The recent identification of a somatically acquired, gain-of-function mutation in Janus kinase-2, JAK2V617F (Tefferi and Gilliland1 and references therein) in varying proportions of patients with sporadic polycythemia vera (PV) (virtually 100%), idiopathic myelofibrosis (IMF) (50%), and essential thrombocythemia (ET) (25%-50%), raises 2 possibilities with regards to disease pathogenesis: (1) JAK2V617F is sufficient for disease, and the precise phenotype is determined by interindividual differences in genetic background and/or environmental influences; and (2) JAK2V617F is the second mutation in a 2-“hit” model that confers selective growth advantage. Here, the first mutation, which may be genetically heterogeneous and possibly inherited, is also required for disease development.
The study of familial myeloproliferative diseases (MPDs) provides a unique opportunity to distinguish between the above possibilities. Families in which first-degree relatives develop MPDs that are indistinguishable from sporadic MPDs have been previously described, and suggest an autosomal dominant inheritance with incomplete penetrance, particularly for expression of the full disease phenotype.2,3 Notably, a study of 6 families with multiple members with PV revealed clonal hematopoiesis in affected females and erythropoietin-independent erythroid progenitors in healthy family members, thus implicating multiple genetic defects in the early pathogenesis of PV.4 Importantly, in this study, linkage analysis suggested that loss of heterozygosity (LOH) of chromosome 9p (where JAK2 is located) is a secondary change and does not target the primary PV locus. In another pedigree with familial PV, where the 2 affected members were linked by an unaffected obligate carrier, all 3 individuals were found to carry the JAK2V617F mutation, thus favoring the second possibility.5
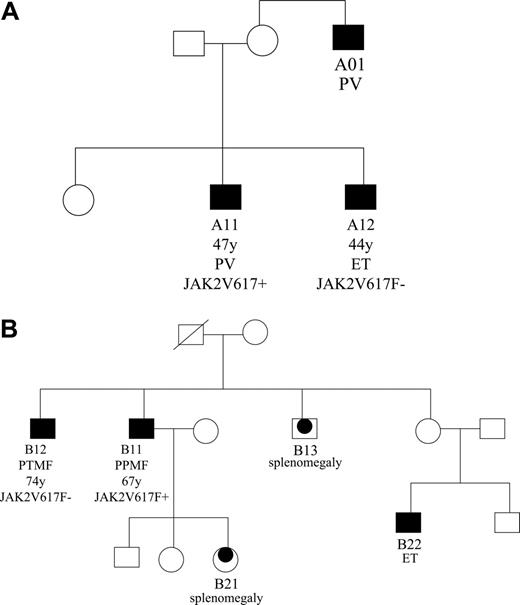
We studied 2 sibling pairs with familial MPDs for the JAK2V617F mutation (Table 1 and Figure 1) using our recently described allele-specific polymerase chain reaction (AS-PCR) methodology, which is sensitive to 0.01% to 0.1%.6 For each pair, the JAK2V617F mutation was found in the sibling (A11 and B11) with PV-related disease (PV and post-PV myelofibrosis, respectively) and not in the sibling (A12 and B12) with non-PV disease (ET and post-ET myelofibrosis, respectively). Notably, the association between the JAK2V617F mutation and PV held true regardless of whether the disease was early stage (siblings A11 and A12 had asymptomatic disease, and the diagnosis was made incidentally), or advanced stage (siblings B11 and B12 had advanced post-PV and post-ET myelofibrosis, respectively, and were being considered for palliative splenectomy).
JAK2V617F mutation status of 2 sibling pairs with familial MPD
Characteristic . | Sibling A11 . | Sibling A12 . | Sibling B11 . | Sibling B12 . |
|---|---|---|---|---|
| Diagnosis | PV | ET | PPVMF | PETMF |
| JAK2V617F | ||||
| Granulocytes | Positive | Negative | Positive | Negative |
| Buccal swab | Positive | Negative | ND | ND |
| Age, y | 47 | 44 | 67 | 74 |
| Sex | M | M | M | M |
| Symptomatic disease | No | No | Yes | Yes |
| Hb level, g/L | 176 | 161 | 118 | 83 |
| WBC count, × 109/L | 8.0 | 19.2 | 36.9 | 1.6 |
| Platelet count, × 109/L | 329 | 660 | 218 | 231 |
| Splenomegaly | + | + | + | + |
Characteristic . | Sibling A11 . | Sibling A12 . | Sibling B11 . | Sibling B12 . |
|---|---|---|---|---|
| Diagnosis | PV | ET | PPVMF | PETMF |
| JAK2V617F | ||||
| Granulocytes | Positive | Negative | Positive | Negative |
| Buccal swab | Positive | Negative | ND | ND |
| Age, y | 47 | 44 | 67 | 74 |
| Sex | M | M | M | M |
| Symptomatic disease | No | No | Yes | Yes |
| Hb level, g/L | 176 | 161 | 118 | 83 |
| WBC count, × 109/L | 8.0 | 19.2 | 36.9 | 1.6 |
| Platelet count, × 109/L | 329 | 660 | 218 | 231 |
| Splenomegaly | + | + | + | + |
Hb indicates hemoglobin; WBC, white blood cell; PV, polycythemia vera; ET, essential thrombocythemia; PPVMF, postpolycythemic myelofibrosis; PETMF, postthrombocythemic myelofibrosis; ND, not done; M, male; and +, present.
Our data supports the hypothesis that the primary locus/loci implicated in familial MPDs remains to be identified, and JAK2V617F may serve as a secondary “patterning” mutation associated with a PV phenotype. Further, as recently shown for an ET cohort,7 acquisition of the JAK2V617F mutation may be independent of disease duration.
Pedigrees of 2 families with familial MPD. (A) Pedigree of family A. (B) Pedigree of family B. Squares indicate males; circles, females. Filled symbols indicate affected individuals; black dots within symbols, possibly affected individuals; strike through, deceased individuals.
Pedigrees of 2 families with familial MPD. (A) Pedigree of family A. (B) Pedigree of family B. Squares indicate males; circles, females. Filled symbols indicate affected individuals; black dots within symbols, possibly affected individuals; strike through, deceased individuals.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal