PIK3CB | 504, 505 | KQPYyyPPFDK | > 10 | No |
| PIK3R1 | 467 | SREYDRLyEEYTR | > 2 | No |
| Lipid phosphatase | ||||
| SHIP | 914 | APPCSGSSITEIINPNyMGVGPFGPPMPLHVK | > 2 | No |
| SHIP-2 | 986 | NSFNNPAyYVLEGVPHQLLPPEPPSPAR | > 5 | Yes* |
| Transcription factor | ||||
| STAT5A | 694 | AVDGyVKPQIK | > 5 | No |
| Other | No | |||
| Calmodulin | 100 | VFDKDGNGyISAAELR | > 2 | No |
| hnRNP-I | 127 | GQPIyIQFSNHK | > 2 | No |
| Peptide intensities remaining unchanged after imatinib treatment | ||||
| Protein kinase | ||||
| Cdc2 | 15 | IGEGTyGVVYK | 1.89 | No |
| DYRK1A | 321 | IYQyIQSR | 1.23 | No |
| GSK3-β | 216 | GEPNVSyICSR | 0.81 | No |
| PRP4 | 849 | LCDFGSASHVADNDITPyLVSR | 0.96 | No |
| Syk | 323 | QESTVSFNPyEPELAPWAADKGPQR | 0.70 | No |
| Lipid kinase | ||||
| PIK3CD | 485 | SNPNTDSAAALLICLPEVAPHPVYyPALEK | 1.39 | No |
| PIK3CD | 524 | GSGELyEHEKDLVWK | 1.20 | No |
| PIK3R1 | 580 | DQyLMWLTQK | 1.12 | No |
| PIK3R2 | 453 | VYHQQyQD | 1.72 | No |
| Protein phosphatase | ||||
| SHP-2 | 62 | IQNTGDyYDLYGGEK | 1.33 | No |
| SHP-2 | 580 | EDSARVyENVGLMQQQK | 1.33 | No |
| Other | ||||
| PAG | 317 | SREEDPTLTEEEISAMySSVNKPGQLVNK | 0.92 | No |
| PAG | 417 | ENDyESISDLQQGR | 0.61 | No |
| PSMB6 | 59 | TTTGSyIANR | 1.25 | No |
| THTP | 30 | LQELGGTLEyR | 0.68 | No |
Peptides common to the Bcr-Abl signature.
Discussion
The role for Bcr-Abl in CML is well established. Many studies have investigated signaling cascades downstream of this oncogenic kinase in a focused, individual protein manner. Using this approach, several sites of tyrosine phosphorylation downstream of Bcr-Abl have been identified. However, using a robust, unbiased phosphoproteomics approach to study 6 different Bcr-Abl cell lines, we identified novel aspects of Bcr-Abl signaling potentially important to understanding the disease mechanism. For example, we identified 2 new Bcr-Abl signature phosphoproteins with previously unknown involvement in Bcr-Abl signaling. Phosphorylation of CD2AP (Y548) may regulate its binding to Cbl37 (another signature protein) and may play a role in the activation of Akt.38 Phosphorylation of the transcriptional repressor of the glucocorticoid receptor GRF1 at (Y1106) was also identified in our Bcr-Abl signature. The gene for GRF1 is found on chromosome 19q13.3, a region that is deleted in several tumor types.39 This site is also phosphorylated in anaplastic lymphoma cells and Jurkat cells,14 but its function is unknown.
An earlier study examined the phosphorylation profile of K562 cells using immunoprecipitation of phosphotyrosine proteins followed by immobilized metal affinity chromatography (IMAC) to enrich for phosphopeptides40 in lieu of directly immunoprecipitating phosphopeptides, as in our study. With the use of this method, 28 phosphotyrosine residues were identified compared with 89 phosphotyrosine sites found in our analysis of the same cells. In the current study, we compared Bcr-Abl signaling in cells of different origin and fusion type to get a global phosphotyrosine signature in Bcr-Abl–containing cells instead of focusing on one fusion or cell type. Many of the phosphorylation sites identified in our study were previously unknown, whereas others are novel in the context of Bcr-Abl signaling. Several of the signature phosphorylation sites found (sites in Abl, Bcr, Shc, and SHIP-2) in our study were also identified by Salomon et al40 (sites found within kinases are recorded in Table 1), but many phosphorylation sites—including the “signature” sites in VASP, CD2AP, and GRF1—are not found in that report. In addition, we determined phosphorylation sites that are responsive to imatinib treatment using the SILAC method to directly compare treated and untreated samples. We have not detected every site of tyrosine phosphorylation that has been previously reported in the context of Bcr-Abl. Several known phosphorylated peptides were missing, including sites within CrkL, Crk, and PI3K. There are several possible explanations for failure to identify a particular phosphopeptide through mass spectrometry. For efficient detection through LC-MS/MS, the tryptic peptide should contain 10 to 30 amino acids. Other potential problems for detection include lack of peptide solubility, poor ionization, poor fragmentation, very low stoichiometry of phosphorylation, and very low copy number. Although not every phosphorylation site will be detected using a mass spectrometric approach, this study has resulted in the most comprehensive description of Bcr-Abl signaling to date.
In addition to the Bcr-Abl kinase signature, we also found phosphorylation site differences attributable to cell lineage and fusion type. Phosphopeptides from the B cell markers CD19 and LAPTM5 were found in the 3 pre-B cell lines (BV173, SD1, and SUP-B15) but not in the myeloid cell lines. The SUP-B15 cell line also contained many phosphopeptides involved in G-protein signaling, such as centaurin delta 1, GDI2, and GIT1. The calcium-regulated membrane protein annexin A2 (Y24) phosphorylation site was found only in the b3a2 cell lines (K562 and KU812). The phosphorylation of (Y77) in centaurin delta 1 was present only in the e1a2 cell lines (SD1 and SUP-B15).
The B cell adaptor protein (BCAP) is involved in the activation of Akt by PI3K through the B cell receptor.36 Accordingly, we show phosphorylation of BCAP (Y570) in the pre-B cell lines BV173 and SUP-B15. Phosphorylation of BCAP (Y570) is also found in KCL22 and KU812 cells, suggesting a previously unrecognized role for BCAP in Bcr-Abl signaling independent of the B cell receptor. We speculate that BCAP could serve as another way of coupling Bcr-Abl to PI3K.
When we compared K562 and EoL-1, we observed marked differences in the phosphorylation of downstream signaling molecules. For example, many phosphopeptides corresponding to PI3K subunits were detected in EoL-1 but not in Bcr-Abl cells. One site, Y580, found in the α regulatory subunit of PI3K, is known to be phosphorylated downstream of the insulin receptor.41 Many of the sites in PI3K are novel tyrosine phosphorylation sites. The role of these novel sites in PI3K activation remains unknown. EoL-1 cells have 2 imatinib-sensitive phosphorylation sites in common with the Bcr-Abl signature, Shc (Y427) and SHIP-2 (Y986). Phosphorylation of Shc is seen in many leukemia cell lines, including anaplastic lymphoma and the T cell acute lymphocytic leukemia (T-ALL) line Jurkat.14 In addition, Shc associates with SHIP-2 in Bcr-Abl–transformed cells42 and may play a role in signaling downstream of PDGFR.43
Several documented kinase mutations in Bcr-Abl develop in patients with imatinib resistance (reviewed in Deininger and Druker4 ). Newer Bcr-Abl kinase inhibitors show differential inhibition, depending on the specific kinase domain mutant.10 Variations in drug resistance of Bcr-Abl mutants could be accounted for by mutant-specific differences in downstream signaling. The phosphotyrosine-profiling method would be suitable to investigate that possibility. The groundwork for these studies requires that we understand the basic phospho-signature of the wild-type kinases and then apply this knowledge to differences we may find in the mutants. Any differences seen between wild-type and mutant kinases could lead to novel diagnostic markers to predict resistance in patients.
We demonstrate that phosphopeptide immunoprecipitation combined with SILAC can quantitatively measure the effects of kinase inhibitors on multiple components of a signaling pathway. Therefore, this approach can be useful for discovering drug-responsive biomarkers. Our results also underscore the importance of applying unbiased discovery methods, such as the one used in this study and in the study by Rush et al,14 to uncover novel and possibly cancer-relevant phosphorylated proteins to help elucidate disease mechanisms.
Prepublished online as Blood First Edition Paper, February 23, 2006; DOI 10.1182/blood-2005-08-3399.
Supported by National Cancer Institute Innovative Molecular Analysis Technologies program Small Business Innovative Research (SBIR) phase I and phase II grants (1R43CA101106 and R44CA101106, respectively; J.R.). Cell Signaling Technology provided research funding to V.G., K.L., A.M., J.N., E.S., J.M., J.R., M.C., and R.P.
V.G., K.L., A.M., J.N., E.S., J.M., J.R., M.C., and R.P. have declared a financial interest in a company (Cell Signaling Technology, Inc) whose products were used in the present work.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal