Abstract
The Stat5 transcription factors Stat5a and Stat5b have been implicated in lymphoid development and transformation. Most studies have employed Stat5a/b-deficient mice where gene targeting disrupted the first protein-coding exon, resulting in the expression of N-terminally truncated forms of Stat5a/b (Stat5a/bΔN/ΔN mice). We have now reanalyzed lymphoid development in Stat5a/bnull/null mice having a complete deletion of the Stat5a/b gene locus. The few surviving Stat5a/bnull/null mice lacked CD8+ T lymphocytes. A massive reduction of CD8+ T cells was also found in Stat5a/bfl/fllck-cre transgenic animals. While γδ T-cell receptor–positive (γδTCR+) cells were expressed at normal levels in Stat5a/bΔN/ΔN mice, they were completely absent in Stat5a/bnull/null animals. Moreover, B-cell maturation was abrogated at the pre–pro-B-cell stage in Stat5a/bnull/null mice, whereas Stat5a/bΔN/ΔN B-lymphoid cells developed to the early pro-B-cell stage. In vitro assays using fetal liver-cell cultures confirmed this observation. Most strikingly, Stat5a/bnull/null cells were resistant to transformation and leukemia development induced by Abelson oncogenes, whereas Stat5a/bΔN/ΔN-derived cells readily transformed. These findings show distinct lymphoid defects for Stat5a/bΔN/ΔN and Stat5a/bnull/null mice and define a novel functional role for the N-termini of Stat5a/b in B-lymphoid transformation.
Introduction
Stat molecules are part of a highly conserved signaling pathway involved in cell-fate decisions like differentiation, proliferation, and apoptosis.1-3 The cytokines interleukin-2, -4, and -7 (IL-2, IL-4, IL-7) regulate important aspects of lymphoid development and are strong activators of the transcription factors Stat5a and Stat5b.4 The importance of Stat5a/b for lymphoid cells is also underlined by the fact that constitutively activated Stat5a/b are found in several forms of lymphoid leukemia in mice and humans.5-10 Gene knockouts have greatly contributed to our knowledge about Stat transcription factors because they allowed exploration of their physiologic and pathophysiologic functions.11 So far, all studies investigating the role of Stat5a/b in lymphopoiesis employed gene-targeted mice still expressing a residual protein corresponding to an N-terminal deletion mutant (Stat5a/bΔN).4,12-14 Stat5a/bΔN/ΔN mice revealed surprisingly mild phenotypes in B- and T-cell development and function.
Characterization of the lymphoid compartment in Stat5a/bΔN/ΔN mice showed a modest reduction of B- and T-lymphoid–cell numbers accompanied by a complete lack of natural killer (NK) cells and CD4+CD25+ suppressor T cells.4,13,15 CD8+ T cells were present but failed to respond to α-CD3 and IL-2.4 Mature B-cell numbers in the periphery were also reduced due to an incomplete block at the early pro-B-cell developmental stage (Hardy fraction B).13,14
Mice lacking IL-7 or the IL-7R have a block at the earliest step of B-cell development at Hardy fraction A and lack mature B-lymphoid cells in the periphery.16,17 Notably, B-cell development can be rescued in these mice by forced expression of a constitutively active Stat5a/b mutant.17 In addition, transgenic mice expressing a constitutively active Stat5b (Stat5b-CA) have increased numbers of pro-B cells.14 As Stat5a/b are critical components in the signaling cascade downstream of IL-7R, abrogation of Stat5a/b was predicted to result in a dramatic phenotype. Thus, the observations in Stat5a/bΔN/ΔN mice were difficult to reconcile with the current understanding of signaling pathways controlling B-cell development.
Moreover, Stat5a/b transcription factors have been shown to play an important role in various T-cell developmental decisions. Transgenic Stat5b-CA mice display increased numbers of CD8+ but not CD4+ T cells.18 This implicates Stat5b as an important regulator of CD4+/CD8+ lineage decision. Moreover, Stat5a/b DNA binding sites were found in regulatory regions of the T-cell receptor γ (TCRγ) gene locus, and Stat5b-CA mice displayed a modest increase in γδ T-cell numbers.18,19 In Stat5a/bΔN/ΔN mice, embryonic γδ T-cell development was severely affected, but numbers were rapidly restored after birth.20 Therefore, the relevance for Stat5a/b in adult γδ thymopoiesis remained elusive.
Another finding in Stat5a/bΔN/ΔN mice was striking. Among many substrates that are phosphorylated downstream of the Abelson oncogene, Stat5a/b were originally described to be among the most strongly activated ones.5,21-23 Therefore, Stat5a/b was considered potential critical factors in Ab-MuLV– and bcr/abl-mediated transformation. This hypothesis was substantiated by a plethora of in vitro data using various forms of dominant-negative Stat5a/b mutants.24-27 Despite this evidence for an essential role for Stat5a/b in Abelson-induced transformation, Stat5a/bΔN/ΔN mice were still susceptible to leukemia when challenged with Abelson oncogenes.13
Because of these inconsistencies and open questions, we set out to unveil the impact of Stat5a/b on lymphopoiesis and on Abelson-induced transformation using mice in which the entire Stat5a/b locus had been deleted (Stat5a/bnull/null mice). Our experiments redefine the role of Stat5a/b in lymphoid development and we can clearly attribute a functional role to the truncated Stat5 proteins present in Stat5a/bΔN/ΔN mice. We furthermore unravel the key role of Stat5a/b in Abelson-induced transformation.
Materials and methods
Mice
Stat5a/bnull/null, Stat5a/bfl/fl, lck-cre (distal promoter) transgenic, Stat5a/bΔN/ΔN, and Rag2–/– mice were described previously12,28-31 and were maintained at the Biomedical Research Institute, Medical University of Vienna, under specifically pathogen-free sterile conditions. The Stat5a/bnull/null, Stat5a/bfl/fl, and Stat5a/bΔN mice were on a mixed 129/C57B/6 background. All animal experiments were carried out in accordance with protocols approved by Austrian law.
Flow cytometric analysis
Single-cell suspensions were preincubated with CD16/CD32 antibodies (BD Biosciences, San Jose, CA) to prevent nonspecific Fc-receptor–mediated binding. Subsequently, 5 × 105 cells were stained with monoclonal antibodies conjugated with fluorescent markers and analyzed by a FACScan flow cytometer using CellQuest Pro software (Becton Dickinson, Heidelberg, Germany). The antibodies used for determination of specific B-lineage maturation stages included the markers B220 (CD45R; RA3-6B2), CD43 (1B11), CD19 (1D3), BP-1 (6C3), IgM (R6-60.2), and IgDb (IgH-5b; 217-170). For surface staining of T-lineage cells, antibodies directed against CD3 (ϵ chain; 145-2C11), CD4 (L3T4), CD8a (Ly-2; 53-6.7), TCRβ chain (H57-597), and γδ TCR (GL3) were used. Hematopoietic stem cell (HSC) staining was performed using a Mouse Lineage Panel kit and anti–sca-1 (Ly-6A/E; D7), anti–c-kit (CD117; 2B8), and anti-CD34 (RAM34) antibodies (all BD Pharmingen, Heidelberg, Germany).
Protein analysis and Western blotting
Splenic T cells were magnetic-activated cell sorted for Thy1.2+ cells according to the manufacturer's instruction (Miltenyi Biotec, Bergisch Gladbach, Germany). Thy1.2+ cells were separated using an autoMACS Instrument (Miltenyi Biotech). Cells were lysed in a buffer containing protease and phosphatase inhibitors (50 mM Hepes, pH 7.5; 0.1% Tween-20; 150 mM NaCl; 1 mM EDTA; 20 mM β-glycero-phosphate; 0.1 mM sodium vanadate; 1 mM sodium fluoride; 10 g/mL aprotinin; leupeptin; and 1 mM PMSF). Protein concentrations were determined using a BCA kit as recommended by the manufacturer (Pierce, Rockford, IL). Proteins (50-100 g) were separated on an 8% SDS polyacrylamide gel and transferred onto Immobilon membranes. Membranes were probed with anti-Stat5a/b (C-17; Santa Cruz Biotechnologies, Santa Cruz, CA) and anti–β-actin (clone Ac-54; Sigma, St Louis, MO) antibodies at dilutions of 1:500 and 1:2000, respectively.
T-cell stimulation
Splenic T cells were isolated from 3 12-week-old Stat5a/bf/f, 3 Stat5a/bf/flck-cre, and 4 Stat5a/bΔN/ΔN mice. Splenic-cell solution was subjected to red blood cell lysis for 5 minutes using a lysis buffer containing 150 mM NH4Cl, 1 mM KHCO3, and 0.1 mM Na2EDTA (pH 7.3) and cultured in T-cell culture medium (RPMI 1640 containing 10% FBS; 10 mM Hepes, pH 7.0; 2 mM L-glutamine; [1 ×] nonessential amino acids; 1 mM sodium pyruvate; and 50 μM 2-mercaptoethanol) in the presence of 1 μg/mL anti-CD3 monoclonal antibody 145.2C11 (Pharmingen) and 1000 units/mL recombinant human IL-2 (Boehringer Mannheim, Mannheim, Germany).4 Pellets were prepared before and after 4 hours of stimulation and subjected to RNA isolation.
RNA isolation and semiquantitative RT-PCR analysis
First-strand cDNA synthesis and PCR amplification were performed using a reverse transcriptase–polymerase chain reaction (RT-PCR) kit (GeneAmp RNA PCR kit; Applied Biosystems, Weiterstadt, Germany) according to the manufacturer's instructions. The following primer sequences were used: pim-1, 5′-ACGTGGAGAAGGACCCGATTTCC-3′ and 5′-GATGTTTTCGTCCTTGATGTCGC-3′; cis, 5′-CTGCTGTGCATAGCCAAGACGTTC-3′ and 5′-CAGAGTTGGAAGGGGTACTGTCGG-3′; cyclin D2, 5′-AGAAGGGGCTAGCAGATGA-3′ and 5′-AGGATGATGAAGTGAACACA-3′; β-actin, 5′-CAGGTCCAGACCCAGGATGGC-3′ and 5′-ACTCCTATGTGGGTGACGAG-3′.
In vitro B-cell differentiation
Single-cell suspensions of fetal liver cells (embryonic day 14 [ED 14]) were prepared. The cells were maintained in RPMI medium containing 10% fetal calf serum (FCS), 100 U/mL penicillin/streptomycin, and 5 μM β-mercaptoethanol on an OP-9 fibroblast feeder layer. IL-7, Flt-3L, and SCF (each 10 ng/mL) were added every other day. Outgrowth of specific B-lineage cells was examined for 12 days by analyzing an aliquot of the suspension cells every other day by FACS.
Tissue culture conditions and virus preparation
Transformed fetal liver, bone marrow cells, and tumor-derived cell lines were maintained in RPMI medium containing 10% FCS, 100 U/mL penicillin/streptomycin, 5 μM β-mercaptoethanol, and 2 mM L-glutamine. GP+E86 cell lines (MSCV-bcr/abl p185-IRES-eGFP producer), A010 cells (Ab-MuLV producer), and mouse embryonic fibroblasts (MEFs) were maintained in DMEM medium containing 10% FCS, 100 U/mL penicillin/streptomycin, 5 μM β-mercaptoethanol, and 2 mM L-glutamine. A010 cells produce an ecotropic replication-deficient form of the Abelson virus and were a generous gift of Dr Naomi Rosenberg. For collection of the viral supernatant, A010 cells were plated in 100-mm dishes precoated with gelatin (1%) and grown to confluency. Supernatant was harvested every 8 hours for 40 hours, pooled, and filtered through a 0.45-μm filter, as described previously.32
Infections, in vitro transformation assays, and establishment of cell lines
For the preparation of fetal liver cells, Stat5a/bnull/+ animals were set up for breeding and vaginal plugs were checked daily. Fourteen days after conception, the pregnant animals were killed and fetal livers were prepared. The tail of the embryo was used for genotyping by PCR. Single-cell suspensions from fetal livers were infected for 1 hour with viral supernatant derived either from A010 cells or from GP+E86 bcr/abl p185-IRES-eGFP producer cell lines in the presence of 7 g/mL polybrene, as described previously.13,32,33 Using the same procedure, single-cell suspensions of bone marrow of tibiae and femora of mice were infected. The cells were then maintained in complete RPMI medium or plated in cytokine-free methylcellulose at a density of 2.5 × 105 cells/mL in 35-mm dishes. After 10 days, cloning efficiency was evaluated by counting colonies by light microscopy (Leica Fluovert microscope, 4 × magnification; Heidelberg, Germany). Photographs of single colonies were taken using an Axiovert 200 microscope (ZEISS, Oberkochen, Germany; 40 ×/0.6 NA objective), a CoolSNAP fx camera (Visitron Systems, Puchheim, Germany), and MetaMorph software (Version 4.6; Molecular Devices, Downingtown, PA). The assays were performed in triplicates. Mock-infected cells did not result in growth factor–independent colonies. As a control, individual clones were picked and analyzed by flow cytometry for the expression of B-lineage markers (CD19, CD43). The ability to form cell lines was tested by transferring an aliquot of the infected cells (1 × 106) to growth factor–free medium. The medium was changed twice a week and the culture was observed for the outgrowth of stable cell lines.
Injection of Abelson-infected cells into Rag2–/– mice
For tail vein injections, 106 cells were resuspended in 200 μL of PBS and injected via tail vein into Rag2–/– mice. Prior to injection, the cells were infected with either Ab-MuLV or pMSCV-bcr/abl p185-IRES-eGFP retrovirus as described under “Infections, in vitro transformation assays, and establishment of cell lines.” Sick mice were killed and analyzed for spleen weights, white blood cell counts, and the presence of leukemic cells in bone marrow, spleen, liver, and blood. The leukemic cells were also analyzed by flow cytometry and expressed the surface markers CD19 and CD43.
[3H]thymidine incorporation
Cells were plated at a density of 2 × 105 cells in 96 round-bottom wells. [3H]thymidine (0.1 μCi/well [0.0037 MBq/well]) was added 18 hours after plating, for another 12 hours.
Statistical analysis
Statistics were carried out using a paired t test, Mann-Whitney test, or a one-way analysis of variance (ANOVA) as appropriate. ANOVA was followed by a Tukey test. Differences in Kaplan-Meier plots were analyzed for statistical significance using the log-rank test.
Results
Stat5a/b are essential for CD8+ and γδ T-cell development
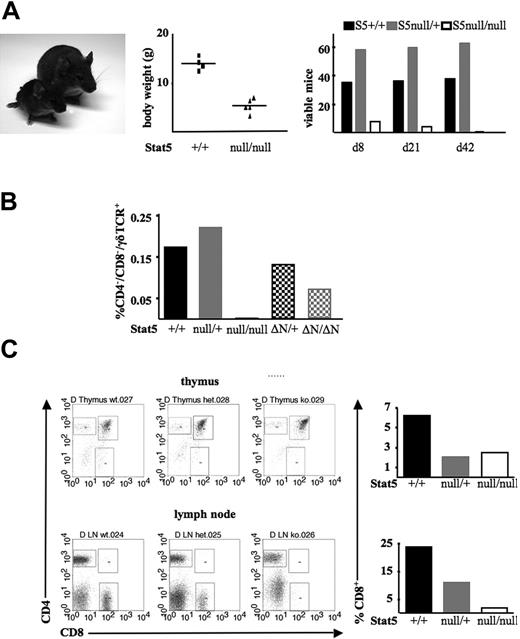
Stat5a/bΔN/ΔN mice on a mixed genetic background are viable and may survive up to 2 years in our mouse colony. However, about 40% of Stat5a/bΔN/ΔN mice die due to an autoimmune phenotype caused by a significant reduction of CD4+CD25+ suppressor T cells within the first few months (Snow et al15 and data not shown). In contrast, Stat5a/bnull/null mice die perinatally28 (Figure 1A). Although the cause of death is not known, severe anemia and reduced lung capacity are possibly contributing factors (Cui et al28 ; L.H., unpublished observation). Approximately 1% of the Stat5a/bnull/null mice reach weaning age. These rare Stat5a/bnull/null survivor mice are much smaller than their littermates, display a significantly reduced body weight, and die within the first 6 weeks after birth (Figure 1A). Stat5a/bΔN/ΔN mice express N-terminally truncated Stat5a/b proteins that are found at significant levels in the lymphoid lineage (Figure S1, available on the Blood website; see the Supplemental Figures link at the top of the online article). We therefore monitored lymphoid development in the survivor mouse population. Five 4-week-old Stat5a/bnull/null mice were killed and thymi and lymph nodes were subjected to flow cytometric analysis. Notably, thymi, spleen, and lymph nodes were smaller than would be expected from the body size of the mice. As depicted in Figure 1B, CD4–CD8–γδTCR+ cells were present in wild-type, Stat5a/bΔN/+, Stat5a/bΔN/ΔN, and Stat5a/bnull/+ mice but were completely absent in the thymic-cell suspension of Stat5a/bnull/null mice. We stress that CD4–CD8–γδTCR+ cells are present at significant levels in Stat5a/bΔN/ΔN mice. Moreover, Stat5a/bΔN/ΔN mice were reported to show a normal distribution of CD4+ and CD8+ T cells in the adult thymus, but Stat5a/bΔN/ΔN-derived CD8+ cells show an inability to proliferate in response to IL-2.4 Again, Stat5a/bnull/null mice revealed a distinct phenotype. The thymus was reduced in size in relation to the body size and age of the mice. We found a significant reduction of CD8+ T cells (2.5-fold). In the lymph nodes, the situation was even more pronounced; Stat5a/bnull/null mice displayed a 12-fold reduction in CD8+ T lymphocytes (Figure 1C).
Impaired CD8+ and γδ T-cell development in Stat5a/bnull/null survivor mice. (A) Picture of a Stat5a/bnull/null survivor mouse (left) compared with a Stat5a/b+/+ littermate at the age of 4 weeks. Representative body weight values and numbers of Stat5a/bnull/null mice and littermates surviving up to day 8, weaning (day 21), or day 42 are depicted. (B) Numbers of CD4–/CD8–/γδTCR+ T lymphocytes in thymi of Stat5a/bnull/null, Stat5a/bΔN/ΔN, and respective littermate controls. (C) Flow cytometric analysis of CD4+, CD8+, and CD4+/CD8+ cells in thymi and lymph nodes of 5 Stat5a/bnull/null survivors and 2 individual Stat5a/bnull/+ and 2 Stat5a/b+/+ littermate controls. Data are summarized in bar graphs. Due to the small size of thymi and lymph nodes, cells were pooled and did therefore not allow generation of error bars (B-C).
Impaired CD8+ and γδ T-cell development in Stat5a/bnull/null survivor mice. (A) Picture of a Stat5a/bnull/null survivor mouse (left) compared with a Stat5a/b+/+ littermate at the age of 4 weeks. Representative body weight values and numbers of Stat5a/bnull/null mice and littermates surviving up to day 8, weaning (day 21), or day 42 are depicted. (B) Numbers of CD4–/CD8–/γδTCR+ T lymphocytes in thymi of Stat5a/bnull/null, Stat5a/bΔN/ΔN, and respective littermate controls. (C) Flow cytometric analysis of CD4+, CD8+, and CD4+/CD8+ cells in thymi and lymph nodes of 5 Stat5a/bnull/null survivors and 2 individual Stat5a/bnull/+ and 2 Stat5a/b+/+ littermate controls. Data are summarized in bar graphs. Due to the small size of thymi and lymph nodes, cells were pooled and did therefore not allow generation of error bars (B-C).
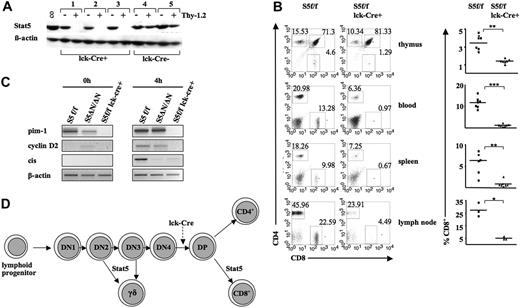
To confirm cell autonomy of Stat5a/b in CD8+ T-cell development, we generated Stat5a/bf/f lck-cre mice. These mice express the Cre-recombinase under the control of the distal promoter of the T-cell receptor–associated kinase Lck, which is first active at the double-negative (CD4–CD8–) stage.30 T-cell lineage–specific deletion of Stat5a/b was confirmed by Western blot analysis of magnetic-activated cell separation (MACS)–sorted Thy1.2+ splenic cells (Figure 2A). Stat5a/bf/f lck-cre mice displayed splenomegaly and lymphoid organ infiltration that was first detected at the age of 4 weeks. This phenotype is most probably due to the expected lack of suppressor T cells that was previously described as a consequence of Stat5a/b deficiency.15 As observed in Stat5a/bnull/null survivors, Stat5a/bf/f lck-cre mice showed a significant reduction of CD8+ T cells in all organs analyzed (thymus, P < .01; peripheral blood, P < .001; spleen, P < .01; lymph node, P < .05; Figure 2B). The selective disappearance of CD8+ cells is also indicated by the increased ratio of CD4+ versus CD8+ cells, as described in Table 1. In order to see whether a different induction of target genes might account for the differences in the phenotype of Stat5a/bΔN/ΔN and Stat5a/bnull/null mice, we stimulated splenic T cells with α-CD3 and IL-2, as depicted in Figure 2C. The Stat5a/b target genes Pim1 and cyclin D2 were clearly expressed in Stat5a/bf/f and Stat5a/bΔN/ΔN cells. Induction of these genes was not found in Stat5a/bnull/null T cells, indicating that Stat5a/bΔN are still capable of inducing some target genes. In contrast, expression of the suppressor of cytokine signaling (SOCS) gene family member CIS was lacking in both Stat5a/bΔN/ΔN and Stat5a/bf/f lck-cre cells. The development of γδ T cells was not altered in these mice (data not shown). This was to be expected because transcription from the distal lck promoter occurs after the junction of γδ TCR+ cells. Taken together, these findings provide evidence that Stat5a/b are indispensable for CD8+ T-cell and γδ TCR+-cell homeostasis (Figure 2D scheme).
Mean ratio of CD4+ to CD8+ T cells in thymi, peripheral blood, spleen, and lymph nodes of Stat5a/bf/f and Stat5a/bf/flck-cre mice
. | Ratio of CD4+/CD8+ cells . | . | |
|---|---|---|---|
. | S5f/f . | S5f/f lck-cre . | |
| Thymus | 4.1:1 | 11:1 | |
| Lymph node | 1.9:1 | 5:1 | |
| Spleen | 2.2:1 | 8:1 | |
| Peripheral blood | 1.5:1 | 6:1 | |
. | Ratio of CD4+/CD8+ cells . | . | |
|---|---|---|---|
. | S5f/f . | S5f/f lck-cre . | |
| Thymus | 4.1:1 | 11:1 | |
| Lymph node | 1.9:1 | 5:1 | |
| Spleen | 2.2:1 | 8:1 | |
| Peripheral blood | 1.5:1 | 6:1 | |
Impaired CD8+ T-cell development in Stat5a/bf/flck-cre mice. (A) Splenic cells of 3 individual Stat5a/bf/flck-cre (nos. 1-3) and 2 Stat5a/bf/f (nos. 4-5) mice were magnetically sorted for Thy1.2+ cells, and Stat5a/b expression was assessed by Western blot analysis. (B) Representative flow cytometric profile of CD4+, CD8+, and CD4+/CD8y cells in thymus, blood, spleen, and lymph nodes of Stat5a/bf/flck-cre mice and Stat5a/bf/f controls. Asterisks denote significant difference as determined by a paired t test. (C) Analysis of transcriptional expression of pim-1, CIS, and cyclin D2 genes by semiquantitative RT-PCR. Splenic T cells were stimulated for 4 hours with α-CD3 (1 μg/mL) and human IL-2 (hIL-2; 1000 U/mL) to induce Stat5a/b target gene transcription. (D) Schematic model for the role of Stat5a/b in T-cell developmental choices. Stages affected in Stat5a/bnull/null survivor mice and/or Stat5a/bf/flck-cre mice are indicated. The time point of Cre-recombinase activation under the control of the distal lck promoter is also depicted.
Impaired CD8+ T-cell development in Stat5a/bf/flck-cre mice. (A) Splenic cells of 3 individual Stat5a/bf/flck-cre (nos. 1-3) and 2 Stat5a/bf/f (nos. 4-5) mice were magnetically sorted for Thy1.2+ cells, and Stat5a/b expression was assessed by Western blot analysis. (B) Representative flow cytometric profile of CD4+, CD8+, and CD4+/CD8y cells in thymus, blood, spleen, and lymph nodes of Stat5a/bf/flck-cre mice and Stat5a/bf/f controls. Asterisks denote significant difference as determined by a paired t test. (C) Analysis of transcriptional expression of pim-1, CIS, and cyclin D2 genes by semiquantitative RT-PCR. Splenic T cells were stimulated for 4 hours with α-CD3 (1 μg/mL) and human IL-2 (hIL-2; 1000 U/mL) to induce Stat5a/b target gene transcription. (D) Schematic model for the role of Stat5a/b in T-cell developmental choices. Stages affected in Stat5a/bnull/null survivor mice and/or Stat5a/bf/flck-cre mice are indicated. The time point of Cre-recombinase activation under the control of the distal lck promoter is also depicted.
Stat5a/b are essential for the pre–pro-B to early pro-B-cell stage transition in vivo
Specific B-cell developmental stages can be distinguished by differential cell-surface expression of B220, CD43, CD19, BP-1, IgM, and IgD (Figure 3C schematic overview). Single fractions can be classified according to the Hardy nomenclature in pre–pro-B (B220+/CD43hi/CD19–/BP-1–; fraction A), early pro-B (B220+/CD43hi/CD19+/BP-1–; fraction B), late pro-B (B220+/CD43hi/CD19+/BP-1+; fraction C), pre-B (B220+/CD43lo/IgM–/IgD–; fraction D), immature (B220+/CD43lo/IgM+/IgD–; fraction E), and mature (B220+/CD43lo/IgM+/IgD+; fraction F) B cells.34,35
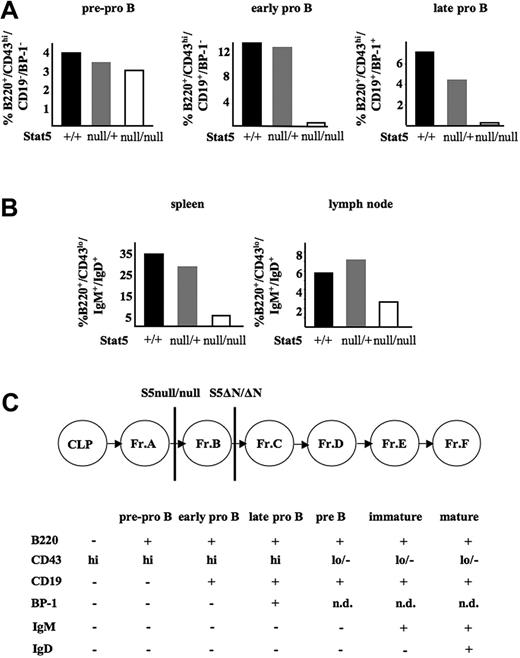
It had been shown in Stat5a/bΔN/ΔN mice that Stat5a/b are required for the transition from the early pro-B (Hardy fraction B) to the late pro-B-cell stage (Hardy fraction C).14 We prepared bone marrow, spleen, and lymph nodes of 5 Stat5a/bnull/null survivor mice and their Stat5a/bnull/+ and wild-type littermates. The numbers of pre–pro-B cells were comparable in all 3 groups. However, we failed to detect early and late pro-B cells in Stat5a/bnull/null mice in the bone marrow (22-fold and 40-fold reduction compared with Stat5a/b+/+, respectively; Figure 3A). Accordingly, the numbers of mature B cells (Hardy fraction F) in spleen and lymph nodes were also significantly reduced compared with Stat5a/b+/+ controls (6.4-fold and 2.2-fold, respectively; Figure 3B). We emphasize that these percentages cannot be directly compared, since the total size and the cellularity of hematopoietic organs differ in wild-type and Stat5a/bnull/null animals. These data strongly argue for a role of Stat5a/b at the earliest steps of B-cell development (pre–pro-B cell to early pro-B-cell transition). However, the developmental block does not appear to be absolute, since few mature B-lymphoid cells are present in the periphery. Again, the phenotype in Stat5a/bnull/null mice is aggravated and very distinct from Stat5a/bΔN/ΔN mice, with a block occurring at an earlier B-cell developmental stage (as indicated in the scheme in Figure 3C).
In vitro B-cell differentiation of Stat5a/bnull/null fetal liver–derived cells
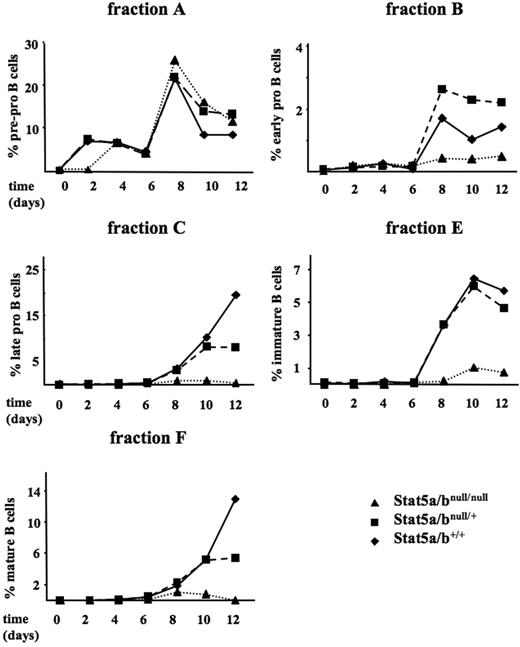
To further investigate the role of Stat5a/b for early B-cell development, we established a protocol that allows following B-cell differentiation of fetal liver–derived cells in vitro. First, we excluded that a reduction of Stat5a/bnull/null HSCs causes any effects and determined the numbers of lin–/c-kit+/Sca-1+ long term-HSCs (CD34–) and short term-HSCs (CD34+). Interestingly, Stat5a/bnull/null fetal livers displayed comparable numbers of both populations (Figure S2A-S2B). We then cultured fetal liver cells (ED 14) of a Stat5a/bnull/+ intercross on an OP-9 fibroblast feeder layer in the continuous presence of IL-7, Flt-3L, and SCF (10 ng/mL each). Outgrowth of B-lineage cells was analyzed every second day by FACS analysis. Outgrowth kinetics of the single Hardy fractions reflected the observations made in the Stat5a/bnull/null survivor mice (Figure 4). Fraction A was found comparable in cells of each genotype. Fraction B, C, E, and F cells were detectable in control cultures from day 6 on but were entirely missing in cultures derived from Stat5a/bnull/null fetal livers. In these cultures, B-cell development was completely abrogated at fraction A and failed to proceed to any further maturation stages. We also performed an in vitro B-cell developmental assay only in the presence of IL-7 using an MEF feeder layer (10 ng/mL), confirming a critical role for Stat5a/b in the transition from Hardy fraction A to B (Figure S3).
B-cell development is arrested at the pre–pro-B-cell stage in Stat5a/bnull/null survivor mice. (A) Percentages of pre–pro-B, early pro-B, and late pro-B cells in bone marrow and (B) mature B cells in spleen and lymph node of 5 Stat5a/bnull/null survivors compared with 2 Stat5a/bnull/+ and 2 Stat5a/b+/+ controls. Due to the small body size, bone marrows were pooled and therefore did not allow generation of error bars. (C) Schematic model for maturation of B-cell developmental fractions A-F. As indicated, individual maturational stages were distinguished by differential surface expression of B220, CD43, CD19, BP-1, IgM, and IgD. The different blocks in Stat5a/bΔN/ΔN and Stat5a/bnull/null mice are indicated by vertical lines. CLP indicates common lymphoid progenitor.
B-cell development is arrested at the pre–pro-B-cell stage in Stat5a/bnull/null survivor mice. (A) Percentages of pre–pro-B, early pro-B, and late pro-B cells in bone marrow and (B) mature B cells in spleen and lymph node of 5 Stat5a/bnull/null survivors compared with 2 Stat5a/bnull/+ and 2 Stat5a/b+/+ controls. Due to the small body size, bone marrows were pooled and therefore did not allow generation of error bars. (C) Schematic model for maturation of B-cell developmental fractions A-F. As indicated, individual maturational stages were distinguished by differential surface expression of B220, CD43, CD19, BP-1, IgM, and IgD. The different blocks in Stat5a/bΔN/ΔN and Stat5a/bnull/null mice are indicated by vertical lines. CLP indicates common lymphoid progenitor.
Taken together, our data show that Stat5a/b are a critical transcription factor for the transition of pre–pro-B cells (fraction A) to the early pro-B-cell stage (fraction B) in adult and fetal hematopoiesis. Moreover, these findings indicate that the N-terminally truncated Stat5a/b proteins present in Stat5a/bΔN/ΔN mice suffice to allow B-lymphoid cells to mature to the early pro-B-cell stage.
Stat5a/b are required for Ab-MuLV– and bcr/abl p185–induced transformation in vitro
A constitutive activation of Stat5a/b is found in a large variety of leukemias and lymphomas,5,9,10,21,36 and constitutive activation of Stat5a/b suffice to induce a multi-lineage leukemia in mice.36 Despite a broad experimental evidence for a role of Stat5a/b in lymphoid leukemia,22,24,26,37,38 we have shown that Stat5a/bΔN/ΔN mice developed Abelson-induced B-lymphoid leukemia with identical properties compared with wild-type littermate controls.13 Bone marrow cells derived from Stat5a/bΔN/ΔN mice were readily transformed by Abelson oncogenes and resulted in the outgrowth of stable cell lines.13 We therefore repeated the transformation experiments with fetal livers and bone marrow from Stat5a/bnull/null mice. Fetal liver cells (ED 14) were infected with Ab-MuLV and plated in growth factor–free methylcellulose. No colonies were detected in any of the Stat5a/bnull/null fetal livers tested. A gene dosage effect was observed in Stat5a/bnull/+ fetal liver–derived cells, where we detected 50% to 60% of the growth factor–independent colonies compared with wild-type controls (Figure 5A-B). The retroviral constructs employed (Ab-MuLV and pMSCV-bcr/abl p185-IRES eGFP) result in the outgrowth of B-lymphoid colonies. This was confirmed by analyzing the colonies by flow cytometry for surface expression of CD43 and CD19. As expected, the transformed colonies were positive for both markers (Figure 5C).
A gene dosage effect was also observed regarding the proliferative capacity of Stat5a/b+/+ and Stat5a/bnull/+ Abelson-transformed cell lines (Figure 5D). Transformation experiments were repeated with bone marrow of Stat5a/bnull/null survivors and confirmed the lack of transformation ability: colony formation was completely abrogated in Stat5a/bnull/null cells (Figure 5E). Stable immortal Ab-MuLV–transformed cell lines were derived from wild-type and Stat5a/bnull/+ mice, but not a single cell line grew out from Stat5a/bnull/null fetal livers or bone marrow (Table 2). These experiments indicate that Stat5a/b are required for Abelson-induced transformation but that Stat5a/bΔN suffice to support the transformation and immortalization process.
Ability of fetal liver and bone marrow cells to form bcr/abl p185- or Ab-MuLV–induced colonies and stable cell lines
. | Colony formation . | Transformed cell lines . |
|---|---|---|
| bcr/abl transformation | ||
| S5ΔN/+, BM | Yes | Yes |
| S5ΔN/ΔN, BM | Yes | Yes |
| S5+/+, FL | Yes | Yes |
| S5null/+, FL | Yes | Yes |
| S5null/null, FL | No | No |
| Ab-MuLV transformation | ||
| S5ΔN/+, BM | Yes | Yes |
| S5ΔN/ΔN, BM | Yes | Yes |
| S5+/+, FL | Yes | Yes |
| S5null/+, FL | Yes | Yes |
| S5null/null, FL | No | No |
| S5+/+, BM | Yes | Yes |
| S5null/+, BM | Yes | Yes |
| S5null/null, BM | No | No |
. | Colony formation . | Transformed cell lines . |
|---|---|---|
| bcr/abl transformation | ||
| S5ΔN/+, BM | Yes | Yes |
| S5ΔN/ΔN, BM | Yes | Yes |
| S5+/+, FL | Yes | Yes |
| S5null/+, FL | Yes | Yes |
| S5null/null, FL | No | No |
| Ab-MuLV transformation | ||
| S5ΔN/+, BM | Yes | Yes |
| S5ΔN/ΔN, BM | Yes | Yes |
| S5+/+, FL | Yes | Yes |
| S5null/+, FL | Yes | Yes |
| S5null/null, FL | No | No |
| S5+/+, BM | Yes | Yes |
| S5null/+, BM | Yes | Yes |
| S5null/null, BM | No | No |
B-cell development is arrested at the pre–pro-B-cell stage in Stat5a/bnull/null fetal liver–derived cultures. Fetal livers of 2 embryos of each genotype were pooled (ED 14) and cocultivated on an OP-9 fibroblast feeder layer in the presence of IL-7, Flt-3L, and SCF (10 ng/mL each). Outgrowth of pro-B-cell stages (fractions A-C), immature (fraction E), and mature (fraction F) B cells over a 12-day period is depicted.
B-cell development is arrested at the pre–pro-B-cell stage in Stat5a/bnull/null fetal liver–derived cultures. Fetal livers of 2 embryos of each genotype were pooled (ED 14) and cocultivated on an OP-9 fibroblast feeder layer in the presence of IL-7, Flt-3L, and SCF (10 ng/mL each). Outgrowth of pro-B-cell stages (fractions A-C), immature (fraction E), and mature (fraction F) B cells over a 12-day period is depicted.
Abelson-induced transformation is dependent on Stat5a/b in vitro. (A) Ab-MuLV–induced colony formation of Stat5a/b+/+, Stat5a/bnull/+, and Stat5a/bnull/null fetal liver cells in methylcellulose. Single-colony pictures of each phenotype are depicted in the bottom panels. Stat5a/bnull/null cells showed no ability to form growth factor–independent colonies. (B) Summary of data obtained from Ab-MuLV–induced colony formation assays represent means ± SEM of 4 embryos per genotype (each performed in triplicates). (C) Surface expression of B-lineage markers was verified by flow cytometric analysis (right; data of one representative CD19+ CD43+ colony is shown). (D) [3H]thymidine incorporation of fetal liver–derived Stat5a/b+/+ and Stat5a/bnull/+ Ab-MuLV–transformed cell lines. Stat5a/b-deficient fetal livers did not give rise to stable transformed cell lines. Data represent means ± SEM of 2 cell lines per genotype. cpm indicates counts per minute. (E) Ab-MuLV–induced colony formation of Stat5a/b+/+ (n = 2), Stat5a/bnull/+ (n = 2), and Stat5a/bnull/null (n = 5; pooled) survivor bone marrow cells in methylcellulose. Stat5a/bnull/null survivor cells showed no ability to form growth factor–free colonies. Experiment was performed in triplicates. Asterisks denote significant differences as determined by a one-way ANOVA followed by a Tukey test (A,C) or a paired t test (B).
Abelson-induced transformation is dependent on Stat5a/b in vitro. (A) Ab-MuLV–induced colony formation of Stat5a/b+/+, Stat5a/bnull/+, and Stat5a/bnull/null fetal liver cells in methylcellulose. Single-colony pictures of each phenotype are depicted in the bottom panels. Stat5a/bnull/null cells showed no ability to form growth factor–independent colonies. (B) Summary of data obtained from Ab-MuLV–induced colony formation assays represent means ± SEM of 4 embryos per genotype (each performed in triplicates). (C) Surface expression of B-lineage markers was verified by flow cytometric analysis (right; data of one representative CD19+ CD43+ colony is shown). (D) [3H]thymidine incorporation of fetal liver–derived Stat5a/b+/+ and Stat5a/bnull/+ Ab-MuLV–transformed cell lines. Stat5a/b-deficient fetal livers did not give rise to stable transformed cell lines. Data represent means ± SEM of 2 cell lines per genotype. cpm indicates counts per minute. (E) Ab-MuLV–induced colony formation of Stat5a/b+/+ (n = 2), Stat5a/bnull/+ (n = 2), and Stat5a/bnull/null (n = 5; pooled) survivor bone marrow cells in methylcellulose. Stat5a/bnull/null survivor cells showed no ability to form growth factor–free colonies. Experiment was performed in triplicates. Asterisks denote significant differences as determined by a one-way ANOVA followed by a Tukey test (A,C) or a paired t test (B).
To control for this somewhat unusual observation, we performed several additional experiments that are summarized in Table 2. First, we repeated the transformation experiments side by side with bone marrow cells derived from Stat5a/bΔN/ΔN and Stat5a/bnull/null survivor mice. Stat5a/bΔN/ΔN cells readily transformed, which resulted in the formation of growth factor–independent colonies and in the outgrowth of stable cell lines, but we failed to see signs of transformation when using Stat5a/bnull/null cells.
We next reasoned that Ab-MuLV–induced transformation might target a distinct subset of B-cell precursors that were absent or present at low numbers in Stat5a/bnull/null fetal liver cells or bone marrow. Hence, we employed a murine stem-cell virus encoding the bcr/abl p185 retrovirus (pMSCV–bcr/abl p185-IRES-eGFP). MSCV infects murine hematopoietic stem cells, which are present at comparable numbers in Stat5a/bnull/null fetal livers and controls (Figure S2). Cells derived from Stat5a/bnull/null (fetal livers) or Stat5a/bΔN/ΔN mice (bone marrow) were infected, the infection was controlled via FACS analysis, and the cells were subsequently plated in growth factor–free methylcellulose or transferred to growth factor–free medium. Again, cells derived from Stat5a/bΔN/ΔN mice formed colonies and gave rise to cell lines, whereas Stat5a/bnull/null cells failed to do so (Table 2).
Abelson-transformed Stat5a/bnull/null cells fail to induce leukemia in vivo
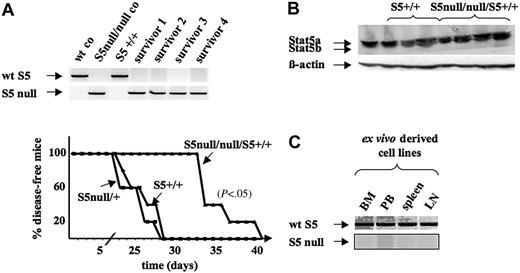
One may speculate that the failure to transform Stat5a/bnull/null cells might be compensated in vivo (eg, via cytokine-dependent activation of redundant signaling pathways). To test this, we first infected fetal livers with pMSCV–bcr/abl p185-IRES-eGFP and injected them via tail vein into 2 Rag2–/– mice each. Mice that had received wild-type bone marrow died from leukemia after 3 months, whereas the Rag2–/– mice that had received Stat5a/bnull/null bone marrow survived in a disease-free state for at least 6 months (data not shown). Uninfected Stat5a/bnull/null bone marrow was also injected into 2 Rag2–/– mice to verify that Stat5a/bnull/null bone marrow did indeed have the capacity to reconstitute hematopoiesis, albeit to a lesser extent than control bone marrow (data not shown; J. O'Shea, NIH, Bethesda, MD, and L.H., oral communication). It is important to mention that Stat5a/bnull/null fetal liver cells allowed the development of a few IgM+ IgD+ cells that were detected in the periphery. We next reasoned that the initial steps of transformation might be cytokine dependent or influenced by surrounding cells and that the environment within Rag2–/– mice only repopulated with Stat5a/bnull/null marrow might prevent transformation in vivo.39-41 To exclude this possibility we performed the following experiment. Bone marrow of 4 Stat5a/bnull/null survivor mice was prepared and mixed with wild-type marrow derived from a littermate control at a ratio of 4:1. The cells were infected with Ab-MuLV retrovirus and again injected via tail vein into recipient Rag2–/– animals. As depicted in Figure 6A, mice that had received either Stat5a/b+/+ or Stat5a/bnull/+ marrow succumbed to a B-lymphoid leukemia within 3 to 4 weeks. When administered mixed bone marrow that contained 80% of Stat5a/bnull/null cells, Rag2–/– mice showed signs of a phenotypically identical disease with latency that was increased by 10 days (P < .05). The animals displayed splenomegaly and elevated white blood cell counts and spleen, bone marrow, and liver were infiltrated with CD19+CD43+ leukemic cells (Figure S4). Western blot analysis showed that all cells expressed Stat5a/b at comparable levels (Figure 6B). In addition, ex vivo–derived cell lines were established and analyzed by PCR (Figure 6C). All leukemic cells expressed Stat5a/b; no transformed cells derived from Stat5a/bnull/null marrow were detectable. These experiments define Stat5a/b as essential transcription factors for Abelson-induced leukemia initiation and exclude the possibility that other signaling pathways compensate in vivo.
Abelson-induced transformation is dependent on Stat5 in vivo. (A) Kaplan-Meier plot of Rag2–/– mice that received a transplant of either Stat5a/b+/+, Stat5a/bnull/+, or a 4:1 mixture of Stat5a/bnull/null and Stat5a/b+/+ freshly Ab-MuLV–transformed bone marrow cells (5 mice/group; 1 × 106 cells each mouse). Genotyping PCR analysis of mice used for 4:1 mixture is depicted. wt indicates wild type. (B) Immunoblotting for Stat5a/b of leukemic cells derived from bone marrow of Rag2–/– mice that received a transplant of either Stat5a/b+/+ or a 4:1 mixture of Stat5a/bnull/null/Stat5a/b+/+ bone marrow. (C) PCR analysis of ex vivo–derived cell lines. Representative data of bone marrow (BM)–, peripheral blood (PB)–, spleen-, and lymph node (LN)–derived leukemic cell lines of one Rag2–/– mouse that received a transplant of a 4:1 mixture of Stat5a/bnull/null/Stat5a/b+/+ bone marrow cells, which was later killed. All cultures derived from Rag2–/– mice that received transplants of Stat5a/bnull/null/Stat5a/b+/+ cells were Stat5a/b+/+ as determined by PCR analysis.
Abelson-induced transformation is dependent on Stat5 in vivo. (A) Kaplan-Meier plot of Rag2–/– mice that received a transplant of either Stat5a/b+/+, Stat5a/bnull/+, or a 4:1 mixture of Stat5a/bnull/null and Stat5a/b+/+ freshly Ab-MuLV–transformed bone marrow cells (5 mice/group; 1 × 106 cells each mouse). Genotyping PCR analysis of mice used for 4:1 mixture is depicted. wt indicates wild type. (B) Immunoblotting for Stat5a/b of leukemic cells derived from bone marrow of Rag2–/– mice that received a transplant of either Stat5a/b+/+ or a 4:1 mixture of Stat5a/bnull/null/Stat5a/b+/+ bone marrow. (C) PCR analysis of ex vivo–derived cell lines. Representative data of bone marrow (BM)–, peripheral blood (PB)–, spleen-, and lymph node (LN)–derived leukemic cell lines of one Rag2–/– mouse that received a transplant of a 4:1 mixture of Stat5a/bnull/null/Stat5a/b+/+ bone marrow cells, which was later killed. All cultures derived from Rag2–/– mice that received transplants of Stat5a/bnull/null/Stat5a/b+/+ cells were Stat5a/b+/+ as determined by PCR analysis.
Discussion
The transcription factors Stat5a/b have been considered key regulators of immune functions and the lymphoid system.10,12,14,15,42,43 Their relevance and importance are underlined by the fact that more than 1700 manuscripts have been published on Stat5a/b since their discovery. A major breakthrough was the generation of the first Stat5a/b knockout mouse in 1998 (Stat5a/bΔN/ΔN mice) that served as a valuable tool for numerous studies and shed light on the multiple roles of Stat5a/b in the organism.4,12,14,44 Despite the key role of IL-7–mediated Stat5a/b activation in lymphoid development, the phenotype of the Stat5a/bΔN/ΔN mice in the lymphoid system is surprisingly moderate.4,20 The most prominent effect is the complete absence of CD4+CD25+ suppressor T cells, leading to an autoimmune disease.15
Our present work provides compelling evidence that the function of Stat5a/b in lymphoid development and immune functions needs redefinition. Our findings prove that Stat5a/b are key regulators of early B-cell development and of CD8+ and γδ T-lymphoid–cell generation. The difference between Stat5a/bΔN/ΔN and Stat5a/bnull/null mice implicitly defines separate roles for the N-terminally truncated Stat5a/b. Since deficiency in Stat5a/b was lethal,28 one might also argue that disturbances at the locus occurred independently of the targeted deletion of Stat5a/b. We do exclude this possibility, since erythroid cells can be genetically complemented by wild-type Stat5a (M.A.K. and H.B., unpublished observations). Our attempts to complement Stat5a/bnull/null HSCs with wild-type Stat5a to rescue lymphoid development continuously failed, most likely based on a severe defect of HSCs upon loss of Stat5a/b.45-47 The vast majority of Stat5a/bnull/null pups died rapidly after delivery. The rare survivors may have reflected outliers in the Gaussian distribution or a compensating adaptive change that occurred at low frequency. Regardless of the underlying basis, it is worth pointing out that the observations in these survivors were reproduced in the Stat5a/bf/flck-cre mice. The analysis of T-cell development in Stat5a/bf/f lck-cre mice also excluded that Stat5a/bnull/null thymic epithelial cells were responsible for the selective lack of CD8+ cells. Finally, B-cell development was recapitulated in vitro by employing fetal hematopoietic progenitors. In this cell-culture system, the absence of Stat5a/b resulted in a complete block of B-cell development at that very stage predicted from the phenotype of the survivors (pre–pro-B-cell stage). Taken together, these data demonstrate that the observations obtained in the rare survivors are not confounded by an undefined adaptive escape phenomenon. They also provide formal proof for the interpretation that deficiency in Stat5a/b affects the lymphoid compartment by a cell-autonomous effect rather than an indirect effect mediated via abnormalities in stromal cells or thymic epithelium.
Hence, our observations fall in place with the predicted role of Stat5a/b in lymphopoiesis and are in perfect agreement with studies performed in other mouse models.17,18,48 Goetz et al49 recently showed that constitutively active Stat5b promotes B-cell development at the expense of early T-cell development in transgenic mice. The authors hypothesize that Stat5a/b serves as a switch, with Stat5a/b activation driving cells into the B-lymphoid lineage whereas a lack of Stat5a/b activation allows for the development of early T-lymphoid cells. Moreover, the block in B-cell development at the earliest step (Hardy fraction A) confirms the original concept that Stat5a/b are the relevant transcription factor downstream of IL-7 in early B-cell development.17 It is also in line with an increased number of pro-B-cells in Stat5b-CA mice.18
Our experiments also lead to another important conclusion: the truncated proteins of Stat5a/b expressed in Stat5a/bΔN/ΔN mice are able to partially rescue B-cell development and to allow for the development of γδTCR+ and CD8+ T cells. It is currently unclear how the truncated Stat5a/b operate but we know that the Stat5ΔN protein enters the nucleus and constitutively binds DNA (data not shown). We also know that, at least in T cells, the Stat5ΔN protein is able to induce some but not all Stat5a/b target genes (eg, cyclin D2, an important mediator of cell proliferation). Further analysis in different cell lineages will finally clarify which target genes can be activated or repressed by Stat5ΔN. In addition, Stat5a/b might act as a scaffold. It was recently shown that constitutively active Stat5a/b assemble in a complex with Gab2 to allow for activation of PI3K.50,51 A potential protein docking function of Stat5a/b would be an alternative hypothesis to explain the differences observed in Stat5a/bΔN/ΔN and Stat5a/bnull/null cells.
Finally, we show that Stat5a/bnull/null cells, in contrast to Stat5a/bΔN/ΔN cells, fail to induce lymphoid leukemia in mice. The truncated Stat5a/b proteins suffice to afford transformation as illustrated for Abelson oncogenes.13 In each approach employed, growth factor–independent clones from Stat5a/bΔN/ΔN cells grew out readily. In contrast, regardless of the experimental setup, we consistently failed to obtain a single colony or growth factor–independent clone from Stat5a/bnull/null cells derived from either fetal livers or bone marrow. This key finding is of high clinical relevance because Stat proteins are potential candidates for drug targeting in the therapy of leukemia and other forms of cancer.52,53 We have recently shown that constitutively active Stat5a/b induced a multi-lineage leukemia and that tetramer formation of Stat5a/b was crucial in this regard.36 Apparently, N-terminally truncated Stat5a/b proteins that lack the tetramerization domain suffice to collaborate with at least some oncogenic tyrosine kinases, as proven here for the Abelson oncogenes. Therefore, a close definition of Stat5a/b functions in cancer progression is urgently needed. Our findings may therefore redirect therapeutic approaches that try to target Stat5a/b signaling in human leukemia. The importance of Stat5a/b in leukemia is further stressed by the fact that constitutively active mutations of Jak2 have recently been characterized as causative oncogenes in human leukemia.54-57 Jak2 is a strong activator of Stat5a/b; hence, it is attractive to speculate that Stat5a/b are essential components in the signaling cascade underlying disease manifestation and progression in these patients. If this can be confirmed, Stat5a/b are likely to emerge as a potential target for therapeutic intervention.
Prepublished online as Blood First Edition Paper, February 21, 2006; DOI 10.1182/blood-2005-09-3596.
Supported by grants of the Austrian Science Foundation (FWF; V.S.; P15865 and SFB F28) and by a grant of the Austrian National Bank (V.S.; 11132).
A.H., B.K., M.A.K., O.S., W.W., R.M., and V.S. designed and performed research; A.H., B.K., M.A.K., L.H., R.M., and V.S. analyzed data; H.B., Y.C., L.H., and R.M. provided vital new reagents and analytic tools; and A.H. and V.S. wrote the paper.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The authors thank Udo Losert and the staff of the Biomedical Research Institute, Medical University of Vienna (MUW) for taking care of mice. We are grateful to Michael Freissmuth, Meinrad Busslinger, John O'Shea, Peter Valent, Christian Sillaber, and Kevin Bunting for helpful discussions in the course of this work.

![Figure 5. Abelson-induced transformation is dependent on Stat5a/b in vitro. (A) Ab-MuLV–induced colony formation of Stat5a/b+/+, Stat5a/bnull/+, and Stat5a/bnull/null fetal liver cells in methylcellulose. Single-colony pictures of each phenotype are depicted in the bottom panels. Stat5a/bnull/null cells showed no ability to form growth factor–independent colonies. (B) Summary of data obtained from Ab-MuLV–induced colony formation assays represent means ± SEM of 4 embryos per genotype (each performed in triplicates). (C) Surface expression of B-lineage markers was verified by flow cytometric analysis (right; data of one representative CD19+ CD43+ colony is shown). (D) [3H]thymidine incorporation of fetal liver–derived Stat5a/b+/+ and Stat5a/bnull/+ Ab-MuLV–transformed cell lines. Stat5a/b-deficient fetal livers did not give rise to stable transformed cell lines. Data represent means ± SEM of 2 cell lines per genotype. cpm indicates counts per minute. (E) Ab-MuLV–induced colony formation of Stat5a/b+/+ (n = 2), Stat5a/bnull/+ (n = 2), and Stat5a/bnull/null (n = 5; pooled) survivor bone marrow cells in methylcellulose. Stat5a/bnull/null survivor cells showed no ability to form growth factor–free colonies. Experiment was performed in triplicates. Asterisks denote significant differences as determined by a one-way ANOVA followed by a Tukey test (A,C) or a paired t test (B).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/107/12/10.1182_blood-2005-09-3596/4/m_zh80120697070005.jpeg?Expires=1767761708&Signature=CgjwLsmix9d8XZ7aiuIcyQZ1w~lMFMwF41x7A1QtT00VvhmtJZewaVl7Uapz5SA~2gkUbVmzMYqJqhyEPnOR6CQR2CgraDWmrYEqDLh6qaNf-2sEWdqsNKB9EpL1ervihifhnXSa3RySzeCqSd4-Pe8mdMGv1nRuKI0qCpkVMaZqkbgNYpGzmBzazZ8PFWkHHiso7u~93A1TOPFgf~rNALPDYtIS71yKbcvg3cF-DGZAa7GA6cm6w3D9AZlolxBADQKkUx0ckeJYeXmFeaqz1lAf6rxOskM-1jt3bQ89OKxvgboehP0mttMqFRgNfc9zFZBs7YHOLT0vvzDTNoZuPA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal