Abstract
There is a 2-fold variation in platelet integrin α2β1 levels among inbred mouse strains. Decreased α2β1 in 4 strains carrying Itga2 haplotype 2 results from decreased affinity of heterogeneous ribonucleoprotein L (hnRNP L) for a 6 CA repeat sequence (CA6) within intron 1. Seven strains bearing haplotype 1 and a 21 CA repeat sequence at this position (CA21) express twice the level of platelet α2β1 and exhibit an equivalent gain of platelet function in vitro. By UV crosslinking and immunoprecipitation, hnRNP L binds more avidly to CA21, relative to CA6. By cell-free, in vitro mRNA splicing, decreased binding of hnRNP L results in decreased splicing efficiency and an increased proportion of alternatively spliced product. The splicing enhancer activity of CA21 in vivo is abolished by prior treatment with hnRNP L–specific siRNA. Thus, decreased surface α2β1 results from decreased Itga2 pre-mRNA splicing regulated by hnRNP L and depends on CA repeat length at a specific site in intron 1.
Introduction
Hui et al1 were the first to show that differential binding of heterogeneous ribonucleoprotein L (hnRNP L) to intronic CA-rich sequences of differing length is the mechanism responsible for regulation of mRNA splicing in the human gene for the endothelial isoform of nitric oxide synthase (eNOS). The largest intron 13 (4.1 kb [kilobase]) in eNOS carries a polymorphic CA-rich region (14-44 repeats), and hnRNP L binds preferentially to the longer CA-rich sequence. Moreover, the length of the polymorphic CA-rich region in the eNOS gene has been correlated with an independent risk factor for coronary artery disease.2
Li et al3 reported a 2-fold decrease in the platelet surface content of α2β1 in the mouse strain FVB, relative to 4 other mouse strains, and showed that this decrease was in linkage disequilibrium with the marker D13mit260 located roughly 2.6 Mb (megabase) up-stream from Itga2 on chromosome 13. In this study, we extend that finding and report the existence of 2 major mouse Itga2 haplotypes. Haplotype 1 is associated with high expression platelet α2β1 and increased platelet reactivity to collagen, whereas haplotype 2 is characterized by roughly one-half the level of platelet α2β1 and a concomitant decrease in platelet function. This phenotype is related to the differential binding of hnRNP L to 2 CA-rich regions within intron 1 that differ in length between the 2 haplotypes.
Hui et al4 had predicted that intronic CA-rich sequences and hnRNP L might mediate splicing regulation in genes other than eNOS. Our study establishes that the murine integrin α2 gene (Itga2) is a further example of this type of regulation. Moreover, in this study, we are the first to demonstrate that knockdown of hnRNP L with specific siRNA inhibits the rate and extent of in vivo pre-mRNA splicing of either a transfected Itga2 minigene or endogenous Itga2.
Materials and methods
Reagents
Fluorescein isothiocyanate (FITC)–conjugated rat monoclonal anti–mouse integrin α2 (Sam.G4), anti–mouse GPVI (Six.E10), and anti–mouse integrin α5 (Tap.A12) were purchased from Emfret Analytics (Wurzburg, Germany). Phycoerythrin (PE)–conjugated rat anti–mouse αIIbβ3 (Leo.D2) and anti–mouse GPIbα (Xia.G5) were also purchased from Emfret Analytics. Polyclonal goat anti–hnRNP L (D17), goat anti-Sp3 (D20), and horseradish peroxidase–conjugated rabbit anti–goat IgG were obtained from Santa Cruz Biotechnology (San Diego, CA). Monoclonal mouse anti-GAPDH (6C5) was purchased from Ambion (Austin, TX) and horseradish peroxidase–conjugated goat anti–mouse IgG from Zymed (San Francisco, CA).
Methoxyflurane was purchased from Medical Developments (Springvale, Australia). Bovine serum albumin (BSA) was obtained from Sigma Chemical (St Louis, MO).
Mice and mouse DNA
Male mice representing 5 inbred strains (BALB/cJ, C57BL/6J, 129 × 1/SvJ, FVB/NJ, and A/J) and genomic DNA from 11 inbred mouse strains (BALB/cJ, C57BL/6J, 129 × 1/SvJ, FVB/NJ, A/J, C3H/HeJ, C57L/J, DBA/2J, SJL/J, SM/J, and SWR/J) were obtained from The Jackson Laboratory (Bar Harbor, ME). At 10 weeks of age, mice were anesthetized with methoxyflurane, and whole blood was collected by retroorbital bleeding into 3.2% sodium citrate as anticoagulant. Platelet count and mean platelet volume were measured with the automated Coulter 9000 apparatus (Mallinckrodt Baker, Phillipsburg, NJ).
Platelet function in whole blood
Pooled whole blood (800 μL) was obtained from 5 to 6 male mice of identical strain. Using either a collagen/epinephrine test cartridge (CEPI) or a collagen/ADP test cartridge (CADP), the ability of platelets in whole blood to adhere to collagen and initiate a thrombus under high shear conditions were measured in the Platelet Function Analyzer-100 (PFA-100; Dade Behring, Wilmington, DE) and recorded as the closure time (CT).5
Cell lines and culture
The human cell line Dami was grown in Iscove modified medium supplemented 10% (vol/vol) horse serum and 1% antimycotic antibiotics (Gibco-Invitrogen, Carlsbad, CA). Dami cells (1 × 107) in 500 μL Iscove modified medium, without serum or antibiotics, were transfected by electroporation (300 V, 1180 μF), in the presence of 20 μg plasmid DNA construct and 20 μg pRL-TK vector. Dual luciferase assays (Promega, Madison, WI) were performed according to the manufacturer's instructions. Luciferase activity was monitored with a Monolight 2010 luminometer (Analytical Luminescence Laboratory, San Diego, CA) during a period of 20 seconds. The murine cell line B16F10, a gift from Dr Brunhilde Felding-Habermann (TSRI), was grown in Eagle Modified Dulbecco Medium supplemented with 10% (vol/vol) fetal bovine serum, 1% (wt/vol) L-glutamine, 1% (wt/vol) MEM nonessential amino acids (Gibco-Invitrogen), 1% (wt/vol) vitamin mix (Cambrex, East Rutherford, NJ), and 1% (wt/vol) sodium pyruvate. Cytoplasmic and nuclear extracts from Dami and B16F10 were prepared as previously described.6
Flow cytometry
Whole blood (2 μL) was mixed with 18 μL Tyrode buffer (12 mM NaHCO3, 2 mM MgCl2, 137.5 mM NaCl, 2.6 mM KCl, pH 7.4) containing 1% (wt/vol) BSA and incubated with 5 μL FITC- or PE-conjugated antibodies for 15 minutes. PBS (400 μL), pH 7.4, was then added, and the amount of bound antibody was measured within 30 minutes using a Becton Dickinson (BD; San Jose, CA) FACSCalibur flow cytometer. Data were analyzed with BD CellQuestPro software, and the amount of bound antibody was reported as the geometric mean fluorescence intensity (GMFI).
In selected experiments, the level of α2 was measured on B16F10 cells following a similar procedure. After the cells were washed in PBS, 1 × 106 cells were resuspended in 19 μL Tyrode buffer containing 1% (wt/vol) BSA, to which was added 5 μL FITC- or PE-conjugated antibodies for 15 minutes. The samples were further exactly processed.
PCR and DNA sequencing
Using genomic DNA from each mouse strain as the template, a 5′ segment of Itga2 encompassing 2-kb sequence upstream from the start codon was amplified by polymerase chain reaction (PCR) using the forward primer 5′-ATGTCTGCTCTGCATCTAT-3′ and the reverse primer 5′-TGTGAGAACATCTCTAAACC-3′. Overlapping portions of the product were sequenced using the following primer pairs: CNS2F, 5′-GCGGCACTAGAAAGGTTG-3′, and CNS2R, 5′-ATCCACCCACAACCAACATC-3′; CNS1F, 5′-AGCCCCTTATCTGACCTGTG-3′, and CNS1R, 5′-TATTCAGCACCCTCGAACCT-3′; and PROMF, 5′-CCGCACTGTCTGTAAGATGC-3′, and PROMR, 5′-GGGGTCCACACAGTATCCTC-3′.
The CA-rich regions of intron 1 were amplified using the following primer pairs: CA1F, 5′-CCCCCAGAGCACTTTAGATT-3′, and CA1R, 5′-TCAGAGCTAGTTCAGCACCTG-3′. Sequencing was performed by the DNA Core Laboratory of the Department of Molecular and Experimental Medicine (The Scripps Research Institute) using an ABI PRISM 3100 Genetic Analyzer via capillary electrophoresis (Applied Biosystems [AB], Foster City, CA). Multiple sequence alignment was accomplished using online Clust-alW analysis at http://searchlauncher.bcm.tmc.edu/multi-align/multi-align.html.7
Plasmid constructs
A segment of BALB/cJ or FVB/NJ genomic DNA sequence corresponding to 2 kb of the Itga2 5′ region (from –2058 to the ATG start codon at –113) was amplified by PCR with KOD hot start DNA polymerase (Novagen, San Diego, CA) using the primer pair A2prombgl2, 5′-GAAGATCTTCAGCCCCTTATCTGACCTGTG-3′, and A2promHind3, 5′-CCCAAGCTTGGGAGCCTGAATGAGAGCA-3′, thereby introducing 5′-BglII and 3′-HindIII restriction sites. Each PCR product was cloned into the multiple cloning site of pGL2-b (Promega), upstream from the luciferase (LUC) reporter gene.
DNA encompassing the CA-rich region of intron 1 (227 bp [base pair]) from FVB/NJ or BALB/cJ genomic DNA was cloned downstream of the T7 RNA polymerase site in PGEM-3z, using 5′-EcoRI and 3′-HindIII restrictions sites, incorporated with the primer pair EcoR1CA1, 5′-CGGAATTCCGATGTCTGCTCTGCATCTAT-3′, and Hind3CA1, 5′-CCCAAGCTTGGGTGTGAGAACATCTCTAAACC-3′.
Radiolabeled RNA probes
The DNA inserted into pGEM-3z or Topo TA was radiolabeled by transcription with T7 polymerase in the presence of 32P-CTP (800 Ci/mmol [2.96 × 1013 Bq/mmol]; Amersham, Piscataway, NJ). Briefly, the linearized vector was incubated with 250 μCi (9.25 MBq) 32P-CTP, T7 polymerase, and a cold nucleotide mix for 2 hours a 37°C. Subsequently, DNA digestion was performed using RQ1 DNAse, and unincorporated nucleotides were removed via a microspin G-50 column (Amersham). Radiolabeled probes were checked for quality by denaturing polyacrylamide Urea gel (Invitrogen) and reveled by autoradiography.
UV crosslinking
UV crosslinking of RNA probes to proteins within nuclear or cytoplasmic extracts was performed as described by Geneste et al.8 One or 5 μg nuclear or cytoplasmic protein was mixed with 105 cpm/min RNA probe in 10 mM HEPES, pH 7.9, containing 3 mM MgCl2, 40 mM KCl, 5% (vol/vol) glycerol, 1 mM dithiothreitol, and 100 ng/μL yeast RNA (Ambion, Austin, TX). After a 5-minute incubation at ambient temperature, the mixture was placed on wet ice and irradiated in a UV Starlinker 1800 (Stratagene, La Jolla, CA) for 30 minutes. RNAse A (2 μg; Ambion) was then added, and the mixture was incubated for 20 minutes at 37°C. Proteins were then separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) using a 12% polyacrylamide gels under nonreducing conditions, and protein bands were revealed by autoradiography.
Immunoprecipitation
After UV crosslinking and RNAse A treatment, an aliquot of the nuclear or cytoplasmic protein extract was incubated for 30 minutes with a 50-μL volume of Protein A-Sepharose beads (Amersham). The beads were then pelleted by centrifugation at 10 000g for 10 minutes, and the supernatant was aspirated and incubated overnight with 2 μg anti-hnRNP L or anti-Sp3. The resultant antibody-antigen-RNA probe complexes were pulled down by the addition of fresh Protein A-Sepharose. The beads were rinsed twice with 3 volumes of PBS, and bound proteins were eluted with 100 mM Tris-HCl 6.8, 4% SDS, 0.2% bromophenol blue, 20% glycerol (Laemmli Buffer; Invitrogen), and 5 mM N-ethyl maleimide (Sigma) for 5 minutes at 95°C, separated by SDS-PAGE using a 12% polyacrylamide gel under nonreducing conditions, and visualized by autoradiography.
Cell-free in vitro mRNA splicing
Two PCR products were generated by using the primer sets ECOEX1, 5′-GGAATTCCAACTCCTCCGTACAGTTTCT-3′, ASCIINT1, 5′-AGGCGCGCCTGAAATAAGAATAGGG-3′, ASCIINT2, 5′-AGGCGCGCCTAGAATCTTACTCTGTAAGCAAA-3′, and HINDEX2, 5′-CCCAAGCTTGGGTGCCTTGTGGGTTCGTAA-3′.
These EcoRI/AscI and AscI/HindIII PCR products were ligated to generate a 1.2-kb construct that includes (from 5′ to 3′) exon 1, a segment of intron 1 flanking the CA-rich region, and exon 2. This EcoRI/HindIII construct was then inserted into pGem-3z using the corresponding restriction sites. The CA-rich sequence was then excised using BamHI sites situated 40 bp upstream and 180 bp downstream from the CA repeats. Transcription was initiated in the presence of m7GpppG cap analog, and in vitro splicing was accomplished as previously described.9 Briefly, 60 ng RNA was incubated with 60 μg extract of B16F10 nuclear proteins for 90 minutes. The splicing product was separated under denaturing conditions using a 6% polyacrylamide gel in the presence of 1 M Tris, 0.9 M boric acid, 0.01 M EDTA, and 7 M urea (TBE/Urea; Invitrogen) and stained with SYBR gold nucleic acid gel stain (Invitrogen) diluted 1:10 000 in 10 mM Tris HCl, 1 mM EDTA pH 8.
In vivo splicing
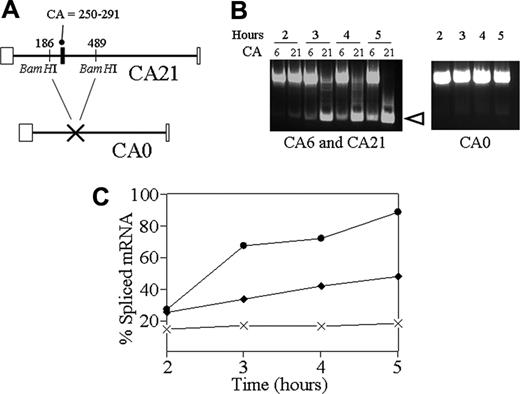
For in vivo splicing assays, cDNA cassettes were created that contain exon 1 and exon 2 plus 0.96-kb intervening intron 1 sequence representing either the haplotype 1 or haplotype 2 CA-rich sequence. To accomplish this, EcoRI/HindIII fragments (1242 bp) were excised from the pGem-3Z constructs used for the in vitro splicing assays and cloned into pcDNA3.1 zeo (–) (Invitrogen). A control DNA segment from which the CA repeat sequence was eliminated (CA0) was created by excising the pcDNA3.1 zeo(–) CA-rich sequence as a 303-bp BamHI/BamHI fragment (nucleotides +186 to +489 from ATG). B16F10 cells (9 × 105) were transfected with 2 μg of each construct using the lipofectamine 2000 reagent (Invitrogen), according to the manufacturer's instructions. The transfection reaction was stopped by extraction of total RNA from the cells using the RNAeasy miniprep kit (Qiagen, Valencia, CA), according to the manufacturer's instructions. Reverse transcriptase and PCR reactions were performed as described for the in vitro splicing assays.
hnRNP-L knockdown using siRNA
Forty-eight hours prior to the onset of the in vivo splicing assay, B16F10 cells were seeded at 4 × 105 cells per well in 6-well microtiter plates. Twenty-four hours later, the cells were transfected with 50 nM siRNA (Dharmacon, Chicago, IL) specific for hnRNP L (5′-TATGGCTTGGATCAATCTA-3′), as previously described by Kim et al,10 using Lipofectamine 2000 (Invitrogen), according to the manufacturer's instructions. As negative control, an equivalent aliquot of cells were transfected with 50 nM siRNA specific for luciferase, 5′-TAAGGCTATGAAGAGATAC-3 (Dharmacon). The decreased expression (knockdown) of HnRNP L was confirmed by Western blot assay, using polyclonal anti-hnRNP L, as described in “Flow cytometry.”
In additional experiments, the effect of the hnRNP L knockdown on expression of endogenous α2β1 by B16F10 cells was measured. B16F10 cells were plated into 6-well microtiter plates (2 × 105 cells per well). Cells were transfected with 50 nM hnRNP L siRNA once, then after 24 hours, a second time with the same amount of siRNA. Seventy-two hours following the second transfection, the surface expression of integrin α2β1 was measured by flow cytometry, using rat monoclonal FITC-conjugated anti–murine α2β1, as described in “Flow cytometry.”
Western blots
Briefly, 20 μg nuclear protein was loaded into the wells of a 4% to 12% linear gradient polyacrylamide gel and subjected to electrophoresis in the presence of sodium dodecyl sulfate, as previously described.11 The separated proteins were then transferred electrophoretically to a nitrocellulose membrane. Membranes were blocked with 5% (wt/vol) powdered milk in 1.5 M NaCl, 0.2 M Tris, 0.1% (vol/vol) Tween 20, pH 7.6 (TBS-T) and immersed in a solution containing 0.5 μg/mL anti-hnRNP L for 2 hours at room temperature. After 3 washes of 15 minutes each in TBS-T with gentle agitation, the membranes were immersed in a solution containing the secondary antibody diluted 1:3000 and incubated for 1 hour at ambient temperature. After 3 additional washes of 15 minutes each in TBS-T, bound antibody was visualized by chemiluminescence. Total protein quantity loaded per well was checked visually by ponceau S staining and blotted with 1 μg/mL anti-GAPDH (Ambion) for 2 hours at room temperature, then revealed as previously described with HRP-conjugated antimouse antibody at a 1:3000 dilution for 45 minutes.
Results
Comparison of genomic sequences from mouse strains
On the basis of a comparison of spatially distant polymorphisms within the 5′ regulatory region and exons 7 and 13 of murine Itga2, we have defined 2 major haplotypes. Haplotype 1 is expressed by 7 inbred strains (C3H/HEJ, C57L/J, DBA2/J, BALB/cJ, C57BL/6J, 129SV/J, and SM/J), whereas haplotype 2 is represented in 4 inbred strains (SJL/J, SWR/J, A/J, and FVB/NJ).
Relation between surface α2β1 level and Itga2 haplotypes
We next used flow cytometry to compare the platelet surface content of α2β1 with Itga2 haplotype in 5 representative inbred strains (Figure 1A). Rat antimouse monoclonal antibodies were used to quantitate the surface level of GPIbα, GPVI, and the integrins αIIbβ3, α5β1, and α2β1. The difference between strains in mean levels of αIIbβ3, GPIbβ, GPVI, and α5β1 were remarkably small, and none of the pair-wise differences were statistically significant, relative to BALB/cJ (P > .1 using ANOVA). In contrast, with regard to mean levels of α2β1, a statistically significant decrease was seen for strains FVB/NJ or A/J, relative to BALB/cJ (P < .01 in both cases). At the same time, the levels of platelet α2β1 in strains C57BL/6J and 129/SvJ were not significantly different from that of BALB/cJ. Because α5β1 shares a common β subunit with α2β1 and the levels of these 2 integrins on platelets are not coregulated, we calculated a normalized level of α2β1 α2β1[(n) using the formula [α2β1(n) = (GMFI α2β1 /GMFI α5β1) × 100]. Relative to strain BALB/cJ, the difference in the mean levels of α2β1(n) in strains FVB/NJ and A/J, but not C57BL/6J or 129/SvJ, were statistically significant (P < .001). The difference is approximately 2-fold, confirming the report of Li et al,3 and correlates exactly with the expression of the 2 major Itga2 haplotypes. FVB/NJ and A/J mice (both haplotype 2) express one-half the level of platelet α2β1 that is found on platelets of the remaining 3 strains, all of which express haplotype 1.
Murine platelet Itga2: surface density and function. (A) Murine platelet glycoprotein levels were determined by flow cytometry. The surface content of platelet glycoproteins αIIbβ3, GPIbα, GPVI, α5β1, and α2β1 was determined in citrated whole blood using rat antimouse monoclonal antibodies. The level of bound FITC- or PE-conjugated rat antibody was expressed as the geometric mean fluorescence intensity and is indicated on the ordinate. Each bar represents the mean ± SD of 6 separate measurements using whole blood pooled from 5 to 6 male mice in each measurement. The results from 5 representative inbred strains are depicted. Three strains bear haplotype 1 (BALB/cJ, C57BL/6J, and 129/SvJ), whereas 2 strains express haplotype 2 (FVB/NJ and A/J). In the bottom right-hand panel, the levels of α2β1 were normalized with respect to the levels of α5β1 where [α2β1(n) = GMFI (α2β1)/GMFI (α5β) × 100]. (B) Platelet adhesion under high shear measured by the PFA-100. Citrated whole blood was pooled from 5 to 6 male mice of each of the 5 representative strains (see panel A) on 3 independent occasions. The CT in seconds was recorded and is indicated on the ordinate. The left-hand side depicts the results when blood was perfused over cartridge membranes impregnated with collagen plus ADP (CT-CADP); for the right-hand side, the findings shown when blood was perfused over cartridge membranes impregnated with collagen plus epinephrine (CT-CEPI). Each entry represents the mean ± SD.
Murine platelet Itga2: surface density and function. (A) Murine platelet glycoprotein levels were determined by flow cytometry. The surface content of platelet glycoproteins αIIbβ3, GPIbα, GPVI, α5β1, and α2β1 was determined in citrated whole blood using rat antimouse monoclonal antibodies. The level of bound FITC- or PE-conjugated rat antibody was expressed as the geometric mean fluorescence intensity and is indicated on the ordinate. Each bar represents the mean ± SD of 6 separate measurements using whole blood pooled from 5 to 6 male mice in each measurement. The results from 5 representative inbred strains are depicted. Three strains bear haplotype 1 (BALB/cJ, C57BL/6J, and 129/SvJ), whereas 2 strains express haplotype 2 (FVB/NJ and A/J). In the bottom right-hand panel, the levels of α2β1 were normalized with respect to the levels of α5β1 where [α2β1(n) = GMFI (α2β1)/GMFI (α5β) × 100]. (B) Platelet adhesion under high shear measured by the PFA-100. Citrated whole blood was pooled from 5 to 6 male mice of each of the 5 representative strains (see panel A) on 3 independent occasions. The CT in seconds was recorded and is indicated on the ordinate. The left-hand side depicts the results when blood was perfused over cartridge membranes impregnated with collagen plus ADP (CT-CADP); for the right-hand side, the findings shown when blood was perfused over cartridge membranes impregnated with collagen plus epinephrine (CT-CEPI). Each entry represents the mean ± SD.
Relationship between platelet function and Itga2 haplotypes
Platelet function was evaluated for the 5 representative strains in whole blood under high shear using the PFA-100 (Figure 1B). From numerous PFA-100 studies of human blood, it has been determined that the CT obtained with the CADP measures the activation of platelets via ADP and the rate of subsequent thrombus formation. This platelet response is not sensitive to the level of platelet collagen receptors, and it was expected that CT-CADP values would not be significantly different between strains, ranging from 62 ± 4 seconds (mean ± SD; n = 3) for 129/SvJ to 76 ± 7 seconds for A/J. However, we have reported that the closure times obtained with the CEPIs in human studies are influenced markedly by variation in platelet α2β1.5
Not surprisingly, the CT-CEPI values for A/J and FVB/NJ are significantly increased to 229 ± 78 and 236 ± 46 seconds, respectively, compared with that of BALB/cJ (124 ± 15; P < .01 in both cases). However, the difference between BALB/cJ and C57BL6/J (138 ± 21) or 129/SvJ (110 ± 5) was not statistically significant. Thus, FVB/NJ and A/J mice (both haplotype 2) exhibit deceased platelet function in response to collagen plus epinephrine.
Comparison of the transcriptional activity of promoter regions
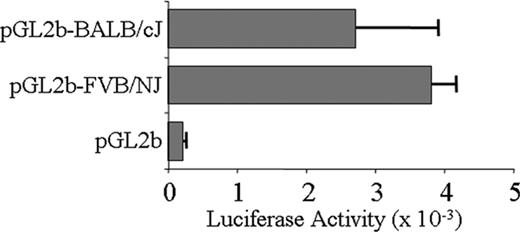
We directly compared the promoter activity of a large segment of haplotype 1 and haplotype 2 5′-regulatory/promoter regions. To accomplish this, we cloned slightly more than 2 kb FVB/NJ and BALB/cJ 5′ genomic sequences (from –2058 to +113) and measured relative promoter activity using a luciferase reporter system. When transfected into Dami cells, the difference in activity of these constructs was not statistically significant (Figure 2). Thus, variation of Itga2 transcription rate is not the likely basis for the variation in expression seen between these 2 haplotypes. We next compared the sequence and potential function of noncoding sequences downstream from the promoter region, beginning with intron 1.
Comparative intron sequences
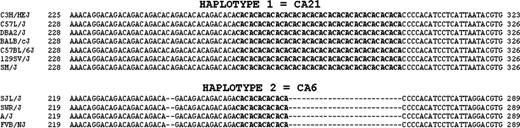
Mouse Itga2 intron 1 is large (31.3 kb) and contains a CA-rich region 250 nucleotides downstream from the 5′ splice site (5′ ss). In the BALB/cJ (haplotype 1) reference genomic sequence (NT_039590) beginning at the ATG start codon (A =+1), the CA-rich region is located at nucleotides 250 through 291. From a comparison of all 11 genomic sequences, strain-related differences in CA repeat length are apparent (Figure 3). In the case of the haplotype 1, the CA-rich region contains a contiguous stretch of 21 CA repeats. As for haplotype 2, a shorter segment with 6 CA repeats is present in the same location. From this point on, the CA-rich region of haplotype 1 will be referred to as CA21 and that of haplotype 2 will be designated CA6.
Transcriptional activity of Itga2 promoter-LUC reporter constructs transfected into Dami cells. The proximal 5′-regulatory region/promoter sequence (from –2058 to the ATG start codon at +113) of Itga2 haplotype 1 (from BALB/cJ) or haplotype 2 (from FVB/NJ) were cloned and assayed for transcriptional activity using a LUC reporter construct Relative luciferase activity is indicated on the abscissa. pGL2b is the LUC reporter vector without an Itga2 insert. Each entry represents the mean ± SD for 3 independent assays.
Transcriptional activity of Itga2 promoter-LUC reporter constructs transfected into Dami cells. The proximal 5′-regulatory region/promoter sequence (from –2058 to the ATG start codon at +113) of Itga2 haplotype 1 (from BALB/cJ) or haplotype 2 (from FVB/NJ) were cloned and assayed for transcriptional activity using a LUC reporter construct Relative luciferase activity is indicated on the abscissa. pGL2b is the LUC reporter vector without an Itga2 insert. Each entry represents the mean ± SD for 3 independent assays.
Role for hnRNP L
Hui et al4 showed that heterogeneous nuclear ribonucleoprotein-L binds specifically to CA repeats in a manner that is influenced by CA repeat length. It then functions as a specific activator/enhancer of mRNA splicing. We next investigated whether hnRNP L might play a similar role in the splicing of Itga2 mRNA.
In pilot experiments (data not shown), we first established by Western blot assay that B16F10 cells contain a significant amount of hnRNP L in nuclear but not cytoplasmic extracts. Secondly, we established that B16F10 express surface α2β1, as determined by flow cytometry. To determine whether the length of the CA repeat has an effect on hnRNP L binding to mRNA, segments of mRNA encompassing CA21 or CA6 were amplified by PCR, cloned into pGem3Z, and radiolabeled. We then mixed the mRNA probes with proteins isolated in nuclear extracts of B16F10 cells and crosslinked any protein-mRNA complexes by UV irradiation, according to the procedure of Hui et al.4
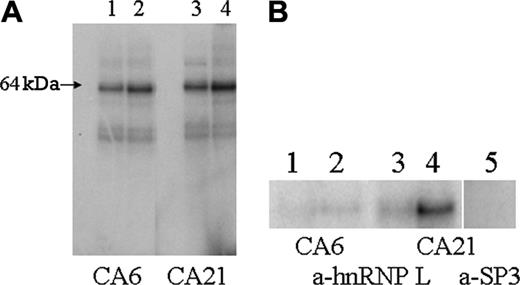
UV crosslinking of CA21 mRNA produced a major complex at an apparent molecular weight (MWApp) of 64 kDa (Figure 4A). This is approximately the MWApp of hnRNP L, reported to be 64 to 68 kDa.12 The affinity of the CA21 sequence was significantly greater than that of the CA6 sequence. On the basis of optical scanning and densitometry, the increase in binding to the CA21 sequence was approximately 50%.
Anti-hnRNP L binds to the 64-kDa protein-mRNA complexes, confirming that hnRNP L is specifically bound by these RNA sequences (Figure 4B). In every case, hnRNP L binds less strongly to the sequence containing the fewer number of CA repeats, as evidenced by significantly less radiolabeled probe associated with hnRNP L. By optical scanning and densitometry, binding of anti-hnRNP L to the CA6 probe is decreased to approximately 20% of that obtained with the CA21 probe. The specificity of hnRNP L for these mRNA probes was confirmed by the finding that the radiolabeled probe was not associated with immunoprecipitates generated with monoclonal antibody specific for the irrelevant nuclear protein, Sp3, also present in the B16F10 nuclear extracts (Figure 4B, lane 5).
Cell-free in vitro mRNA splicing
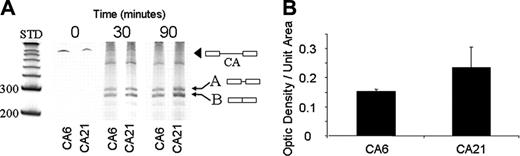
In our cell-free in vitro mRNA splicing studies, the total mRNA product is consistently greater in the presence of CA21 relative to CA6 (Figure 5A). This result suggests that increased binding of hnRNP L to the CA21 increases the efficiency of the proper splicing pattern. Alternatively spliced products were present for both CA6 and CA21, but in the presence of CA21, more correctly spliced product is produced. Thus, the presence of CA6 both decreases the efficiency of correct splicing and increases the incidence of incorrect splicing with a concomitant shift to different splice site.
The relative amount of correctly spliced mRNA product in the case of the CA6 and CA21 mRNA probes was determined by optical densitometry (Figure 5B). A significant increase in the correctly spliced product (band B) was obtained with the CA21 probe (mean = 0.23) relative to the CA6 probe (mean = 0.15).
In vivo minigene splicing
We next determined whether in vitro mRNA splicing enhancement stimulated by CA21 could be confirmed in vivo in the murine cell line B16F10. We used a series of minigene cassettes carrying Itga2 exons 1 and 2 separated by a truncated intron 1 segment bearing the CA21 or CA6 sequences. These constructs were expressed using pcDNA3.1 and correspond to the same constructs used in in vitro splicing.
A comparison of intron 1 CA-rich sequences. Intron 1 sequences that encompass the CA-rich regions from the 11 inbred strains are aligned for comparison. Two haplotypes can be distinguished: Haplotype 1 (represented by the top 7 sequences) contains a contiguous stretch of 21 CA repeats (nucleotides 250-291, based on the BALB/cJ sequence, setting the A of the ATG start codon to 1). Haplotype 2 (represented by the bottom 4 sequences) contains no more than 6 consecutive CA repeats within the same region. The sequence within haplotype 1 is designated CA21; that within haplotype 2, CA6.
A comparison of intron 1 CA-rich sequences. Intron 1 sequences that encompass the CA-rich regions from the 11 inbred strains are aligned for comparison. Two haplotypes can be distinguished: Haplotype 1 (represented by the top 7 sequences) contains a contiguous stretch of 21 CA repeats (nucleotides 250-291, based on the BALB/cJ sequence, setting the A of the ATG start codon to 1). Haplotype 2 (represented by the bottom 4 sequences) contains no more than 6 consecutive CA repeats within the same region. The sequence within haplotype 1 is designated CA21; that within haplotype 2, CA6.
After transient transfection of B16F10, we analyzed the splicing efficiency of these Itga2 constructs by reverse transcriptase (RT)–polymerase chain reaction using primers specific for the flanking exons. For these studies, an additional cDNA probe was constructed (CA0) in which the CA-rich region and flanking sequence of CA21 was excised as a 303-bp BamH1/BamH1 fragment (Figure 6A). In the absence of a CA-rich sequence (CA0), the basal splicing activity was very low (10%-15%) (Figure 6B, right; Figure 6C, solid crosses). In the presence of CA6, splicing activity increased nearly 3-fold, to approximately 40% after 5 hours (Figure 6C, solid diamonds). The presence of the CA21 sequence resulted in a further significant increase in splicing activity to roughly 80% to 90%, as shown in Figure 6C (solid circles). The efficiency of in vivo splicing with either C6 or C21 is significantly greater than that seen in vitro, a finding that is consistent with the observations of Hui et al13 who analyzed different genes.
Identification of hnRNP L as the major protein in nuclear extracts of B16F10 cells that binds CA6 or CA21 mRNA probes. (A) A 32P-labeled mRNA probe representing either the FVB/NJ sequence (CA6) (lane 1, 1 μg; lane 2, 5 μg) or the BALB/cJ sequence (CA21) (lane 3, 1 μg; lane 4, 5 μg) was mixed with a nuclear extract from B16F10 cells and subjected to UV irradiation. The mRNA-protein complexes were then separated by SDS-PAGE, and proteins that had bound each probe were visualized by autoradiography. The major complex has a MWApp of 64 kDa, the mobility expected for hnRNP L. On the basis of optical scanning and densitometry, the intensity of the analogous band obtained with the FVB/NJ CA6 probe is approximately 50% of that obtained with the BALB/cJ CA21 probe. (B) Immunoprecipitation of mRNA-protein complexes with anti-hnRNP L. A 32P-labeled CA6 mRNA probe (lane 1, 1 μg; lane 2, 5 μg) or CA21 mRNA probe (lane 3, 1 μg; lane 4, 5 μg) was mixed with a nuclear extract from B16F10 cells and subjected to UV irradiation. mRNA-protein complexes were then precipitated with anti-hnRNP L. The immunoprecipitate were isolated and washed, and the components of the precipitate were solubilized in SDS and separated by SDS-PAGE. Proteins that had bound probe were visualized by autoradiography. Once again a single major band with a MWApp of 64 kDa is clearly visible in lane 4. As a negative control, the procedure was repeated with anti-Sp3 (lane 5). In this case, no labeled protein bands were detected in the immunoprecipitate. The results depicted were derived from 1 experiment that is representative of 3 independent experiments.
Identification of hnRNP L as the major protein in nuclear extracts of B16F10 cells that binds CA6 or CA21 mRNA probes. (A) A 32P-labeled mRNA probe representing either the FVB/NJ sequence (CA6) (lane 1, 1 μg; lane 2, 5 μg) or the BALB/cJ sequence (CA21) (lane 3, 1 μg; lane 4, 5 μg) was mixed with a nuclear extract from B16F10 cells and subjected to UV irradiation. The mRNA-protein complexes were then separated by SDS-PAGE, and proteins that had bound each probe were visualized by autoradiography. The major complex has a MWApp of 64 kDa, the mobility expected for hnRNP L. On the basis of optical scanning and densitometry, the intensity of the analogous band obtained with the FVB/NJ CA6 probe is approximately 50% of that obtained with the BALB/cJ CA21 probe. (B) Immunoprecipitation of mRNA-protein complexes with anti-hnRNP L. A 32P-labeled CA6 mRNA probe (lane 1, 1 μg; lane 2, 5 μg) or CA21 mRNA probe (lane 3, 1 μg; lane 4, 5 μg) was mixed with a nuclear extract from B16F10 cells and subjected to UV irradiation. mRNA-protein complexes were then precipitated with anti-hnRNP L. The immunoprecipitate were isolated and washed, and the components of the precipitate were solubilized in SDS and separated by SDS-PAGE. Proteins that had bound probe were visualized by autoradiography. Once again a single major band with a MWApp of 64 kDa is clearly visible in lane 4. As a negative control, the procedure was repeated with anti-Sp3 (lane 5). In this case, no labeled protein bands were detected in the immunoprecipitate. The results depicted were derived from 1 experiment that is representative of 3 independent experiments.
Effect of hnRNP L knockdown
To establish a direct role for hnRNP L in the in vivo enhancement of mRNA splicing, we repeated the minigene cassette splicing studies with and without prior transfection of B16F10 with hnRNP L–specific siRNA. B16F10 cells were transfected with either hnL1066 siRNA against hnRNP L10 or siRNA specific for the irrelevant gene LUC (firefly luciferase gene). After 24 hours, the level of endogenous hnRNP-L had decreased to no greater than 40% of the initial level, based on a Western blot assay (not shown). At this stage, the Itga2 minigenes (CA6 or CA21) were transfected, and splicing efficiency was measured by RT-PCR (Figure 7A-B). After transfection with the control LUC-specific siRNA, the rate and extent of minigene splicing was not different from that seen in untreated B16F10 cells (compare Figure 7A, right, to Figure 6B, left). However, prior transfection with hnRNP L siRNA abolished any splicing above basal levels (decreased to roughly 10%) (Figure 7A, left, and Figure 7B).
We next extended this observation by analyzing the effect of hnRNP L siRNA treatment on the surface expression of endogenous B16F10 integrin α2β1. Prior treatment with hnRNP L siRNA caused a 30% decrease in surface α2β1 (Figure 7C), as determined by fluorescence-activated cell sorting (FACS; GMFI decreased from 38 to 27). However, prior treatment with the control LUC-specific siRNA had little if any effect on either surface α2β1 expression. In the representative experiment depicted in Figure 7C, the GMFI for untreated cells was 38 and that for LUC siRNA treated cells was 39.
Discussion
In this study we have obtained conclusive evidence that hnRNP L regulates murine α2β1 expression by controlling the rate and extent of Itga2 pre-mRNA splicing. Previous studies had shown that hnRNP L binds preferentially to longer CA repeats within CA-rich sequences in various pre-mRNA molecules.1,4,10,13 In this report, we have confirmed that this is also the case with Itga2 pre-mRNA. Murine Itga2 haplotypes 1 and 2 differ most notably in the number of CA repeats within a segment of intron 1 that is immediately downstream from the 5′ splice site. Haplotype 1, bearing 21 CA repeats at this position, is associated with a 2-fold increase in the rate of Itga2 pre-mRNA splicing and a corresponding increase in surface α2β1 expression, relative to haplotype 2, which contains only 6 CA repeats in the same position. Thus, our results add murine Itga2 to the list of genes whose expression is regulated by hnRNP L through this CA repeat-dependent mechanism. Murine Itga2 is the first example of an integrin subunit gene that is regulated by this mechanism.
In vitro splicing of Itga2 pre-mRNA. (A) Itga2 pre-mRNA containing CA6 or CA21 was incubated under splicing conditions with a nuclear extract from B16F10 cells for 0, 30, or 90 minutes. The spliced RNA products were then separated on a 6% denaturing polyacrylamide gel, and RNA bands were detected by staining with SYBR gold. The positions of size standards representing 200 or 300 nucleotides is indicated to the left of the gel. The position of pre-mRNA (black arrowhead) and spliced products (arrows A and B) are indicated. Product B represents the correctly spliced exon 1-exon 2 product. The yield of product B is significantly greater in the presence of CA21, relative to CA6. (B) In vitro splicing efficiency. The relative amounts of the correctly spliced exon1-exon2 products (band B in panel A) were determined by optical densitometry using image J.38 Optical densities divided by unit area (square pixels) for 3 independent assays were obtained, and the mean ± SD is depicted. The mean density obtained in the case of the CA21 probe (0.23) is significantly higher than that obtained for the CA6 probe (0.15).
In vitro splicing of Itga2 pre-mRNA. (A) Itga2 pre-mRNA containing CA6 or CA21 was incubated under splicing conditions with a nuclear extract from B16F10 cells for 0, 30, or 90 minutes. The spliced RNA products were then separated on a 6% denaturing polyacrylamide gel, and RNA bands were detected by staining with SYBR gold. The positions of size standards representing 200 or 300 nucleotides is indicated to the left of the gel. The position of pre-mRNA (black arrowhead) and spliced products (arrows A and B) are indicated. Product B represents the correctly spliced exon 1-exon 2 product. The yield of product B is significantly greater in the presence of CA21, relative to CA6. (B) In vitro splicing efficiency. The relative amounts of the correctly spliced exon1-exon2 products (band B in panel A) were determined by optical densitometry using image J.38 Optical densities divided by unit area (square pixels) for 3 independent assays were obtained, and the mean ± SD is depicted. The mean density obtained in the case of the CA21 probe (0.23) is significantly higher than that obtained for the CA6 probe (0.15).
More importantly, our study is the first to provide direct evidence that a decrease in intracellular hnRNP L levels, induced by a specific siRNA knockdown approach, results in a dramatic decrease not only in the in vivo splicing of a transfected Itga2 minigene but also in the surface expression of endogenous α2β1. Thus, our findings provide further direct evidence to support for the hypothesis that regulation of pre-mRNA splicing by hnRNP L is a major mechanism controlling the expression of diverse genes.
CA repeats are the most common dinucleotide polymorphism within microsatellite DNA regions, representing as much as 0.25% of the human genome. By binding to AC-rich RNA sequences, hnRNP L contributes to various aspects of mRNA metabolism, including CA repeat-dependent splicing enhancement,1 internal translation initiation of hepatitis C virus (HCV),14 stability of the human vascular endothelial growth factor (VEGF) mRNA under hypoxic growth conditions,15 and intron-independent mRNA export.16 The first gene in which hnRNP L was shown to regulate mRNA splicing dependent on CA-repeat length was eNOS.4,17 Murine Itga2 represents the second instance, as reported here. There are many additional genes with polymorphic intronic CA repeats, including the human interferon gamma gene,18 the epidermal growth factor receptor gene,19 the cellular stress response gene,20 and the cystic fibrosis transmembrane conductance regulator gene.21 The extent to which hnRNP L might be involved in CA repeat-associated differences in the expression of these genes remains to be determined.
In vivo minigene splicing. (A) For the in vivo minigene splicing studies, an additional probe was constructed. To generate CA0, having no CA repeats, a BamH1/BamH1 segment representing nucleotides 186 to 489 was excised from the CA21 probe. The resulting construct lacks the CA repeat region at nucleotide 250 to 291. (B) B16F10 cells were transfected with 2 μg pcDNA3.1(–) containing either the CA6, CA21, or CA0 cassettes. Total RNA was then extracted at 2, 3, 4, and 5 hours after transfection. Spliced mRNA was amplified by RT-PCR, and separated by agarose gel electrophoresis. The amount of spliced mRNA (open arrowhead) was calculated using image J38 and is expressed as the percentage of total mRNA (nonspliced plus spliced mRNA). (C) The percentage of spliced mRNA is plotted as a function of time (hours) after transfection with the minigene cassette. The graph depicts the levels obtained with the CA21 (•), CA6 (♦), or CA0 (X-X) minigene cassettes. Each data entry represents the mean of triplicate measurements in a single experiment that is representative of 4 independent experiments.
In vivo minigene splicing. (A) For the in vivo minigene splicing studies, an additional probe was constructed. To generate CA0, having no CA repeats, a BamH1/BamH1 segment representing nucleotides 186 to 489 was excised from the CA21 probe. The resulting construct lacks the CA repeat region at nucleotide 250 to 291. (B) B16F10 cells were transfected with 2 μg pcDNA3.1(–) containing either the CA6, CA21, or CA0 cassettes. Total RNA was then extracted at 2, 3, 4, and 5 hours after transfection. Spliced mRNA was amplified by RT-PCR, and separated by agarose gel electrophoresis. The amount of spliced mRNA (open arrowhead) was calculated using image J38 and is expressed as the percentage of total mRNA (nonspliced plus spliced mRNA). (C) The percentage of spliced mRNA is plotted as a function of time (hours) after transfection with the minigene cassette. The graph depicts the levels obtained with the CA21 (•), CA6 (♦), or CA0 (X-X) minigene cassettes. Each data entry represents the mean of triplicate measurements in a single experiment that is representative of 4 independent experiments.
hnRNP L binds with high affinity not only to simple CA repeats but also CA-rich sequences and can mediate either enhancement or repression of pre-mRNA splicing, depending in part on the proximity of the CA-rich sequence to the 5′ ss.13 On the basis of studies of an eNOS minigene, moving the CA-rich sequence to a more downstream location in the target intron resulted in activation of cryptic 5′ ss and a reduction in use of the normal 5′ ss. The location of the CA-rich sequence was more important than the exact sequence of the ss and its homology to consensus ss. Thus, hnRNP L binds to the CA-rich sequence and activates the 5′ ss directly upstream from its location. In murine Itga2, the position of the polymorphic CA repeat sequence in intron 1 is ideally situated for hnRNP L–mediated enhancement of mRNA splicing at the appropriate 5′ ss.
Integrin α2β1 is a ubiquitous cell-surface receptor with a restricted specificity for collagens when expressed by megakaryocytes or platelets.22,23 Platelet α2β1 together with platelet glycoprotein VI (GPVI) contribute to the adhesion of platelets to collagens within the matrix of the blood vessel wall, a pivotal stage in the formation of a platelet thrombus.24-27 Genetically based differences in platelet α2β1 expression have been described in both humans28-30 and mice.3 The surface content of human platelet α2β1 can vary up to 6-fold, depending on the method of measurement,28,31 and this difference correlates with the inheritance of 6 major haplotypes of the α2 subunit gene Itga2.28,32 The totality of the mechanisms underlying expression differences remains to be determined, but transcriptional regulation controlled by Sp1 during megakaryocytic differentiation22 and polymorphisms within the core promoter region at positions –92 and –5229,30 have been shown to contribute to the wide variability in expression in humans. Increased α2β1 expression in man, associated with inheritance of Itga2 haplotype 1 (807T), has also been implicated in risk of thrombosis.33-37
On the basis of preliminary comparisons (T.J.K., unpublished results, September 2004), we have found that similar but not identical CA repeats are present within human Itga2. The number and variation of these repeats in the human gene is more complex than that of the mouse. However, preliminary indications are that longer CA repeats are in linkage disequilibrium with allelic markers of human Itga2 that have already been associated with increased expression of platelet α2β1. The exact nature of human Itga2 CA repeats and their influence on pre-mRNA splicing and α2β1 expression are currently under investigation.
hnRNP L siRNA knockdown. (A) B16F10 cells were transfected for 24 hours with (i) siRNA control or (ii-v) 5 nM, 10 nM, 20 nM, or 50 nM, respectively, of HNRNP L siRNA. Nuclear protein was extracted and separated by SDS-PAGE (30 μg per lane). The presence of hnRNP L was detected by Western blot with anti-hnRNP L. (B) B16F10 cells were transfected with 50 nM hnRNP L siRNA (left) or control siRNA (right) for 24 hours. The cells were then transfected with 2 μg pcDNA3.1 (–) C6 or C21 exactly as described in panel B. The amount of spliced mRNA (open arrowheads) was calculated using image J38 and expressed as the percentage of total mRNA (nonspliced plus spliced mRNA). (C) The percentage of spliced mRNA obtained following transfection with hnRNP L siRNA or control siRNA. Following treatment with hnRNP L siRNA, the levels obtained for CA21 (○) or CA6 (⋄) are dramatically reduced. After treatment with the siRNA control, the levels obtained for CA21 (•) or CA6 (♦) are not different from that observed in the absence of siRNA. Each data entry represents the mean of triplicate measurements in a single experiment that is representative of 3 independent experiments. (D) Effect of hnRNP L siRNA on B16F10 surface expression of endogenous α2β1. B16F10 cells were transfected twice with 50 nM HNRNP L siRNA (gray curve), 50 nM control siRNA (solid black line), or Lipofectamine 2000 alone (dotted line) over a 72-hour period. Twenty-four hours after the second transfection, flow cytometry was used to quantitate surface α2β1. The results, reported as the GMFI ± SD, are as follows: hnRNP L siRNA, 19.28 ± 0.8; control siRNA, 28.99 ± 1.77; lipofectamine, 31.12 ± 1.46. These results were obtained in 1 representative experiment of 4 independent experiments.
hnRNP L siRNA knockdown. (A) B16F10 cells were transfected for 24 hours with (i) siRNA control or (ii-v) 5 nM, 10 nM, 20 nM, or 50 nM, respectively, of HNRNP L siRNA. Nuclear protein was extracted and separated by SDS-PAGE (30 μg per lane). The presence of hnRNP L was detected by Western blot with anti-hnRNP L. (B) B16F10 cells were transfected with 50 nM hnRNP L siRNA (left) or control siRNA (right) for 24 hours. The cells were then transfected with 2 μg pcDNA3.1 (–) C6 or C21 exactly as described in panel B. The amount of spliced mRNA (open arrowheads) was calculated using image J38 and expressed as the percentage of total mRNA (nonspliced plus spliced mRNA). (C) The percentage of spliced mRNA obtained following transfection with hnRNP L siRNA or control siRNA. Following treatment with hnRNP L siRNA, the levels obtained for CA21 (○) or CA6 (⋄) are dramatically reduced. After treatment with the siRNA control, the levels obtained for CA21 (•) or CA6 (♦) are not different from that observed in the absence of siRNA. Each data entry represents the mean of triplicate measurements in a single experiment that is representative of 3 independent experiments. (D) Effect of hnRNP L siRNA on B16F10 surface expression of endogenous α2β1. B16F10 cells were transfected twice with 50 nM HNRNP L siRNA (gray curve), 50 nM control siRNA (solid black line), or Lipofectamine 2000 alone (dotted line) over a 72-hour period. Twenty-four hours after the second transfection, flow cytometry was used to quantitate surface α2β1. The results, reported as the GMFI ± SD, are as follows: hnRNP L siRNA, 19.28 ± 0.8; control siRNA, 28.99 ± 1.77; lipofectamine, 31.12 ± 1.46. These results were obtained in 1 representative experiment of 4 independent experiments.
Prepublished online as Blood First Edition Paper, February 2, 2006; DOI 10.1182/blood-2005-12-4822.
Supported by the National Heart, Lung, and Blood Institute (grant R01 HL-075821) (T.J.K.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Drs Daniel Salomon and Jerry Ware (TSRI) for their advice and assistance in selected mouse experiments. We thank Dr Brunhilde Felding-Habermann (TSRI) for providing us with the B16F10 cell line.
This manuscript is no. 17437-MEM from The Scripps Research Institute.

![Figure 1. Murine platelet Itga2: surface density and function. (A) Murine platelet glycoprotein levels were determined by flow cytometry. The surface content of platelet glycoproteins αIIbβ3, GPIbα, GPVI, α5β1, and α2β1 was determined in citrated whole blood using rat antimouse monoclonal antibodies. The level of bound FITC- or PE-conjugated rat antibody was expressed as the geometric mean fluorescence intensity and is indicated on the ordinate. Each bar represents the mean ± SD of 6 separate measurements using whole blood pooled from 5 to 6 male mice in each measurement. The results from 5 representative inbred strains are depicted. Three strains bear haplotype 1 (BALB/cJ, C57BL/6J, and 129/SvJ), whereas 2 strains express haplotype 2 (FVB/NJ and A/J). In the bottom right-hand panel, the levels of α2β1 were normalized with respect to the levels of α5β1 where [α2β1(n) = GMFI (α2β1)/GMFI (α5β) × 100]. (B) Platelet adhesion under high shear measured by the PFA-100. Citrated whole blood was pooled from 5 to 6 male mice of each of the 5 representative strains (see panel A) on 3 independent occasions. The CT in seconds was recorded and is indicated on the ordinate. The left-hand side depicts the results when blood was perfused over cartridge membranes impregnated with collagen plus ADP (CT-CADP); for the right-hand side, the findings shown when blood was perfused over cartridge membranes impregnated with collagen plus epinephrine (CT-CEPI). Each entry represents the mean ± SD.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/107/11/10.1182_blood-2005-12-4822/4/m_zh80110696190001.jpeg?Expires=1765911961&Signature=FQYGyiFRZiczyABTZ62n46ifo4v1CRgNnlWif3xGTa~jBWYxk7IBAPMinFoQC4oeYxEtopKBnOgBSydXusoR6hEXH1XlEA~eYuoCy-dAT-5HWNGnkB6C8sr-Z-HU2jc4Ms4xidrjMQH365v120i~0PCXbWJiQBoxiGtgK8Q019db1fyCRF5aJKYJRxCLpPbd92Ns9Y0rNAMsxAMAROyUlh20ptiqikc42eCVaDdes5UzGJ0mrOLS7DxB2vZfytnRsoMyWVAgwk3GCBAAmWvjDD~og-bEDkgp40Kpr3TuZCWmn~CtyEglAk48lpJdVZFuPLel~lkYYT9kGXyfd5g76A__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal