Abstract
Molecular mechanisms for the developmental stage and tissue-specific regulation of the erythropoietin (EPO) gene are poorly understood. Recent findings indicate a role of the Wilms tumor suppressor, Wt1, in the formation of the hematopoietic system. Herein, we tested the hypothesis that Wt1 is a transcriptional regulator of the EPO gene. Binding of the transcriptionally competent Wt1(–KTS) isoform to the minimal EPO promoter was demonstrated by electrophoretic mobility shift assay and chromatin immunoprecipitation. Under normoxia, EPO expression was significantly increased in HEK 293 and HepG2 cells with forced expression of Wt1(–KTS). A reporter construct harboring the 117-bp minimal human EPO promoter was activated up to 20-fold by transient cotransfection of Wt1(–KTS) in different cell lines. Mutation of the Wt1 binding site in the EPO promoter abrogated this stimulatory effect of the Wt1(–KTS) protein. Hepatic Epo mRNA expression was significantly reduced in embryonic mice with homozygous Wt1 deletion. Furthermore, Wt1 and EPO were colocalized in hepatocytes of the liver and in neuronal cells of the dorsal root ganglia in developing mice. Both proteins were also detected in Sertoli cells of the adult murine testis. In conclusion, we identified Wt1(–KTS) as a novel transcriptional activator for the tissue-specific expression of the EPO gene.
Introduction
Circulating erythropoietin (EPO), which is required for red blood cell production, is primarily produced in the fetal liver and adult kidney.1-3 In addition, EPO is also expressed in a variety of other tissues, including the nervous system and the reproductive organs (for reviews, see Masuda et al,4 Gassmann et al,5 and Jelkmann and Wagner6 ). Although a reduced PO2 level in the arterial and venous blood is the predominant stimulus for EPO mRNA expression, it has become increasingly clear that the EPO gene is also regulated in a developmental-stage and tissue-specific manner under both normoxia and hypoxia.4,7-9 Understanding of the molecular mechanisms for the tissue-specific and developmental expression of EPO is of particular interest because locally released EPO acts as a potent cell protective and trophic factor in various organs, such as the heart and the nervous system (for reviews, see Gassmann et al5 and Jelkmann and Wagner6 ).
Regulatory DNA sequences in the EPO gene have been initially identified with the use of transgenic mice. Hypoxia-inducible elements for expression in the liver and kidney are located within 0.7 kb 3′ and between 9.5 and 14 kb 5′ to the EPO gene, respectively. In addition, a specific negative regulatory liver element was identified between 2.2 and 7 kb 3′ to the gene (for a review, see Fandrey10 ). Detailed in vitro analysis of cis-acting elements allowed delineation of a 50-bp hypoxia-responsive enhancer, which is located downstream of the 3′ EPO polyadenylation signal and which plays a crucial role in EPO regulation by the local oxygen tension. EPO expression in response to hypoxia is mediated by proteins that interact with the binding site for hypoxia-inducible factor (HIF), a CACA element, and a direct repeat DR-2 site in the 3′ EPO enhancer, which allows binding of hepatic nuclear factor 4α (HNF-4α). Factors binding to the EPO enhancer are additionally stabilized by the coactivator CBP/p300 (for reviews, see Fandrey10 and Ebert and Bunn11 ).
The minimal EPO promoter, which encompasses a 117-bp fragment in the 5′ untranslated region (5′UTR) of the human EPO gene, acts synergistically with the enhancer to achieve up to 100-fold stimulation of EPO mRNAexpression in vitro.12,13 This effect seems to be driven by the transcription factors hypoxia-associated factor (HAF) and Sp1, which bind to a 17-bp sequence within the promoter and which operate in concert with transcription factors bound to the 3′ enhancer.14,15 In addition, the promoter is crucial for the tissue-restricted expression of the EPO gene. We have recently shown that GATA-4 functions as a tissue-specific transcriptional activator of EPO in fetal hepatocytes.9 After the developmental down-regulation of GATA-4 in fetal hepatocytes, binding of GATA-2 may become predominant and may inhibit hepatic EPO expression.16
Despite these recent advances, the molecular mechanisms for the developmental expression of the EPO gene remain incompletely understood. To obtain novel insights into the tissue-specific regulation of EPO, we sought to determine whether the EPO gene is controlled by the Wilms tumor transcription factor Wt1. This hypothesis is based on the following rationale: Defects in the hematopoietic system of Wt1-deficient mice suggest that Wt1 plays a role in regulating genes that have a significant impact on the function of hematopoietic stem cells and the proliferation and differentiation of progenitor cells, in particular of erythroid colony-forming units.17 During embryogenesis, Wt1 is expressed where hematopoietic stem cells are known to reside.18-20 Furthermore, detailed sequence analysis of the EPO promoter indicated several predicted binding motifs for Wt1 (GnGGGnGnG) that are highly conserved in mice and humans. Finally, Wt1, like EPO, is expressed in a spatially and temporally restricted manner during development.
Wt1 was originally identified as a tumor-suppressor gene because of its inactivation in a subset of pediatric renal carcinomas (nephroblastomas, Wilms tumors). Subsequent studies demonstrated that Wt1 plays an essential role in the formation of certain organs, including the genitourinary system,21,22 mesothelial tissues,21,23 the spleen,24 the retina,25 and the olfactory epithelium.26 Wt1 encodes a zinc finger protein of the Cys2His2 type, which can function as a transcription factor. Among the 4 alternative Wt1 splice variants, insertion of the lysine-threonine-serine (KTS) tripeptide into the zinc finger domain reduces the DNA binding affinity of Wt1. The Wt1(+KTS) forms presumably fulfill a role in RNA processing rather than in gene transcription. On the other hand, Wt1 proteins, which lack the KTS splice insertion (Wt1[–KTS]), bind to GC- and TC-rich DNA sequences and regulate a variety of target genes (for a review, see Lee and Haber27 ).
Herein, we describe Wt1 as a novel transcriptional activator of the EPO gene under normoxia. It is suggested that induction by Wt1 could be an important regulatory mechanism for the tissue-restricted expression of EPO.
Materials and methods
Cell culture
U2OS human osteosarcoma cells (ATCC HTB-96), human hepatocellular carcinoma Hep3B (ATCC HB-8064) and HepG2 cells (ATCC HB-8065), and Drosophila SL2 Schneider cells (ATCC CRL-1963) were obtained from the American Type Culture Collection and were grown as described.9,28,29 Generation and culture of human embryonic kidney (HEK) 293 cells with forced expression of Wt1 has been reported elsewhere.30
Electrophoretic mobility shift assay
Assays were performed with purified recombinant GST-Wt1 protein, as previously described.30 To analyze the in vitro binding of different Wt1 variants to the human EPO promoter, DNA binding reactions were carried out on ice for 15 minutes with 20 μg purified GST-Wt1(+KTS) or 20 μg GST-Wt1(–KTS) protein in 20 μL of a 1 × reaction buffer (10 mM Tris-HCl, pH 7.5, 50 mM KCl, 50 mM NaCl, 1 mM MgCl2, 1 mM EDTA, 5 mM DTT, 5% glycerol, 0.05 mg/mL herring sperm DNA). Double-stranded oligonucleotides, which are located within the proximal EPO promoter, were chosen on the basis of predicted Wt1 binding sites (Table 1). DNA fragments were end-labeled with 32P. Competition experiments were performed with 5- and 100-fold molar excess amounts of a 21-bp DNA sequence containing the previously identified Wt1(–KTS) binding site in the vitamin D receptor gene promoter (5′-TGAACTTAGTGGGCGTGGTTG-3′).30 Binding reactions were run for 3 hours on a nondenaturing 6% polyacrylamide gel at 4°C. The gels were dried, and complex formation was visualized by autoradiography.
Oligonucleotides used for EMSA experiments based on the presence of predicted Wt1 binding sites in the minimal human EPO promoter
Oligonucleotide . | Position of mutations within the oligonucleotides . | Single-strand sequence . |
|---|---|---|
| Oligo -116/-87 | — | 5′-TGCTCTGACCCCGGGTGGCCCCTACCCCTG-3′ |
| Oligo -86/-67 | — | 5′-GCGACCCCTCACGCACACAG-3′ |
| Oligo -66/37 | — | 5′-CCTCTCCCCCACCCCCACCCGCGCACGCAC-3′ |
| Oligo -66/-37 | ||
| Mutant A | -60 to -58 | 5′-CCTCTCAAACACCCCCACCCGCGCACGCAC-3′ |
| Mutant B | -54 to -52 | 5′-CCTCTCCCCCACTTTCACCCGCGCACGCAC-3′ |
| Mutant C | -60 to -58 and -54 to -52 | 5′-CCTCTCAAACACTTTCACCCGCGCACGCAC-3′ |
Oligonucleotide . | Position of mutations within the oligonucleotides . | Single-strand sequence . |
|---|---|---|
| Oligo -116/-87 | — | 5′-TGCTCTGACCCCGGGTGGCCCCTACCCCTG-3′ |
| Oligo -86/-67 | — | 5′-GCGACCCCTCACGCACACAG-3′ |
| Oligo -66/37 | — | 5′-CCTCTCCCCCACCCCCACCCGCGCACGCAC-3′ |
| Oligo -66/-37 | ||
| Mutant A | -60 to -58 | 5′-CCTCTCAAACACCCCCACCCGCGCACGCAC-3′ |
| Mutant B | -54 to -52 | 5′-CCTCTCCCCCACTTTCACCCGCGCACGCAC-3′ |
| Mutant C | -60 to -58 and -54 to -52 | 5′-CCTCTCAAACACTTTCACCCGCGCACGCAC-3′ |
Oligonucleotide numbers indicate the positions of the nucleotides relative to the transcription start site. Transcription start site is given here at nucleotide 387 of the EPO gene (NCBI accession number M11319.1), whereas Blanchard et al12 describe the transcription start site at nucleotide 386.59 The underlined letters in the single-stranded sequences indicate the nucleotide mutations.
Additional electrophoretic mobility shift assay (EMSA) experiments were performed to analyze the DNA binding of Wt1(–KTS) and Wt1(+KTS) proteins to the EPO promoter in the presence of the transcription factor Sp1. DNA binding reactions were carried out exactly as described with purified recombinant GST-Wt1 protein (in increasing concentrations from 100 ng to 1 μg) and recombinant Sp1 protein (in increasing concentrations from 100 ng to 1 μg; E6391; Promega, Mannheim, Germany).
Chromatin immunoprecipitation
Chromatin immunoprecipitation assay (ChIP) was performed in Hep3B cells cultured under normoxia, as described.9 For immunoprecipitation, antibodies (2 μg) against the following proteins were used: acetylated histone 3 (rabbit polyclonal) (06-599; Upstate, Lake Placid, NY), Wt1 (C-19; Santa Cruz Biotechnology, Santa Cruz, CA), and Sp1 (PEP-2; Santa Cruz Biotechnology). One aliquot (no antibody) served as negative control, and the input sample served as positive control. After immunoprecipitation, purified DNA was eluted in 50 μL UltraPure DNase, RNase free ddH2O (10 977-015; Gibco/Invitrogen, Carlsbad, CA). For amplification of purified DNA fragments by polymerase chain reaction (PCR), 0.5 μL input DNA (diluted in a total volume of 5 μL ddH2O), 5 μL of the “no antibody” sample, and 5 μL immunoprecipitated DNA were mixed with primers, PCR buffer, dNTPs, and GoTaq DNA Polymerase (Promega), as previously described. The following primer pairs were used: EPO 5′flanking region, 5′-ACTCAGCAACCCAGGCATCTCTGA-3′ (forward primer) and 5′CGACCCCTCACGCACACAGCCTCT-3′ (reverse primer); EPO 3′ enhancer region, 5′-CCAGGTCCGGGAAATGAGGGGTGG-3′ (forward primer) and 5′-TTGGCAGCTGCCTTACTGCGGTGA-3′ (reverse primer). All samples were processed in the same PCR reaction (32 cycles). The annealing temperature was 60°C. PCR products were electrophoresed on a 1.5% agarose gel.
Plasmids
The pGL2 reporter vector containing the 117-bp sequence of the proximal human EPO gene promoter or the 126-bp sequence of the EPO enhancer (nt 3575-3449; National Center for Biotechnology Information [NCBI] accession no. M11319.1), or both (p117e126), were kindly provided by Kerry L. Blanchard (Lilly Research Laboratories, Indianapolis, IN) and Joachim Fandrey (Department of Physiology, University of Duisburg-Essen, Germany).12 The Wt1 expression constructs (Wt1 cDNA +/–KTS in pCB6+) were the gift of Daniel A. Haber (Massachusetts General Hospital Cancer Center and Harvard Medical School, Charlestown, MA).31 The plasmid for the expression of human Sp1 in Schneider cells (pPacSp1) was kindly provided by Guntram Suske.32 A Wt1 expression construct for use in Schneider cells was generated by blunt-end ligation of the Wt1(–KTS) cDNA into the XhoI restriction site of plasmid pPac.
A PCR-based protocol was applied to introduce site-directed mutations into 2 identified Wt1-binding elements in the EPO promoter. For this purpose, the following PCR primers were used, with p117 serving as DNA template: 5′-TAAGAGCTCTGCTCTGACCCCGGGT-3′ (forward primer), 5′-ATTAAGCTTGCTCTGGCCGGGGGTCG-3′ (reverse primer), 5′-TCAAACACTTTCACCCGCGCACG-3′ (mutant forward primer), 5′-CGCGGGTGAAAGTGTTTGAGAGG-3′ (mutant reverse primer). (Underlining indiates nucleotide mutations.) The mutant EPO promoter sequence was ligated into the HindIII and SacI restriction sites of the pGL2 basic reporter vector (Promega). Correct identity of the amplified DNA was verified by dideoxy sequencing of both strands.
Cell transfections and reporter gene assays
Reporter constructs (0.5 μm each), together with 0.25 μg cytomegalovirus (CMV)–driven β-galactosidase plasmid and 1.25 μg expression constructs encoding Wt1(–KTS) or Wt1(+KTS), were transiently cotransfected into different cell lines. At the time of transfection, the cells had reached approximately 60% confluence in 60-mm dishes. Transfections were carried out using FuGENE 6 (Roche Diagnostics, Mannheim, Germany) transfection reagent according to the manufacturer's instructions at a 10:1 (FuGENE 6/DNA) volume ratio. Appropriate control experiments were performed by transfection of the “empty” pCB6+ expression and pGL2-basic reporter vectors. Transfected cells were kept in a humidified incubator either at normoxia (20% O2/5% CO2) or at hypoxia (1% O2/5% CO2). Cells were lysed 16 hours after transfection, and luciferase activities were measured in a luminometer (Microlite TLX1; MGM Instruments, Hamden, CT) with beetle luciferin as substrate. β-Galactosidase activities were determined spectrophotometrically using the Dual-Luciferase Reporter Assay System (Promega) according to the protocol provided. Values are given as relative light units, normalized to β-galactosidase activities for internal control of transfection efficiencies. Results shown are averages of 5 transfection experiments, each performed in duplicate. Drosophila SL2 cells were transfected with Effectene reagent (Qiagen, Hilden, Germany) according to the manufacturer's instructions using 5 μL transfection reagent and 8 μL enhancer per microgram DNA.
RT-PCR analysis
Total RNA was prepared from HEK293 cells that had been stably transfected with the Wt1(–KTS) expression construct, with the Wt1(+KTS) expression construct, or with the pCB6+ vector backbone using Trizol reagent (Invitrogen). The RNA pellet was dissolved in diethyl pyrocarbonate–treated H2O at a concentration of 1 μg/μL. First-strand cDNA synthesis was performed with 2 μg total RNA using oligo(dT) primers and superscript II reverse transcriptase (Invitrogen), as previously described.3 One tenth of the reaction product was taken for PCR amplification in a thermal cycler (GeneAmp PCR System 2400; Perkin Elmer, Wellesley, MA). PCR was carried out under the following conditions: DNA denaturation at 94°C, primer annealing at 58°C, extension of double-stranded DNA at 72°C (30 cycles, 30 seconds each phase). The cDNA was diluted with H2O at a 1:10 ratio for amplification of β-actin transcripts. Reverse transcription–-PCR (RT-PCR) was performed accordingly, with total RNA isolated from transiently transfected HepG2 cells and with total RNA from the livers of wild-type and Wt1-deficient mouse embryos. The following primer sets were used for PCR amplification: human EPO, 5′-TAGAATGTCCTGCCTGGCTGT-3′ (forward primer), 5′-CCTCCATCCTCTTCCAGGCAT-3′ (reverse primer); human β-actin, 5′-TTCTACAATGAGCTGCGTGTG-3′ (forward primer), 5′-CGTCACACTTCATGATGGAGT-3′ (reverse primer); mouse Wt1, 5′-ATCAGATGAACCTAGGAG-3′ (forward primer), 5′CTGGGTATGCACACATGA-3′ (reverse primer)26 ; mouse Epo, 5′CTGGGAGCTCAGAAGGAATTGATG-3′ (forward primer), 5′-CAGTCTGTCCCATGGACACTCCAG-3′ (reverse primer)9 ; mouse Gapdh, 5′-ACGACCCCTTCATTGACCTCA-3′ (forward primer), 5′-TTTGGCTCCACCCTTCAAGTG-3′ (reverse primer). Amplified DNA sequences were 494 bp (hEPO), 591 bp (hβ-actin), 269 bp (mWt1), 298 bp (mEpo), and 251 bp (mGAPDH) long, electrophoresed in a 1.5% agarose gel, and stained with ethidium bromide.
Quantitative RT-PCR analysis of EPO mRNA expression in the livers of wild-type and Wt1-deficient mouse embryos
Mouse breeding pairs (C57BL/6 strain) with heterozygous Wt1 mutation (Wt1+/–), originally obtained from the Jackson Laboratory (Bar Harbor, ME), were mated. The embryos were collected on embryonic day 12.5 (E12.5; morning of vaginal plug was considered E0.5) and genotyped by PCR analysis.22 Total RNA was prepared from isolated embryonic mouse livers with the Trizol reagent and reverse transcribed (1 μg/20 μL) using oligo(dT) primers and superscript II reverse transcriptase (Invitrogen). One twentieth of the reaction volume was used for PCR quantification with the GeneAmp 5700 Sequence Detection System (Applied Biosciences, Foster City, CA), as described previously.33 The following primer combinations were used for PCR amplification: mouse Epo, 5′-CATCTGCGACAGTCGAGTTCTG-3′ (forward primer), 5′-TGCACAACCCATCGTGACAT-3′ (reverse primer); mouse β-actin, 5′-AACTGCAGAGGACTCCTATGTGGGTGACG-3′ (forward primer), 5′-CGGGATCCGATGGCTACGTACATGGCTGG-3′ (reverse primer); mouse α-fetoprotein, 5′-AGCCATGAAGTGGATCACACC-3′ (forward primer), 5′-AACACCCATCGCCAGAGTTT-3′ (reverse primer); mouse albumin, 5′-ACACCATGCCTGCTGCTCTG-3′ (forward primer), 5′-AGGCTGAAATTCAGCAAGCAC-3′ (reverse primer). Epo, α-fetoprotein, and albumin transcript levels were compared in the livers of wild-type and Wt1–/– embryos on the basis of differences in the Ct values. Data were normalized against β-actin mRNA.33
Western blot analysis
Immunoblot analysis of Wt1, EPO, and β-actin in HEK293 and HepG2 cells that had been transfected with Wt1 expression constructs or with “empty” vector was performed as described elsewhere.9,28 The following primary antibodies were used for protein detection: polyclonal anti-Wt1 antibody from rabbit (C-19; Santa Cruz Biotechnology; 1:100 dilution in PBS, 5% Blotto, 0.05% Tween-20), polyclonal goat anti-EPO antibody (N-19; Santa Cruz Biotechnology; 1:100 dilution), goat polyclonal anti–β-actin antibody (C-11; Santa Cruz Biotechnology; 1:500 dilution).
Immunohistochemistry
For double-immunofluorescence analysis of Wt1 and EPO expression patterns, tissue from staged mouse embryos (C57Bl6) was harvested and fixed at 4°C in 4% paraformaldehyde for 1 hour (spinal cord, embryonic/fetal liver) to 3 hours (testis). Approval for the animal study was given by the Local Committee on Research Animal Care. After fixation, the tissue specimens were embedded in Tissue-Tek OCT medium (Sakura Finetek Europe, Zoeterwonde, The Netherlands). Ten micrometer–thick cryosections were cut and transferred onto superfrost glass slides (Langenbrinck, Teningen, Germany). Tissue sections were permeabilized and incubated in blocking solution (1 × PBS, 1% mouse serum, 0.1% Triton X-100) for 30 minutes at room temperature and rinsed with 1 × PBS. Incubation of the tissue sections was performed overnight at 4°C with the following primary antibodies (double labeling): rabbit anti-Wt1 polyclonal antibody (C-19; Santa Cruz Biotechnology) and goat anti-EPO polyclonal antibody (N-19; Santa Cruz Biotechnology), each diluted 1:100 in blocking solution. Negative controls were prepared for each sample by following the same staining procedure in the absence of primary antibodies. After one rinse in PBS and three 10-minute washes with blocking solution, the slides were incubated for 1 hour at room temperature with a donkey Cy3-conjugated antirabbit antibody (1:10 000 dilution in blocking solution; 711-166-152; Jackson ImmunoResearch Laboratories, West Grove, PA) and donkey Cy2-conjugated anti–goat IgG (1:500 dilution; 705-226-147; Jackson ImmunoResearch). After one short wash with PBS, three 10-minute washes with blocking solution, and a final wash with PBS, slides were mounted using medium containing DAPI to visualize the nuclei (H-1200; Vectashield Mounting Medium with DAPI, Vector Laboratories, Burlingame, CA). Proteins were detected in the tissue sections by the emission of fluorescence of the Cy2 (green) and Cy3 (red) signals. Microscopic analysis was performed under an epifluorescence microscope (AxioPlan 2 Imaging System; Carl Zeiss, Jena, Germany), using PL 10 × 25 oculars. For 100 × magnification (as indicated by 100-μm scale bars in images), Achroplan 10 × 0.25 numeric aperture (NA) Ph1 objective lenses were used; for 200 × magnification (as indicated by 50-μm scale bars in images), Plan-Apochromat 20 × 0.75 NA objective lenses; and for 630 × magnification (as indicated by 20-μm scale bars in images), Plan-Neofluar 63 × 1.25 NA oil objective lenses. Photographs were taken with a connected digital camera (AxioCAM MRc; Carl Zeiss) using AxioVision 4.2 software (Carl Zeiss).
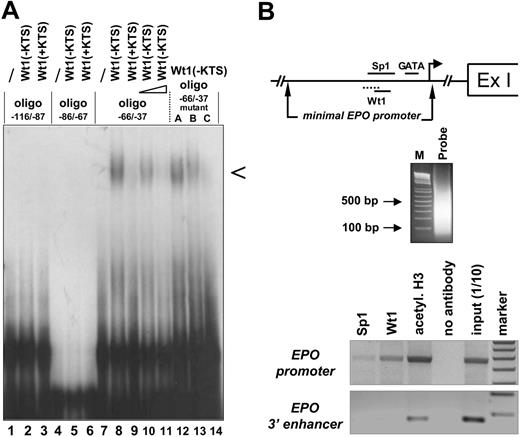
Binding of the Wt1(–KTS) isoform to the minimal human EPO promoter. (A) EMSA demonstrating binding of the Wt1(–KTS) protein to a DNA fragment (oligo –66/–37) of the minimal human EPO promoter. Arrowhead indicates the specific retardation band. The binding oligonucleotide (for detailed base pair sequence, see Table 1) harbors nt –66 to –37 relative to the transcription start site of the EPO gene. Note that the specific retardation signal (lane 8) can be competed with a 5-fold (lane 10) and a 100-fold (lane 11) molar excess of a 21-bp DNA sequence containing the previously identified Wt1(–KTS) binding site in the vitamin D receptor gene promoter. For comparison, Wt1(+KTS) protein binds to the same oligo –66/–37 with much lower affinity (lane 9). Introducing different mutations (mutants A, B, C; Table 1) into oligo –66/–37 reduces Wt1(–KTS) binding. Note that both Wt1 proteins (±KTS) failed to interact with 2 other GC-rich oligonucleotides (oligo –116/–87 and oligo –86/–67) of the human EPO promoter. EMSA experiments were performed with 20 μg recombinant Wt1 protein in each reaction. (B) Chromatin-immunoprecipitation analysis of protein interactions with the EPO gene. Note illustration of the potential binding sites of Sp1, GATA, and Wt1 transcription factors within the minimal EPO promoter, relative to the transcription start site (arrow) and the first exon (Ex I). Sizes of the DNAfragments obtained in Hep3B cells after sonification were revealed by electrophoresis in an ethidium bromide–stained 1.6% agarose gel. PCR-amplified products of the immunoprecipitates were electrophoresed in a 1.5% agarose gel and stained with ethidium bromide; for better visualization, the gel photograph is presented as a negative of the original. Apparently, under normoxia, Wt1 and Sp1 bind to the 5′ promoter but not to the hypoxia-responsive 3′ enhancer of the EPO gene.
Binding of the Wt1(–KTS) isoform to the minimal human EPO promoter. (A) EMSA demonstrating binding of the Wt1(–KTS) protein to a DNA fragment (oligo –66/–37) of the minimal human EPO promoter. Arrowhead indicates the specific retardation band. The binding oligonucleotide (for detailed base pair sequence, see Table 1) harbors nt –66 to –37 relative to the transcription start site of the EPO gene. Note that the specific retardation signal (lane 8) can be competed with a 5-fold (lane 10) and a 100-fold (lane 11) molar excess of a 21-bp DNA sequence containing the previously identified Wt1(–KTS) binding site in the vitamin D receptor gene promoter. For comparison, Wt1(+KTS) protein binds to the same oligo –66/–37 with much lower affinity (lane 9). Introducing different mutations (mutants A, B, C; Table 1) into oligo –66/–37 reduces Wt1(–KTS) binding. Note that both Wt1 proteins (±KTS) failed to interact with 2 other GC-rich oligonucleotides (oligo –116/–87 and oligo –86/–67) of the human EPO promoter. EMSA experiments were performed with 20 μg recombinant Wt1 protein in each reaction. (B) Chromatin-immunoprecipitation analysis of protein interactions with the EPO gene. Note illustration of the potential binding sites of Sp1, GATA, and Wt1 transcription factors within the minimal EPO promoter, relative to the transcription start site (arrow) and the first exon (Ex I). Sizes of the DNAfragments obtained in Hep3B cells after sonification were revealed by electrophoresis in an ethidium bromide–stained 1.6% agarose gel. PCR-amplified products of the immunoprecipitates were electrophoresed in a 1.5% agarose gel and stained with ethidium bromide; for better visualization, the gel photograph is presented as a negative of the original. Apparently, under normoxia, Wt1 and Sp1 bind to the 5′ promoter but not to the hypoxia-responsive 3′ enhancer of the EPO gene.
Statistical analysis
Analysis of variance (ANOVA) with Bonferroni test as post hoc test calculations and Student t test were performed as indicated to reveal statistical significances. P values less than .05 were considered statistically significant.
Results
Binding of Wt1 to the human EPO promoter
Sequence analysis revealed that the GC-rich minimal human EPO promoter contained several predicted Wt1 consensus elements. To examine whether Wt1 directly interacts with one of these potential binding sites in the EPO promoter, EMSA was performed with recombinant Wt1 protein (Figure 1A). The Wt1(–KTS) form, which functions as a transcription factor, bound with high affinity to the oligonucleotide extending from nt –66 to –37 relative to the transcription start site. In contrast, the Wt1(+KTS) protein, which has been implicated in mRNA processing,34,35 bound to the same DNA fragment with very low affinity. Within the DNA probe spanning oligonucleotides nt –66 to –37, 2 potential Wt1 consensus elements were analyzed in detail through the introduction of mutations to abrogate binding of the Wt1(–KTS) protein. As shown in Figure 1, triple base-pair mutations at positions nt –60 to –58 in the oligonucleotide (Table 1) did not significantly affect Wt1 binding, whereas mutations in positions nt –54 to –52 clearly reduced the binding affinity of Wt1(–KTS). Thus, Wt1(–KTS) has the highest binding affinity for the Wt1 consensus element located most 3′ in the minimal human EPO promoter. Binding of Wt1 to the human EPO promoter under in vivo conditions was confirmed by ChIP assay in Hep3B cells (Figure 1B). This assay also shows weak binding of Sp1 to the EPO promoter. The specificity of these results is confirmed by the lack of PCR signals if the same samples were amplified with primers detecting a DNA fragment of the EPO 3′ enhancer. Of note, these experiments were performed under normoxia, a condition in which we do not assume a physical interaction between EPO promoter and enhancer binding activities, as was postulated by Sanchez-Elsner et al15 for experiments performed under hypoxia.
Activation of the EPO promoter by Wt1(–KTS)
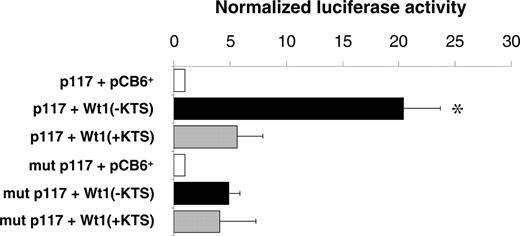
Reporter gene assays were performed in U2OS cells to examine whether binding of the Wt1(–KTS) protein could activate the human EPO promoter. A reporter gene construct containing the 117-bp sequence of the minimal promoter (pGL2p117)12 was transiently cotransfected with Wt1(–KTS), Wt1(+KTS), or empty expression plasmid (pCB6+), respectively. The Wt1(–KTS) variant significantly enhanced EPO promoter activity, which is indicated by the 20.4-fold (± 3.3-fold; P < .05) increase of normalized luciferase activity (Figure 2). For comparison, Wt1(+KTS) stimulated EPO promoter activity only 5.4-fold (± 2.3-fold; P = NS). Mutations in the pGL2p117 reporter construct at positions nt –60 to –58 and nt –54 to –52 relative to the transcription start site (corresponding to mutant C; Table 1) clearly reduced EPO promoter activity in the presence of Wt1(–KTS). The remaining slight increase in normalized luciferase activity suggests that additional relevant elements may exist in the minimal EPO promoter that could mediate, to a minor extent, the effect of Wt1. Significant 9- and 5-fold increases in reporter activity by Wt1(–KTS) was also seen in transiently transfected HEK293 and HepG2 cells, respectively (data not shown). Notably, cotransfection of hepatoma-derived Hep3B cells, an established model for studying oxygen-dependent EPO gene expression,36 with Wt1(–KTS) under hypoxia (1% O2, 16 hours) had no additional stimulatory effect on the activity of reporter constructs harboring the EPO promoter, the EPO 3′ enhancer, or both elements (data not shown).
Activation of the minimal human EPO promoter by Wt1(–KTS) in U2OS cells. Relative luciferase activities measured in lysates of human osteosarcoma U2OS cells. The cells were transiently transfected with a pGL2 reporter vector containing the minimal human EPO promoter either as wild-type (p117, top 3 boxes) or with a mutated (mutant C in Table 1) Wt1(–KTS) binding site (mut p117; bottom 3 boxes). Wt1 expression constructs encoding different splice variants—Wt1(–KTS) or Wt1(+KTS)—were cotransfected along with a cytomegalovirus promoter–driven β-galactosidase expression vector for normalization of transfection efficiencies. pCB6+ is the empty expression vector. Values are shown as mean ± SEM of 5 experiments, each performed in duplicate. *P < .05; ANOVA.
Activation of the minimal human EPO promoter by Wt1(–KTS) in U2OS cells. Relative luciferase activities measured in lysates of human osteosarcoma U2OS cells. The cells were transiently transfected with a pGL2 reporter vector containing the minimal human EPO promoter either as wild-type (p117, top 3 boxes) or with a mutated (mutant C in Table 1) Wt1(–KTS) binding site (mut p117; bottom 3 boxes). Wt1 expression constructs encoding different splice variants—Wt1(–KTS) or Wt1(+KTS)—were cotransfected along with a cytomegalovirus promoter–driven β-galactosidase expression vector for normalization of transfection efficiencies. pCB6+ is the empty expression vector. Values are shown as mean ± SEM of 5 experiments, each performed in duplicate. *P < .05; ANOVA.
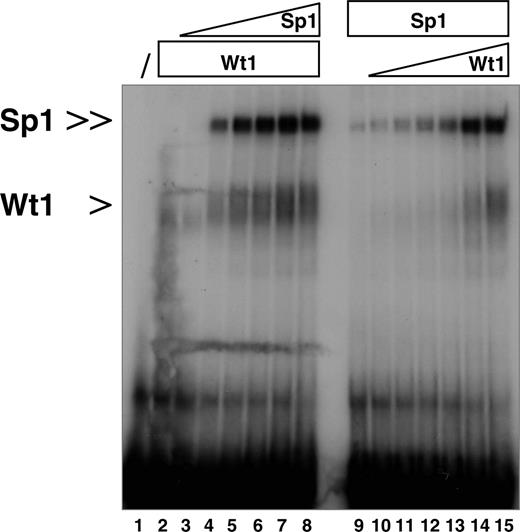
Mutually enhanced binding affinities of Sp1 and Wt1(–KTS) to the human EPO promoter. EMSA demonstrating that Wt1 and Sp1 enhance the binding of each protein to the identified EPO promoter element (oligonucelotide –66/–37). Lane 1, oligonucelotide alone. In lane 2, 1 μg recombinant Wt1(–KTS) isoform was added to the reaction. Note that increasing concentrations of recombinant Sp1 protein (100 ng, 200 ng, 300 ng, 400 ng, 500 ng, 1 μg; lanes 3-8, respectively) enhanced Wt1(–KTS) binding to the DNA. Conversely, Sp1/DNA complex formation was improved with increasing concentrations of Wt1(–KTS) protein (100 ng, 200 ng, 300 ng, 400 ng, 500 ng, 1 μg; lanes 10-15, respectively). Protein complexes were resolved on a 5% polyacrylamide gel, and complex formation was visualized after 48 hours by autoradiography.
Mutually enhanced binding affinities of Sp1 and Wt1(–KTS) to the human EPO promoter. EMSA demonstrating that Wt1 and Sp1 enhance the binding of each protein to the identified EPO promoter element (oligonucelotide –66/–37). Lane 1, oligonucelotide alone. In lane 2, 1 μg recombinant Wt1(–KTS) isoform was added to the reaction. Note that increasing concentrations of recombinant Sp1 protein (100 ng, 200 ng, 300 ng, 400 ng, 500 ng, 1 μg; lanes 3-8, respectively) enhanced Wt1(–KTS) binding to the DNA. Conversely, Sp1/DNA complex formation was improved with increasing concentrations of Wt1(–KTS) protein (100 ng, 200 ng, 300 ng, 400 ng, 500 ng, 1 μg; lanes 10-15, respectively). Protein complexes were resolved on a 5% polyacrylamide gel, and complex formation was visualized after 48 hours by autoradiography.
Increased binding affinity of Wt1(–KTS) to the EPO promoter in the presence of Sp1
Detailed sequence analysis revealed that Wt1(–KTS) binds exactly to that region in the minimal EPO promoter, which also interacts with the ubiquitous transcription factor Sp1 and the hypoxia-associated factor (HAF).14,15 To address whether the presence of Sp1 affects binding of Wt1(–KTS) to the oligonucleotide nt –66 to –37, EMSA experiments were performed with recombinant Wt1(–KTS) and Sp1 proteins. The 2 proteins were added either alone or at different concentration ratios. Increasing amounts of recombinant Sp1, from 100 ng to 1 μg, facilitated the binding of Wt1(–KTS), which was added at 1 μg throughout (Figure 3). Similarly, the addition of increasing amounts of Wt1(–KTS) protein enhanced Sp1 binding to the EPO promoter fragment. Unlike the Wt1(–KTS) protein, the addition of the Wt1(+KTS) variant did not change the binding affinity of Sp1 (data not shown). Thus, Sp1 and Wt1(–KTS) each increased the binding affinity of the other protein to the EPO promoter.
We also addressed whether Wt1(–KTS) and Sp1 would potentiate each other in stimulating EPO promoter activity. For this purpose, transient cotransfections using Wt1(–KTS) and Sp1 expression plasmids were performed in U2OS, HEK293, and Drosophila SL2 Schneider cells, which do not contain endogenous Sp1.37 The 2 expression constructs were used at a concentration that increased EPO promoter activity 2- to 3-fold when the plasmids were added separately. However, simultaneous cotransfection of the Sp1 and Wt1(–KTS) expression plasmids had no potentiating effect, either at normoxia (20% O2) or at hypoxia (1% O2), on the activity of different EPO promoter reporter constructs with or without the EPO 3′ enhancer (data not shown).
Induction of EPO mRNA and protein by Wt1(–KTS) in vitro
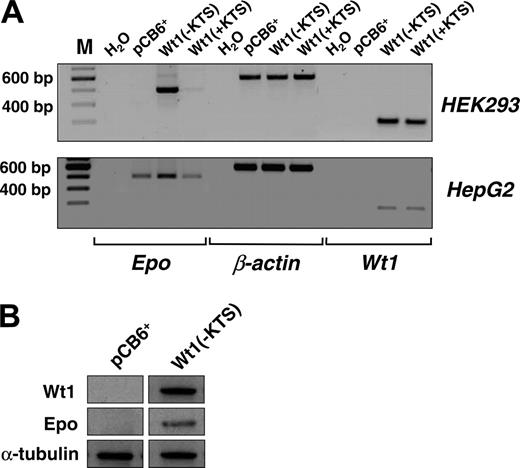
Next we examined the effect of Wt1(–KTS) on the expression of the endogenous EPO gene. For this purpose, EPO transcripts and protein were analyzed in our previously established human embryonic kidney (HEK) 293 cells that were stably transfected with a Wt1(–KTS) expression construct, a Wt1(+KTS) expression plasmid, or an empty pCB6+ vector.30 Although EPO transcripts were not detected in wild-type HEK293 cells, stable overexpression of Wt1(–KTS) resulted in a significant induction of EPO mRNA expression (Figure 4A). Western blot analysis confirmed the translation of EPO mRNA into protein (Figure 4B). Similarly, transient transfection with the Wt1(–KTS) construct, but not with the Wt1(+KTS) variant, increased EPO transcripts in human hepatoma-derived HepG2 cells (Figure 4A). HepG2 cells were chosen for these experiments because of their higher transfection efficiency compared with Hep3B cells. These data indicate that Wt1(–KTS) is a potent activator of EPO gene expression under normoxia.
Lack of Wt1 causes significant reduction of Epo gene expression in the fetal liver
As a prerequisite for transcriptional regulation of the EPO gene by Wt1 in vivo, one would expect a similar cellular distribution of both proteins in different tissues. Immunohistologic analysis indicates that EPO and Wt1 are indeed coexpressed in hepatocytes of the murine embryonic liver at the stage of primitive hematopoiesis (Figure 5A). Consistent with the nested distribution of hematopoietic progenitor cells in the developing liver, colocalization of both proteins was pronounced in hepatocytes at the periphery of the liver at E10.5 of gestation (Figure 5A). To find out whether Wt1 is necessary for normal Epo gene expression in vivo, we quantified Epo transcripts by real-time RT-PCR in the livers of wild-type and Wt1-deficient (Wt1–/–) mouse embryos (E12.5) of the same littermate. At this stage of gestation, circulating EPO is primarily produced in the embryonic/fetal liver of mice.9,38 As shown by standard RT-PCR technique and real-time amplification, Epo mRNA expression was significantly lower (P < .01; Student t test) in the livers of Wt1–/– compared with wild-type embryos. For comparison, α-fetoprotein and albumin transcripts were similar (P > .05) in the livers of Wt1–/– and wild-type embryos (Figure 5B-C). These data indicate that Wt1 is critical for normal hepatic Epo gene expression during development.
Induction of EPO expression by Wt1(–KTS) in human embryonic kidney (HEK293) and hepatoma-derived HepG2 cells. (A, top row) Transcripts of EPO, Wt1, and β-actin were detected by reverse transcription PCR (amplification with 32 cycles) in human embryonic (HEK293) kidney cells stably transfected with Wt1(–KTS), Wt1(+KTS), or empty expression vector (pCB6+). (A, bottom row) Expression of EPO, Wt1, and β-actin in transiently transfected HepG2 cells. cDNA was diluted 1:10 with H2O for amplification of β-actin. RT-PCR products were run on 1.5% agarose gels and stained with ethidium bromide; for better visualization, gel photographs are presented as negatives of the originals. (B) Induction of EPO expression by Wt1(–KTS) was confirmed by immunoblotting of EPO protein in whole cell extracts of HEK293 cells. Wt1 immunoblotting reveals significant Wt1(–KTS) expression. α-Tubulin was detected as internal control of equal amounts of protein.
Induction of EPO expression by Wt1(–KTS) in human embryonic kidney (HEK293) and hepatoma-derived HepG2 cells. (A, top row) Transcripts of EPO, Wt1, and β-actin were detected by reverse transcription PCR (amplification with 32 cycles) in human embryonic (HEK293) kidney cells stably transfected with Wt1(–KTS), Wt1(+KTS), or empty expression vector (pCB6+). (A, bottom row) Expression of EPO, Wt1, and β-actin in transiently transfected HepG2 cells. cDNA was diluted 1:10 with H2O for amplification of β-actin. RT-PCR products were run on 1.5% agarose gels and stained with ethidium bromide; for better visualization, gel photographs are presented as negatives of the originals. (B) Induction of EPO expression by Wt1(–KTS) was confirmed by immunoblotting of EPO protein in whole cell extracts of HEK293 cells. Wt1 immunoblotting reveals significant Wt1(–KTS) expression. α-Tubulin was detected as internal control of equal amounts of protein.
Epo expression analysis in wild-type and Wt1-deficient mouse embryos. (A) Double-immunofluorescence labeling for Wt1 (red) and EPO (green) in sections of the murine embryonic liver (E10.5). Wt1 is expressed predominantly in hepatocytes at the periphery of the liver because the typical lobular architecture at this early developmental stage is not yet established (Ai; higher magnification, Avii; negative control, Aii). Nuclear localization of Wt1 is confirmed by counterstaining with DAPI (Aviii,Aix). Hematoxylin-eosin staining reveals nests of hematopoietic progenitor cells in the developing liver (Av, arrow). Labeling of Epo is shown in Aiii, Avi, and Avii at higher magnification (Aiv, negative control). Note the colocalization of Wt1 and EPO in hepatocytes at the periphery of the liver (Avi, Avii, Aix). (B) Representative RT-PCR results for Epo mRNA expression in the liver of a mouse embryo at E12.0 with homozygous Wt1 deletion (Wt1-/-) in comparison with a wild-type littermate (Wt1+/+). Note the reduced Epo mRNA level in the liver of the Wt1-deficient embryo. Gapdh signal revealed a similar efficiency of RNA preparation and cDNA synthesis in both livers. For better visualization, the gel photograph is presented as a negative of the original. M = 100 bp DNA ladder as a marker for the size of PCR products. (C) Real-time RT-PCR analysis of Epo expression in the livers of 9 murine embryos (E12.0) lacking Wt1 (Wt1–/– ▪) compared with 9 wild-type littermates (Wt1+/+ □). EPO expression, which was defined as 100% in wild-type embryos, was normalized to β-actin transcripts. For comparison, albumin and α-fetoprotein (Afp) mRNA levels were not significantly different (P > .05) in the liver specimens of wild-type and Wt1-deficient embryos (n = 5 each). Values are mean ± SEM. *Statistical significance (P < .01; Student t test).
Epo expression analysis in wild-type and Wt1-deficient mouse embryos. (A) Double-immunofluorescence labeling for Wt1 (red) and EPO (green) in sections of the murine embryonic liver (E10.5). Wt1 is expressed predominantly in hepatocytes at the periphery of the liver because the typical lobular architecture at this early developmental stage is not yet established (Ai; higher magnification, Avii; negative control, Aii). Nuclear localization of Wt1 is confirmed by counterstaining with DAPI (Aviii,Aix). Hematoxylin-eosin staining reveals nests of hematopoietic progenitor cells in the developing liver (Av, arrow). Labeling of Epo is shown in Aiii, Avi, and Avii at higher magnification (Aiv, negative control). Note the colocalization of Wt1 and EPO in hepatocytes at the periphery of the liver (Avi, Avii, Aix). (B) Representative RT-PCR results for Epo mRNA expression in the liver of a mouse embryo at E12.0 with homozygous Wt1 deletion (Wt1-/-) in comparison with a wild-type littermate (Wt1+/+). Note the reduced Epo mRNA level in the liver of the Wt1-deficient embryo. Gapdh signal revealed a similar efficiency of RNA preparation and cDNA synthesis in both livers. For better visualization, the gel photograph is presented as a negative of the original. M = 100 bp DNA ladder as a marker for the size of PCR products. (C) Real-time RT-PCR analysis of Epo expression in the livers of 9 murine embryos (E12.0) lacking Wt1 (Wt1–/– ▪) compared with 9 wild-type littermates (Wt1+/+ □). EPO expression, which was defined as 100% in wild-type embryos, was normalized to β-actin transcripts. For comparison, albumin and α-fetoprotein (Afp) mRNA levels were not significantly different (P > .05) in the liver specimens of wild-type and Wt1-deficient embryos (n = 5 each). Values are mean ± SEM. *Statistical significance (P < .01; Student t test).
Coexpression of Wt1 and EPO in other organs
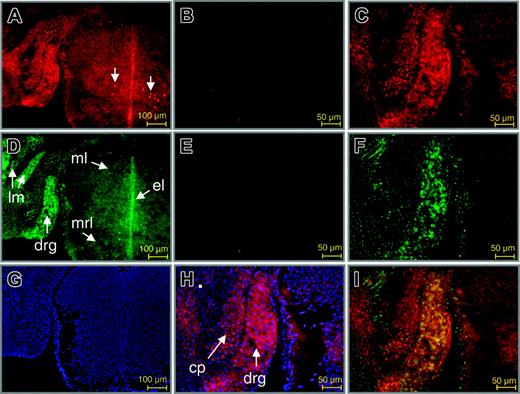
To further establish a role for Wt1 in the control of EPO gene expression in vivo, double-immunofluorescence labeling of Wt1 and EPO proteins was performed at different stages in developing and adult mice. Strikingly, an overlapping expression of both proteins was detected in neurons of the dorsal root ganglion (DRG; Figure 6). Thus, as reported by others earlier,39,40 nuclear staining of Wt1 was found in neurons of the DRG (Figure 6A, C, H), and the ventral mantle layer of the spinal cord. EPO was detected in the cytoplasm of neurons of the DRG and the spinal cord, particularly in the mantle, ependymal, and marginal layers (Figure 6D, F). Furthermore, EPO was found in myocytes of the long muscles of the neck. EPO protein has been previously detected in the DRG of adult rat. In that study, a different pattern of EPO and EPO receptor (EPO-R) staining indicated that EPO expression is bona fide and not simply caused by ligand binding in the DRG.41 As shown by merging the Cy3 and Cy2 signals for labeling of antibodies directed against Wt1 and EPO protein, respectively, colocalization of both proteins was restricted to neuronal cells of the DRG (Figure 6I).
Colocalization of Wt1 and EPO proteins in the developing murine dorsal root ganglion. Double-immunofluorescence labeling of Wt1 (red) and EPO (green) in sections of the murine spinal cord and dorsal root ganglion. (A,C) Staining of Wt1. (B) Negative control. Wt1 is contained in the nuclei of neuronal cells in the dorsal root ganglion and in a subtype of neurons in the mantle layer of the spinal cord at E14.0 of gestation (A, arrows). Nuclear localization of Wt1 is confirmed by counterstaining with DAPI (H). (D,F) Staining of EPO. (E) Negative control. Note the colocalization of Wt1 and EPO in neuronal cells of the developing dorsal root ganglion (drg), which becomes evident by the intense yellow fluorescence upon merging the red and green signals (I). Cy2 fluorescence in the primordial cartilage of the neural cord represents unspecific staining (F). Counterstaining with DAPI is shown (G). ml indicates mantle layer; el, ependymal layer; mrl, marginal layer; lm, long muscles of the neck; cp, cartilage primordium of neural cord.
Colocalization of Wt1 and EPO proteins in the developing murine dorsal root ganglion. Double-immunofluorescence labeling of Wt1 (red) and EPO (green) in sections of the murine spinal cord and dorsal root ganglion. (A,C) Staining of Wt1. (B) Negative control. Wt1 is contained in the nuclei of neuronal cells in the dorsal root ganglion and in a subtype of neurons in the mantle layer of the spinal cord at E14.0 of gestation (A, arrows). Nuclear localization of Wt1 is confirmed by counterstaining with DAPI (H). (D,F) Staining of EPO. (E) Negative control. Note the colocalization of Wt1 and EPO in neuronal cells of the developing dorsal root ganglion (drg), which becomes evident by the intense yellow fluorescence upon merging the red and green signals (I). Cy2 fluorescence in the primordial cartilage of the neural cord represents unspecific staining (F). Counterstaining with DAPI is shown (G). ml indicates mantle layer; el, ependymal layer; mrl, marginal layer; lm, long muscles of the neck; cp, cartilage primordium of neural cord.
In adult mice, double-immunofluorescence labeling of Wt1 and EPO proteins was seen in Sertoli cells of the testis (Figure 7I). Consistent with previous reports,42 Sertoli cells were the only Wt1-positive cell type in the adult testis (Figure 7B-C,H-I). In contrast, prominent expression of EPO was detected also in peritubular myoid cells, Leydig cells, and spermatogonia in addition to Sertoli cells (Figure 7E-F). Epo mRNA expression has been reported in Sertoli cells and peritubular myoid cells in the testis of adult rat but excluded in Leydig cells and spermatogonia.43 Because functional EPO-R is expressed on non-EPO–producing cells, in particular on Leydig cells,44 staining of these cell types may result from EPO bound to EPO-R.
Discussion
EPO gene expression is primarily regulated at the transcriptional level by cis-acting elements located in the 5′ promoter and the 3′ enhancer.10,11 In this study, we describe a novel mechanism for the regulation of EPO expression by the Wilms tumor protein Wt1. This is supported by the fact that the Wt1(–KTS) isoform, which functions as a transcription factor,34,35 induces EPO promoter activity by specific binding to a GC-rich domain (nt –63 to –52 relative to the transcription start site; Figures 1, 2), which is highly conserved in humans and mice (NCBI accession nos. M11319 and M26652). Importantly, forced expression of Wt1(–KTS), but not of the Wt1(+KTS) form, results in a significant increase of EPO gene expression in human embryonic kidney (HEK293) and HepG2 cells (Figure 4). The in vivo relevance of Wt1 in regulating EPO is confirmed by the reduced Epo expression in the liver of murine Wt1–/– embryos compared with their wild-type littermates (Figure 5). These findings indicate that, in addition to GATA-4, which has been previously identified as a basal activator of EPO in fetal hepatocytes,9 Wt1 is necessary for normal EPO production in the fetal liver in vivo. It appears unlikely, however, that Wt1 regulates EPO expression in the kidney, which is the major site of EPO formation in adults, because Wt1 is absent in peritubular/interstitial fibroblasts, which have been identified by in situ hybridization as the EPO-producing cells.11,45-47 Instead, Wt1 expression is restricted to the glomerular podocytes in the adult kidney.21,23,48 Our findings raise the possibility that Wt1, by promoting EPO expression in the fetal liver, could be a target for the developmental switch of the EPO production site from the liver to the kidneys. The role of Wt1 in regulating hematopoiesis is still controversial,17,18 but evidence indicates that Wt1 is important in the lineage-specific differentiation of hematopoietic cells and leukemogenesis.49-51 Because Wt1 does not affect the up-regulation of EPO in response to hypoxia, or at least it did not in our cotransfection experiments, a lower hepatic Epo expression, as shown on E12.5 of gestation (Figure 4), is not primarily responsible for later embryonic lethality of Wt1–/– mice. Indeed, the hematopoietic system appears to be grossly normal in Wt1–/– mice at the time of death (E13.5 in the C57/B16 Wt1–/– strain).17 However, deficits in hematopoietic stem cell function, as indicated by the reduced number and proliferation of Wt1-deficient, early lineage–specific hematopoietic progenitors,17 may be caused by impaired expression of hematopoietic receptors or other factors than by changes in EPO expression in the local hematopoietic environment, such as the fetal liver.
Colocalization of Wt1 and EPO in the adult murine testis. Double-immunofluorescence labeling for Wt1 (red) and EPO (green) in sections of the adult testis. (B-C) Staining of Wt1. (A) Negative control. Wt1 is present only in the nuclei of Sertoli cells. (E-F) Staining of EPO. (D) Negative control. EPO is detected in various cell types of the testis, but colocalization with Wt1 is restricted to Sertoli cells (yellow fluorescence, I). Counter-staining with DAPI is also shown in slides used for negative controls. SeC indicates Sertoli cell; LeyC, Leydig cell; SpeG, spermatogonia; SP, sperm.
Colocalization of Wt1 and EPO in the adult murine testis. Double-immunofluorescence labeling for Wt1 (red) and EPO (green) in sections of the adult testis. (B-C) Staining of Wt1. (A) Negative control. Wt1 is present only in the nuclei of Sertoli cells. (E-F) Staining of EPO. (D) Negative control. EPO is detected in various cell types of the testis, but colocalization with Wt1 is restricted to Sertoli cells (yellow fluorescence, I). Counter-staining with DAPI is also shown in slides used for negative controls. SeC indicates Sertoli cell; LeyC, Leydig cell; SpeG, spermatogonia; SP, sperm.
Wt1(–KTS) binds to a highly conserved GC-rich fragment that overlaps with previously reported binding sites of HAF (nt –61 to –45) and Sp1 (nt –59 to –47) in the EPO promoter.14,15 Because HAF and Sp1 act synergistically with factors binding to the 3′ enhancer to increase the hypoxia-responsiveness of EPO expression, we addressed whether Wt1(–KTS) also facilitates such an effect under hypoxia. Although Wt1 itself is up-regulated under hypoxia in certain tissues and cell types,33 Wt1(–KTS) does not potentiate the activity of the EPO promoter or enhancer in reporter gene assays under hypoxia. Thus, the combined data indicate that Wt1(–KTS) is an activator of basal EPO expression under normoxia rather than a direct mediator of the effect of hypoxia. However, we cannot exclude the possibility that promoter activation by Wt1 is necessary for enhanced EPO gene expression in response to hypoxia.
The question becomes whether binding of Wt1(–KTS) to the 17-bp fragment (nt –61 to –45 relative to the transcription start site) competes or inhibits the interaction of other factors, particularly Sp1, with the same fragment.15 This is relevant because Sp1 is critically involved in the cooperative effect of TGF-β and hypoxia on EPO transcription.15 This regulatory mechanism is most likely associated with a conformational change in the EPO gene locus mediated by Smad3, which may act as a bridge between HNF-4α, bound to the 3′ enhancer, and Sp1, bound to the 5′ promoter.15 However, the binding affinity of Sp1 alone to the human EPO promoter is relatively weak, as shown by us (using EMSA and chromatin-immunoprecipitation assay) and by others.15 Interestingly, recombinant Sp1 and Wt1(–KTS) mutually increase their binding affinities to the EPO promoter (Figure 3). Although the increased binding affinity is not encompassed with a significant potentiation of the EPO promoter activity, Wt1(–KTS), acting synergistically with Sp1, may be important to achieve and to stabilize a conformational change in the EPO locus.15
Besides its role in hematopoiesis, EPO is required for normal organ development, particularly of the heart and the central nervous system (CNS).52 Because EPO has significant cell-protective effects in various tissues after ischemic or hypoxic injury, detailed analysis of the developmental stage and tissue-specific regulation of EPO under normoxia and hypoxia is of particular interest.5,6,10 Therefore, we investigated the distribution of Wt1 and EPO in various organs during murine embryonic development. In addition to hepatocytes, Wt1 and EPO were coexpressed in neuronal cells of the DRG (Figure 6). This finding is consistent with a previous study describing EPO in the cytoplasm of neuronal cells of the DRG in rat.42 Wt1 immunoreactivity has previously been observed in the CNS, including the eye and the olfactory system, and most recent work indicates that Wt1 is involved in neurogenesis.25,26,39,40,42,53,54 The presence of Wt1 in sensory neurons of the DRG suggests a further, yet unknown, role of Wt1 in neuron formation in the peripheral nervous system. The biologic mechanism, which is activated on Wt1-dependent Epo expression in the DRG, remains to be elucidated. Considering the prevention of sensory axonal degeneration by EPO in the DRG,55 it appears plausible that EPO and Wt1 act as trophic factors through limiting apoptosis.25,26,56
Wt1 and EPO were also colocalized in Sertoli cells of the adult testis (Figure 7). Wt1 is required for proper development of the somatic cell lineages and the germ cell lineages of the testis.57 After development, Sertoli cells are the only cell population that maintains Wt1 expression in the adult testis.20,39,58 Data on the biology of EPO in the testis are limited. EPO mRNA expression has been previously reported in Sertoli cells and peritubular myoid cells in the testis of adult rat.43 In our study, EPO protein was detected in most cells in the adult testis. Given that functional EPO-R is expressed on non–EPO-producing cells, in particular in the plasma membrane of Leydig cells,44 receptor-bound EPO might have been detected in these cells by immunohistochemical staining. The biologic significance of EPO production in Sertoli cells still awaits elucidation. Male mice with heterozygous deficiency of Wt1 or EPO show reduced fertility (C.D. and H.S, unpublished observations, spring 2001 and summer 2005). Thus, the activation of EPO expression by Wt1 could be relevant for normal spermiogenesis, such as through paracrine effects of EPO released from Sertoli cells. Interestingly, adult Sertoli cells also express GATA-4,57 the other known tissue-specific activator of EPO expression,9 suggesting that basal EPO synthesis in these cells may have a specific long-term implication for non–self-renewing Sertoli cells, perhaps by inhibiting apoptosis.
In summary, our data demonstrate that the Wilms tumor transcription factor Wt1(–KTS) is a potent activator of the EPO gene under normoxia. Furthermore, Wt1 is required for normal Epo expression in the fetal liver in vivo. Both proteins are colocalized in sensory neurons of the dorsal root ganglion and Sertoli cells of the testis, suggesting that Wt1 may regulate paracrine EPO synthesis in a tissue-specific manner. This study extends our previous work9,16 by providing additional evidence that the tissue-specific regulation of EPO is mediated by cis-acting elements in the minimal EPO promoter.
Prepublished online as Blood First Edition Paper, February 7, 2006; DOI 10.1182/blood-2005-07-2889.
Supported by the Deutsche Forschungsgemeinschaft (grants Da 484/2-1 and Scho 634/5-1) and the Bundesministerium für Bildung und Forschung (grants NGFN, KGCV1, and 01GS0416). Supported also by the Sonnenfeld Stiftung (Berlin), the Sanitätsrat Emil Alexander Hübner und Frau Gemahlin Stiftung (Essen), and the Verein für das Frühgeborene Kind e.V. (Berlin) for the microscope and equipment used for immunohistochemical analysis and images.
C.D. and H.S. designed the research. Each author performed at least one experiment. C.D. wrote the manuscript. H.S. performed statistical analysis. Each author contributed to the interpretation and discussion of the results.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank I. Grätsch and A. Richter for their expert technical assistance. We thank Guntram Suske for providing the expression plasmid pPacSp1. We also thank Iwona Palaszewski, Joachim Fandrey, and Jörg Bungert for critical reading and discussion of the manuscript.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal