Abstract
l-asparaginase (l-Asp) is an effective drug for treatment of children with acute lymphoblastic leukemia (ALL). The effectiveness is generally thought to result from a rapid depletion of asparagine in serum and cells. Asparagine synthetase (AS) opposes the action of l-Asp by resynthesis of asparagine. In vitro, resistance to l-Asp has been associated with up-regulation of AS mRNA expression. We monitored AS mRNA levels in leukemic cells before and during 5 days after intravenous administration of 1000 IU/m2 pegylated l-asparaginase (PEG-Asp) in a therapeutic window in children with ALL at initial diagnosis. Within 24 hours, AS mRNA levels increased by 3.5-fold and remained stable in the following 4 days. Baseline and l-Asp–induced expression levels of AS did not differ between clinically good, intermediate, and poor responders to PEG-Asp. No significant difference of AS mRNA up-regulation was found between precursor B- and T-ALL or between hyperdiploids, TEL/AML1 rearranged ALL or absence of genetic abnormalities. In 3 of 12 patients with T-ALL even a slight down-regulation of AS mRNA expression upon l-Asp exposure was found. In conclusion, although l-Asp exposure induces the expression of AS mRNA, the up-regulated gene expression does not correlate with an early clinical poor response to this drug in children with ALL.
Introduction
l-asparaginase (l-Asp) is an effective drug for the treatment of children with acute lymphoblastic leukemia (ALL).1,2 In newly diagnosed patients with ALL, 25% to 60% will reach a complete remission after monotherapy with l-Asp.1 The efficacy of this drug is generally thought to result from a rapid and complete depletion of asparagine in plasma by hydrolyzing this amino acid to aspartic acid.
l-Asp resistance has been attributed to high levels of intracellular asparagine synthetase (AS).3 Cell line studies showed that l-Asp–sensitive leukemic cells have low intracellular AS activity and are dependent on the availability of extracellular asparagine.4 Andrulis et al demonstrate that complete asparagine depletion in vitro results in an amino acid–dependent up-regulation of mRNA, protein, and activity of AS.5 Resistance to l-Asp in cell lines is in vitro–mediated by an up-regulation of AS expression in response to asparagine depletion of culture medium.6,7 Whereas these cell line studies suggest that up-regulation of AS expression is an important mechanism of l-Asp resistance, clinical evidence is lacking for this assumption. In recent studies we found evidence that a high baseline intracellular AS gene expression is related to in vitro l-Asp resistance in children with TEL/AML1–negative ALL,8 but not in TEL/AML1–positive children.9 This suggests that the genotype plays an important role in the cause of l-Asp resistance. However, it is yet unknown whether baseline and/or l-Asp–induced AS mRNA levels are linked to the clinical response to this drug given as a therapeutic window upfront of combination chemotherapy.
In the present in vivo study we investigate whether baseline and/or l-Asp–induced AS mRNA levels are related to the clinical response to a therapeutic window with l-Asp in children with newly diagnosed ALL.
Patients, materials, and methods
Patients and therapeutic window with PEG-Asp
In close collaboration between our institution and the Dutch Childhood Oncology Group (DCOG; the former Dutch Childhood Leukemia Study Group), a window study with pegylated Escherichia colil-Asp (PEG-Asp) upfront to the ALL-9 treatment schedule was initiated in July 2000. The DCOG ALL-9 study was implemented in the Netherlands to confirm the good results of the ALL-6 study,10 which was originally based on German ALL-BFM strategy. The aim of our study is to determine the clinical response as well as molecular determinants of l-Asp response in ALL. Children with ALL at initial diagnosis and presenting with white blood count (WBC) greater than 10 × 109/L were eligible. Similar to a study from the Dana Farber Cancer Institute,11 we assessed a 5-day investigational window. A complete and persistent depletion of asparagine is considered to be the mechanism of action of l-Asp treatment. Boos et al12 showed that a plasma E coli Asp activity of more than 100 IU/L leads to an asparagine depletion of less than 0.2 μM in plasma. Muller demonstrated that one dose of 1000 IU/m2 PEG-Asp resulted in more than 100 U/L serum enzyme activity of l-Asp for 3 weeks.13 In a previous study we confirmed that 1000 IU/m2 PEG-Asp given as a therapeutic window at day –5 (ie, 5 days before starting combined chemotherapy) results in more than 100 U/L l-Asp activity for at least 10 days in children with ALL at initial diagnosis.14 In the present study patients received a single dose of 1000 IU/m2 PEG-Asp in a 1-hour infusion 5 days before starting the DCOG ALL-9 combination chemotherapy treatment schedule. PEG-Asp, kindly provided by Medac (Hamburg, Germany), was used mainly because of its lower immunogenicity than native (unpegylated) l-Asp.15 This lower immunogenicity is important since these patients will be treated with unpegylated l-Asp as part of their regular combination chemotherapy hereafter.
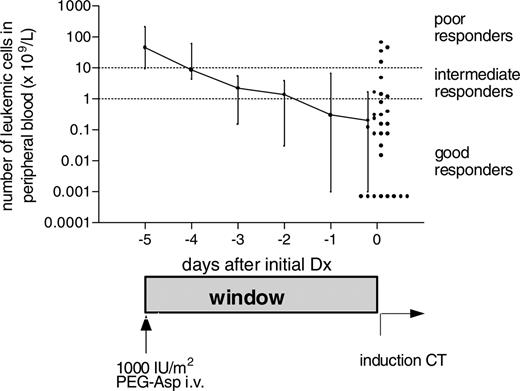
We decided to use the same definition for clinical response that is used for response to prednisone16 : more than 1 × 109 leukemic blasts per liter (1000/μL) of peripheral blood has been shown to be highly predictive for an inferior outcome. So, the clinical response on day 0 (5 days after the PEG-Asp infusion) was defined as good when the number of leukemic cells had declined to less than 1 × 109/L of peripheral blood, as intermediate when leukemic cells were 1 × 109/L to 10 × 109/L, and as poor when leukemic cells were greater than 10 × 109/L.
Between July 2000 and October 2002, 31 patients with ALL were enrolled in the study. Of these, 25 children were diagnosed in the Erasmus MC-Sophia Children's Hospital, Rotterdam, and 6 children were diagnosed in 3 other university hospitals in the Netherlands. Patients' characteristics are shown in Table 1. Because pro–B-ALL is not frequently found in children older than 1 year, we additionally measured the baseline AS mRNA expression in 23 infants with pro–B-ALL (Interfant-99) to compare data of this type of leukemia with those of other subtypes of patients with ALL enrolled in the window study.
Characteristics of 31 patients treated with one dose of PEG-Asp before the DCOG-ALL9 study
Characteristic . | Value . |
|---|---|
| Male/female | 17/14 |
| Age, median, y (range) | 4.2 (1.2-13.1) |
| Median WBC count at diagnosis, × 109/L (range) | 47 (11.4-417) |
| Immunophenotype | |
| Pro-B-ALL | 1 |
| Common ALL | 9 |
| Pre-B-ALL | 9 |
| T-ALL | 12 |
| Cytogenetic characteristics | |
| Hyperdiploid | 9* |
| TEL-AML 1 fusion | 5 |
| BCR-ABL fusion | 1* |
| MLL gene rearranged | 0 |
| Others | 17 |
| CNS involvement† | |
| Yes | 0 |
| No | 31 |
Characteristic . | Value . |
|---|---|
| Male/female | 17/14 |
| Age, median, y (range) | 4.2 (1.2-13.1) |
| Median WBC count at diagnosis, × 109/L (range) | 47 (11.4-417) |
| Immunophenotype | |
| Pro-B-ALL | 1 |
| Common ALL | 9 |
| Pre-B-ALL | 9 |
| T-ALL | 12 |
| Cytogenetic characteristics | |
| Hyperdiploid | 9* |
| TEL-AML 1 fusion | 5 |
| BCR-ABL fusion | 1* |
| MLL gene rearranged | 0 |
| Others | 17 |
| CNS involvement† | |
| Yes | 0 |
| No | 31 |
Patients were administered one intravenous dose of 1000 IU/m2 pegylated l-asparaginase before undergoing combination chemotherapy as part of the DCOG (Dutch Childhood Oncology Group) ALL-9 study.
One patient had both a hyperdiploidy and a BCR-ABL fusion.
CNS involvement is defined as more than 5 cells/μL with blasts in the cerebrospinal fluid.
The immunophenotyping was performed at reference laboratories of the participating groups. The B-lineage ALLs (CD19+, HLA-DR+) were classified into the following differentiation stages: pro–B-ALL cells were CD10–, cytoplasmic μ chain–negative (cμ–), and surface immunoglobulin–negative (sIg–); cALLs were CD10+/cμ–/sIg–; pre–B-ALLs were CD10+/–/cμ+/sIg–. B-ALL cells characterized by CD10–/cμ–/sIg+ were excluded from the study.
The window study on PEG-Asp and the Interfant study on infants with pro–B-ALL were approved by the local ethical committee and by the institutional research board of the DCOG. The patient and/or the parents and guardians have given informed consent for these studies in accordance with the Declaration of Helsinki.
Patient samples
Bone marrow and peripheral blood samples were obtained at initial diagnosis of ALL (day –5) before the administration of PEG-Asp. To perform daily analyses of asparagine synthetase (AS) expression in leukemic cells we decided for ethical reasons that daily collection of bone marrow was unacceptable. Stams et al9 have shown that purified leukemic cells out of peripheral blood revealed comparable AS mRNA levels compared with leukemic cells isolated out of bone marrow. Therefore, blood samples were collected during 5 consecutive days until the start of combination chemotherapy at day 0. Within 24 hours after sampling, mononuclear cells were isolated by density gradient centrifugation using Lymphoprep (density 1.077 g/mL; Nycomed Pharma, Oslo, Norway) and centrifuged at 480g for 15 minutes at room temperature. The collected mononuclear cells were washed twice and kept in culture medium consisting of RPMI 1640 medium (Dutch modification without l-glutamine; Gibco BRL, Life Technologies, Breda, MD), 20% fetal calf serum (FCS; Integro, Zaandam, The Netherlands), 2 mM l-glutamine (Gibco BRL, Life Technologies), 5 μg/mL insulin, 5 μg/mL transferrin, 5 ng/mL sodium selenite (ITS media supplement; Sigma, St Louis, MO), 100 IU/mL penicillin, 100 μg/mL streptomycin, 0.125 μg/mL amphotericin B (Life Technologies), and 0.2 mg/mL gentamycin (Life Technologies). Contaminating nonleukemic cells in the ALL samples were removed by immunomagnetic beads as described by Kaspers et al.17 All samples contained more than 90% leukemic cells, as determined morphologically on May-Grünwald-Giemsa–stained (Merck, Darmstadt, Germany) cytospins.
RNA extraction and cDNA synthesis
Total cellular RNA was extracted from a minimum of 5 × 106 cells using Trizol reagent (Life Technologies) according to the manufacturer's protocol, with minor modifications as reported previously.18 The concentration of RNA was quantified spectrophotometrically and the quality was checked on agarose gels. Following a denaturation step of 5 minutes at 70°C, 1 μg RNA was reverse-transcribed into single-stranded cDNA. The reverse transcription (RT) was performed in a total volume of 25 μL, containing 2.5 nM random hexamers and 20 nM oligo dT primers (Amersham Pharmacia Biotech, Piscataway, NJ), 200 U Moloney murine leukemia virus reverse transcriptase (Promega, Madison, WI), and 25 U RNAsin (Promega), and was incubated at 37°C for 30 minutes, 42°C for 15 minutes, and 94°C for 5 minutes. The obtained cDNA was diluted to a final concentration of 8 ng/μL and stored at –80°C.
Quantitative real-time PCR
The mRNA expression levels of AS and the endogenous housekeeping gene encoding for glyceraldehyde-3-phosphate dehydrogenase (GAPDH), as a reference, were quantified using real-time polymerase chain reaction (RTQ-PCR) analysis (TAQMAN chemistry) on an ABI Prism 7700 sequence detection system (PE Applied Biosystems, Foster City, CA). Amplification of specific PCR products was detected using dual-fluorescent nonextendable probes labeled with 6-carboxyfluorescein (FAM) at the 5′ end and 6-carboxytetramethylrhodamine (TAMRA) at the 3′ end. The primers and probe combinations were designed using OLIGO 6.22 software (Molecular Biology Insights, Cascade, CO) and have been published elsewhere.18 Because all PCRs were performed with equal efficiencies (> 95%), relative mRNA expression levels of AS for each patient could be normalized for input RNA using the GAPDH expression of the patient. The relative mRNA expression level of the target gene in each patient was calculated using the comparative cycle time (Ct) method.19
Briefly, the target PCR Ct values (ie, the cycle number at which emitted fluorescence exceeds 10× the standard deviation [SD] of baseline emissions, as measured from cycles 3 to 12) are normalized by subtracting the GAPDH Ct value from the target PCR Ct value, which gives the ΔCt value. From the ΔCt value, the relative expression level to GAPDH for AS is calculated using the following equation: relative mRNA expression= 2–ΔCt × 100%.
Statistics
Differences in mRNA expression levels measured at different days were analyzed using the Wilcoxon matched-pairs signed rank test. The relationship between AS mRNA expression and in vivo PEG-Asp response and between AS expression and immunophenotype or cytogenetic subtype was analyzed with the Mann-Whitney U test.
Results
Children with newly diagnosed ALL and WBC greater than 10 × 109/L were consecutively enrolled into the study. As is shown in Table 1, 31 children were eligible at the moment of analysis: 1 with pro–B-ALL, 9 with common ALL, 9 with pre–B-ALL, and 12 with T-ALL.
Similar to day 7 for a prednisone window response, we evaluated the in vivo response to PEG-Asp by counting the number of leukemic blasts in the peripheral blood at day 0, 5 days after PEG-Asp was given. As can be seen in Figure 1, the leukemic cells in the peripheral blood dropped continuously over 5 days. The number of leukemic cells reduced 224-fold, from a median of 44.7 × 109/L at day –5 to a median of 0.2 × 109/L at day 0. This was more than a 2-log decrease in leukemic cell burden. There were 21 (68%) children who were PEG-Asp good responders (blast number < 1 × 109/L at day 0), 6 (19%) who were intermediate responders (blasts 1 × 109/L to 10 × 109/L at day 0), and 4 (12%) who were poor responders (blasts > 10 × 109/L at day 0) (Figure 1).
Clinical response to pegylated l-asparaginase (PEG-Asp) in pediatric acute lymphoblastic leukemia (ALL). Clinical response to 1000 IU/m2 intravenous PEG-Asp, measured as the decrease in the absolute number of leukemic cells in the peripheral blood of 31 children with ALL. The final values at day 0 are shown for each individual by dots. The clinical response line shows the median and 25th and 75th percentiles. A good clinical response is defined by less than 1 × 109/blasts/L at day 0, an intermediate response by 1 × 109/blasts/L to 10 × 109/blasts/L on day 0, and poor response by more than 10 × 109/blasts/L at day 0. Dotted lines indicate the cut-off values for these clinical responses.
Clinical response to pegylated l-asparaginase (PEG-Asp) in pediatric acute lymphoblastic leukemia (ALL). Clinical response to 1000 IU/m2 intravenous PEG-Asp, measured as the decrease in the absolute number of leukemic cells in the peripheral blood of 31 children with ALL. The final values at day 0 are shown for each individual by dots. The clinical response line shows the median and 25th and 75th percentiles. A good clinical response is defined by less than 1 × 109/blasts/L at day 0, an intermediate response by 1 × 109/blasts/L to 10 × 109/blasts/L on day 0, and poor response by more than 10 × 109/blasts/L at day 0. Dotted lines indicate the cut-off values for these clinical responses.
Asparagine synthetase (AS) mRNA expression induced by pegylated l-asparaginase (PEG-Asp) in time. Time-response curves of AS mRNA expression in the leukemic cells of 31 children after a single dose of PEG-Asp (1000 IU/m2 given intravenously at day –5). *A 3-fold increase in AS mRNA from day –5 to day –4 (P < .001).
Asparagine synthetase (AS) mRNA expression induced by pegylated l-asparaginase (PEG-Asp) in time. Time-response curves of AS mRNA expression in the leukemic cells of 31 children after a single dose of PEG-Asp (1000 IU/m2 given intravenously at day –5). *A 3-fold increase in AS mRNA from day –5 to day –4 (P < .001).
The baseline expression level of AS mRNA relative to GAPDH was a median of 0.26% (range, 0.05%-2.5%) in leukemic cells (> 90% purity). This was in the range of healthy controls as described in our previous study.18 The expression levels of AS in leukemic cells relative to GAPDH increased significantly a median of 3-fold, from 0.26% (basal expression) to 0.75% (24 hours later; P < .001; Figure 2). During the following 4 days the expression of AS mRNA remained stable at the level of 24 hours (Figure 2; Table 2).
Asparagine synthetase (AS) mRNA expression values in time
. | Day . | . | . | . | . | . | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
. | -5 . | -4 . | -3 . | -2 . | -1 . | 0 . | |||||
| Expression of AS compared with GAPDH, % | |||||||||||
| Median | 0.26 | 0.75 | 0.75 | 0.87 | 1.05 | 0.99 | |||||
| 25th—75th percentile | 0.16-0.46 | 0.45-1.4 | 0.52-1.18 | 0.47-1.17 | 0.48-1.05 | 0.53-1.8 | |||||
| Wilcoxon signed rank test, P | |||||||||||
| Compared between successive days | NA | < .001 | .4 | .4 | .06 | .4 | |||||
| Day -5 compared with days -4, -3, -2, -1, and 0 | NA | < .001 | .001 | < .001 | .001 | .002 | |||||
. | Day . | . | . | . | . | . | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
. | -5 . | -4 . | -3 . | -2 . | -1 . | 0 . | |||||
| Expression of AS compared with GAPDH, % | |||||||||||
| Median | 0.26 | 0.75 | 0.75 | 0.87 | 1.05 | 0.99 | |||||
| 25th—75th percentile | 0.16-0.46 | 0.45-1.4 | 0.52-1.18 | 0.47-1.17 | 0.48-1.05 | 0.53-1.8 | |||||
| Wilcoxon signed rank test, P | |||||||||||
| Compared between successive days | NA | < .001 | .4 | .4 | .06 | .4 | |||||
| Day -5 compared with days -4, -3, -2, -1, and 0 | NA | < .001 | .001 | < .001 | .001 | .002 | |||||
Median expression values (and 25th—75th percentiles of AS mRNA compared with GAPDH in time in leukemic cells of 31 children induced by pegylated L-asparaginase (PEG-Asp). Wilcoxon signed rank test compared between successive days, and day -5 values compared with levels of day -4, day -3, day -2, day -1, and day 0.
NA indicates not applicable.
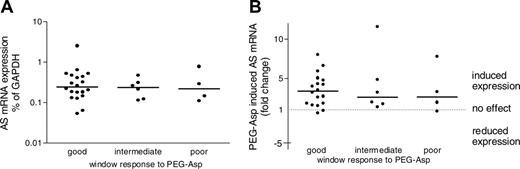
The baseline expression level of AS mRNA did not differ between good and intermediate responders (P = .614), good and poor responders (P = .852), and intermediate and poor (P = 1.0) responders, nor did the up-regulated AS levels after 24 hours of PEG-Asp differ between good and intermediate (P = .614) responders, good and poor (P = .737) responders, and intermediate and poor (P = .914) responders (Figure 3A-B). The fold-change in AS mRNA expression levels was also not related to the relative (P = .997) or absolute (P = .804) decrease in leukemic cells in all 31 patients.
The AS expression for the different immunophenotypic ALL subgroups at diagnosis is shown in Figure 4. The median levels of AS mRNA relative to GAPDH mRNA for c/pre–B-ALL patients (0.21%) and for the T-ALL patients (0.28%) did not significantly differ (P = .376). One window patient had a pro–B-ALL for which the baseline expression of AS mRNA was 3-fold higher than the other c/pre–B-ALL patients. To explore whether pro–B-ALL is associated with a high AS mRNA expression we analyzed the AS expression of 23 infant pro–B-ALL cases. Infants with pro–B-ALL had a median 0.15% (range, 0.07%-1.43%) AS mRNA expression level, which was not significantly different from the baseline AS expression values in non-infants with c/pre–B- or T-ALL (Figure 4A). The c/pre–B-ALL group had a median baseline AS expression level of 0.21%, which rose significantly to a median of 0.72% 1 day later (median 3.99-fold individual up-regulation, P < .001). Patients with T-ALL demonstrated a significant increase from basal 0.28% to 0.68% 1 day later (median 1.94-fold individual up-regulation, P = 0.012). Patients with T-ALL tended to have a lower individual up-regulation of AS mRNA compared with the children with c/pre–B-ALL, but this was not statistically different (P = .107; Figure 4B). Only 3 cases had a slight down-regulation of the AS mRNA expression (–1.6-, –1.2-, and –1.05-fold). These 3 cases were all patients with T-ALL, of whom 2 were clinically good responders.
Relationship between asparagine synthetase (AS) mRNA expression induced by pegylated l-asparaginase (PEG-Asp) and clinical response. (A) Baseline AS mRNA expression levels. (B) PEG-Asp–induced changes in AS mRNA expression levels measured after 24 hours of in vivo exposure to PEG-Asp compared with baseline expression levels. Dots represent individual expression values; solid lines, the median expression value per group. For definition of clinical response, see Figure 1.
Relationship between asparagine synthetase (AS) mRNA expression induced by pegylated l-asparaginase (PEG-Asp) and clinical response. (A) Baseline AS mRNA expression levels. (B) PEG-Asp–induced changes in AS mRNA expression levels measured after 24 hours of in vivo exposure to PEG-Asp compared with baseline expression levels. Dots represent individual expression values; solid lines, the median expression value per group. For definition of clinical response, see Figure 1.
The baseline and Asp-induced expression levels of AS mRNA did not differ between hyperdiploid (n = 9), TEL/AML1–positive (n = 5), and other B-lineage ALL (n = 5). For infants with MLL gene–rearranged ALL, no data were available for the effect of l-Asp on AS mRNA levels, because these patients were not eligible for the PEG-Asp window study.
Discussion
Studies on putative causes of l-Asp resistance have been performed most extensively in mouse cell lines.5,6 l-Asp–sensitive tumor cells that did not contain detectable levels of AS developed resistance to l-Asp through exposure of cells to sublethal concentrations of the drug.20 Resistant cells up-regulated AS expression and activity by 60-fold. It is well known that AS plays a crucial role in maintaining amino acid homeostasis in cells.21 A rapid transcriptional control of the AS gene occurs following deprivation of any single essential amino acid.6,22 In 1997, Hutson et al6 demonstrated that depletion of the intracellular asparagine pool by l-Asp was sufficient to activate in vitro AS expression in human leukemic cell lines. The increase in AS mRNA expression also resulted in a simultaneous up-regulation of AS protein levels and AS enzyme activity.6,7 The direct correlation among mRNA, protein, and activity levels was confirmed by Aslanian et al.23 The activity of AS was inversely related to the sensitivity to l-Asp in human leukemia cell lines.6,7 These studies suggest that the expression of the AS gene is linked to resistance to l-Asp. However, in addition to the fact that these studies dealt not with primary patient samples but with cell lines, Wagner and Boos24 argued that the test conditions in Hutson's experiments were not comparable with in vivo situations, where various products and substrates (such as aspartate, glutamate, glutamine, and ammonia, among others) are all part of metabolic pathways and equilibrium conditions.5
Relationship between immunophenotype and asparagine synthetase (AS) mRNA expression. (A) Baseline AS mRNA expression levels. (B) Pegylated L-asparaginase (PEG-Asp)–induced changes in AS expression levels in acute lymphoblastic leukemia (ALL) cells. Dots represent individual expression values; solid lines, the median expression value per group.
Relationship between immunophenotype and asparagine synthetase (AS) mRNA expression. (A) Baseline AS mRNA expression levels. (B) Pegylated L-asparaginase (PEG-Asp)–induced changes in AS expression levels in acute lymphoblastic leukemia (ALL) cells. Dots represent individual expression values; solid lines, the median expression value per group.
In 1969 Haskell et al studied in vivo AS activity in 18 patients with leukemia.3 Prior to therapy, AS activity was nearly undetectable in leukemic cells. Patients were treated with 200 IU/kg E colil-Asp daily for 3 days to 3 weeks. A 7-fold increase in AS activity was found in 5 l-Asp–resistant patients compared with 4 l-Asp–sensitive patients (mixed cohort of ALL, acute myeloid leukemia [AML], chronic myelogenous leukemia [CML], and chronic lymphocytic leukemia [CLL]). Haskell et al3 suggested that l-Asp resistance was related to the capacity of leukemic cells to up-regulate AS expression for asparagine biosynthesis. However, besides the limited number of patients in a very heterogeneous group, the criteria used to determine whether the patient was resistant or sensitive to l-Asp were not described.
In order to study the effect of monotherapy with l-Asp on leukemic blasts we used PEG-Asp. The effectivity of different l-ASP products like Erwinase, E coli ASP, or PEG-Asp is the same if the serum enzyme activity of l-Asp is higher than 100 U/L.13 We studied whether baseline levels or up-regulated levels of AS mRNA expression in leukemic cells after an in vivo treatment with PEG-Asp monotherapy were associated with short-term clinical response to this drug in children with ALL. The baseline AS expression levels were in the same range as healthy controls, as reported before.18 Up-regulation of AS mRNA occurred within 24 hours after PEG-Asp exposure and thereafter no further changes were found. Because the drop in leukemic cells was seen during the whole window period (Figures 1 and 2), it is unlikely that only leukemic cells resistant to PEG-Asp with intrinsic higher AS expression levels were left over on day –4. Baseline and l-Asp–induced AS mRNA expression levels did not differ between patients with good, intermediate, or poor response (Figure 3). So, l-Asp–induced up-regulation of AS mRNA is not related to early in vivo blast reduction in childhood ALL and thus is not predictive for the short-term clinical response to l-Asp. As mentioned earlier, cell line studies showed that mRNA, protein, and activity levels of AS are correlated,6,7,23 but at present it is unknown whether this is also the case for clinical samples because only limited amounts of patients' samples can be obtained. Immunophenotypic and genetic abnormalities are related to drug resistance and outcome in childhood ALL.25,27 T-ALL cells from children are, in vitro, more resistant to l-Asp than are cells from children with precursor B-lineage ALL.28 The relative resistance to l-Asp of T-ALL cases cannot be explained by altered expression of the AS gene, since both baseline and l-Asp–induced changes in AS mRNA expression did not differ between patients with T- and c/pre–B-ALL (Figure 4). Remarkable was the finding that 3 of 12 children with T-ALL even demonstrated a slight AS mRNA down-regulation, which would, in vitro, even point to sensitivity for l-Asp. Hyperdiploidy and the TEL/AML1 fusion are related to favorable outcome in childhood ALL,26,29 and are both in vitro–sensitive to l-Asp.18,30,31 In a previous study in TEL/AML1–positive ALL, Stams et al8 showed that TEL/AML1–positive children expressed 5-fold more AS mRNA compared with TEL/AML1–negative patients and healthy controls. In the present study, TEL/AML1 and hyperdiploid cases do not show an impaired in vivo up-regulation of AS that might have explained their high sensitivity to l-Asp. Taken together, both studies suggest that sensitivity to l-Asp as found in TEL/AML1–positive and hyperdiploid cells is not linked to decreased AS mRNA expression.
AS mRNA up-regulation in ALL cells occurs very rapidly (< 24 hours) after cellular asparagine depletion following PEG-Asp administration. Amino acids are required for protein synthesis, but they also play a role in the control of gene expression.6,21 The promotor of AS contains a nutrient-sensing response unit (NSRU) that is responsible for the induction of AS gene transcription upon amino acid deprivation.32 Iiboshi et al showed that withdrawal of asparagine and glutamine by l-Asp resulted in a rapid inactivation of p70 S6 kinase.33 P70 S6 kinase participates in the mammalian target of rapamycin (mTOR) protein synthesis by controlling translational initiation and elongation factors as well as protein kinases that affect ribosomal assembly. Recently, gene expression profiling revealed that l-Asp–resistant ALL cells overexpressed several ribosomal protein-encoding genes as well as initiation factors.34 Using gene expression profiling, Fine et al showed that l-Asp–resistant cell lines expressed more baseline AS mRNA than sensitive leukemic cell lines, whereas no such association was found for primary pediatric ALL samples.35 This study emphasizes the fact that leukemic cell lines and primary samples from leukemic patients are different from each other and cell line data cannot be extrapolated to a primary patient's cells that easily. Exposure to l-Asp altered in a primary patient's samples the expression of a number of genes related to protein synthesis (ie, tRNA synthetases and amino acid transporters). However, no genes discriminative for l-Asp resistance in patient samples were found. These data point to a consistent coordinated response to amino acid starvation, which occurs irrespective of the level of resistance to l-Asp in a patient's cells. Therefore, AS up-regulation may be a consequence of amino acid deprivation by l-Asp but is not the limiting key factor explaining resistance to l-Asp in pediatric ALL.
We conclude that up-regulation of AS mRNA in childhood ALL cells occurs within 24 hours after in vivo exposure to PEG-Asp, but these up-regulated levels are not associated with an early (poor) response to PEG-Asp in this small group of children.
Prepublished online as Blood First Edition Paper, February 23, 2006; DOI 10.1182/blood-2005-06-2597.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal