Abstract
A rare genetic disease, Fanconi anemia (FA), now attracts broader attention from cancer biologists and basic researchers in the DNA repair and ubiquitin biology fields as well as from hematologists. FA is a chromosome instability syndrome characterized by childhood-onset aplastic anemia, cancer or leukemia susceptibility, and cellular hypersensitivity to DNAcrosslinking agents. Identification of 11 genes for FA has led to progress in the molecular understanding of this disease. FA proteins, including a ubiquitin ligase (FANCL), a monoubiquitinated protein (FANCD2), a helicase (FANCJ/BACH1/BRIP1), and a breast/ovarian cancer susceptibility protein (FANCD1/BRCA2), appear to cooperate in a pathway leading to the recognition and repair of damaged DNA. Molecular interactions among FA proteins and responsible proteins for other chromosome instability syndromes (BLM, NBS1, MRE11, ATM, and ATR) have also been found. Furthermore, inactivation of FA genes has been observed in a wide variety of human cancers in the general population. These findings have broad implications for predicting the sensitivity and resistance of tumors to widely used anticancer DNA crosslinking agents (cisplatin, mitomycin C, and melphalan). Here, we summarize recent progress in the molecular biology of FA and discuss roles of the FA proteins in DNA repair and cancer biology.
Introduction
Many hematologists are familiar with Fanconi anemia (FA) as a cause of childhood-onset aplastic anemia. However, this rare genetic disease now attracts broader attention from cancer biologists and basic researchers. FA is a rare chromosome instability syndrome characterized by aplastic anemia, cancer and leukemia susceptibility, and cellular hypersensitivity to interstrand DNA crosslinking agents, such as cisplatin, mitomycin C (MMC), diepoxybutane (DEB), and melphalan.1,3 Treatment with these agents causes increased chromosome breakage in cells derived from patients with FA.4 Because of this hypersensitivity to DNA crosslinking agents, many researchers have speculated that the primary defect of FA cells is in DNA damage response or DNA repair, although until recently direct evidence of a connection between FA and DNA repair had not been shown.
Recent identification of the responsible genes for FA has changed our view of the molecular pathogenesis of the disease. FAcan be divided into at least 12 complementation groups (A, B, C, D1, D2, E, F, G, I, J, L, and M) defined by cell fusion studies, and 11 of the 12 responsible FA genes have been identified (Table 1; Figure 1).5,8 Proteins encoded by FA genes (FA proteins) include a ubiquitin ligase (FANCL/PHF9/POG),9 a monoubiquitinated protein (FANCD2),10,11 a helicase (FANCJ/BACH1/BRIP1),7,8,12 a protein with helicase motifs and a nuclease motif (FANCM), and a well-known breast/ovarian cancer susceptibility protein (FANCD1/BRCA2).13 FA proteins (including BRCA2) and another well-known breast/ovarian cancer susceptibility protein, BRCA1, cooperate in a common DNA repair process that is required for cellular resistance to DNA interstrand crosslinks (ICLs), and a concept of the Fanconi anemia-BRCA pathway14 (or FA-BRCA network15 ) has been proposed. Furthermore, molecular and functional interaction of FA proteins with proteins [ATM (ataxia-telangiectasia mutated),16 MRE11,17 BLM,18,20 NBS1,17,21 and ATR (ATM and Rad3-related)22,23 ] responsible for other rare genetic chromosome instability syndromes [ataxia telangiectasia,24 AT-like disorder,25 Bloom syndrome,26 Nijmegen breakage syndrome (NBS),27 and Seckel syndrome, respectively] in DNA damage response is now evident. Therefore, the FA pathway appears to regulate DNA repair.
Twelve complementation groups and responsible genes for Fanconi anemia
Subtype . | Responsible gene . | Patients with FA, estimated, %* . | Chromosome location . | Protein, kDa . | Requirement for FANCD2 monoubiquitination . | Main function of the protein . | Comments . |
|---|---|---|---|---|---|---|---|
| A | FANCA | 57 | 16q24.3 | 163 | + | FA core complex | NA |
| B | FANCB (FAAP95) | 0.3 | Xp22.31 | 95 | + | FA core complex | NA |
| C | FANCC | 15 | 9q22.3 | 63 | + | FA core complex | Cytoplasmic functions |
| D1 | FANCD1/BRCA2 | 4 | 13q12-13 | 380 | - | RAD51 recruitment | NA |
| D2 | FANCD2 | 3 | 3p25.3 | 155, 162 | + | ? | Monoubiquitinated protein |
| E | FANCE | 1 | 6p21-22 | 60 | + | FA core complex | Direct binding to FANCD2 |
| F | FANCF | 2 | 11p15 | 42 | + | FA core complex | NA |
| G | FANCG/XRCC9 | 9 | 9p13 | 68 | + | FA core complex | NA |
| I | Not identified | Rare | ? | ? | + | ? | NA |
| J | FANCJ/BACH1/BRIP1 | 1.6 | 17q22-q24 | 130 | - | 5′>3′ DNA helicase/ATPase | Binding to BRCA1 |
| L | FANCL/PHF9/POG (FAAP43) | 0.1 | 2p16.1 | 43 | + | FA core complex, ubiquitin ligase | NA |
| M | FANCM/Hef (FAAP250) | Rare | 14q21.3 | 250 | + | FA core complex, ATPase/translocase | Helicase/nuclease motif |
Subtype . | Responsible gene . | Patients with FA, estimated, %* . | Chromosome location . | Protein, kDa . | Requirement for FANCD2 monoubiquitination . | Main function of the protein . | Comments . |
|---|---|---|---|---|---|---|---|
| A | FANCA | 57 | 16q24.3 | 163 | + | FA core complex | NA |
| B | FANCB (FAAP95) | 0.3 | Xp22.31 | 95 | + | FA core complex | NA |
| C | FANCC | 15 | 9q22.3 | 63 | + | FA core complex | Cytoplasmic functions |
| D1 | FANCD1/BRCA2 | 4 | 13q12-13 | 380 | - | RAD51 recruitment | NA |
| D2 | FANCD2 | 3 | 3p25.3 | 155, 162 | + | ? | Monoubiquitinated protein |
| E | FANCE | 1 | 6p21-22 | 60 | + | FA core complex | Direct binding to FANCD2 |
| F | FANCF | 2 | 11p15 | 42 | + | FA core complex | NA |
| G | FANCG/XRCC9 | 9 | 9p13 | 68 | + | FA core complex | NA |
| I | Not identified | Rare | ? | ? | + | ? | NA |
| J | FANCJ/BACH1/BRIP1 | 1.6 | 17q22-q24 | 130 | - | 5′>3′ DNA helicase/ATPase | Binding to BRCA1 |
| L | FANCL/PHF9/POG (FAAP43) | 0.1 | 2p16.1 | 43 | + | FA core complex, ubiquitin ligase | NA |
| M | FANCM/Hef (FAAP250) | Rare | 14q21.3 | 250 | + | FA core complex, ATPase/translocase | Helicase/nuclease motif |
+ indicates that the gene is required for FANCD2 monoubiquitination; NA, not applicable; -, gene not required for FANCD2 monoubiquitination; ?, unknown.
Unpublished data from the International Fanconi Anemia Registry (IFAR) (kindly provided by Dr Arleen D. Auerbach).
Ubiquitination is a posttranslational modification in which a 76-residue small protein, ubiquitin, is covalently attached to a target protein (reviewed in Mani and Gelmann28 ). Polyubiquitination is well known for its function in targeting proteins for degradation by the 26 S proteasome. In contrast, monoubiquitination (conjugation of one ubiquitin molecule onto a protein) is a reversible signal regulating either protein targeting, membrane trafficking, histone function, transcription regulation, or DNA repair.29 Monoubiquitination of one of the FA proteins, FANCD2, is a critical step in the activation of the FA-BRCA pathway, and most of the other FA proteins are required for this monoubiquitination event.11
Importantly, defects of the FA genes have been found in a wide variety of human cancers in the general population (patients without FA).30,40 Defects of DNA repair and cell-cycle checkpoints, such as the defects of the FA pathway, are possible mechanisms of genomic instability in cancer and may also be responsible for the hypersensitivity of cancer cells to certain types of chemotherapeutic drugs and radiation. Disruption of FA genes may become a useful predictor of sensitivity to chemotherapy with widely used anticancer DNA crosslinking agents (cisplatin, MMC, and melphalan).30,38,41
Thus, understanding the molecular biology of FA is important, not only for hematologists who manage patients with FA but also for cancer biologists interested in the mechanism of drug sensitivity and resistance and for basic scientists working on DNA repair, ubiquitin biology, or both.
Clinical course of FA
FA is a rare autosomal (all complementation groups except FA-B group) or X-linked (FA-B group) recessive disease, clinically characterized by multiple congenital abnormalities, bone marrow (BM) failure, and cancer susceptibility. The clinical course of FA has been extensively reviewed.2,3,42 The prevalence of FA is estimated to be 1 to 5 per million, and heterozygous carrier frequency is estimated to be 1 in 300, although the true frequency is probably higher.1,2
Patients with FA show extreme clinical heterogeneity.2,3 The median age at diagnosis is 6.5 years for male patients, and 8 years for female patients, although the age at diagnosis ranges from birth to 48 years. The male-female ratio is 1.24. The median survival age has improved to 30 years in patients reported between 1991 and 2000.3 Survival was 19 years in those reported between 1981 and 1990.3 The common congenital defects seen in patient with FA includes short statue (51%); abnormalities of the skin (55%), upper extremities (43%), head (26%), eyes (23%), kidneys (21%), and ears (11%); and developmental disability (11%).3 Thirty-two percent of male patients with FA show abnormal gonads, although abnormal gonads have been described in only 3% of female patients with FA.3 A significant percentage (25%-40%) of the patients with FA were reported to be physically normal.3
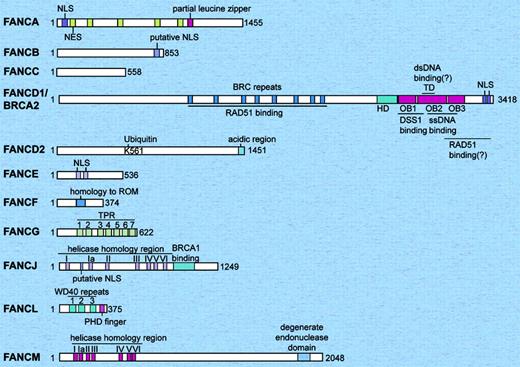
Schematic representation of the 11 human Fanconi anemia proteins. The relative sizes of the FA proteins are shown to scale. The FANCF and FANCL proteins are the smallest, and the FANCM and FANCD1/BRCA2 proteins are the largest. The only FA proteins with known enzymatic activity are FANCJ (helicase), FANCM (DNA translocase), and FANCL (E3 ubiquitin ligase). FANCD2 and FANCD1/BRCA2 have been shown to have direct DNA binding activity. dsDNA indicates double strand DNA; HD, helical domain; NES, nuclear export sequences; NLS, nuclear localization signals; OB, oligonucleotide/oligosaccharide-binding folds; ssDNA, single-strand DNA; TD, tower domain; TPR, tetratricopeptide repeat.
Schematic representation of the 11 human Fanconi anemia proteins. The relative sizes of the FA proteins are shown to scale. The FANCF and FANCL proteins are the smallest, and the FANCM and FANCD1/BRCA2 proteins are the largest. The only FA proteins with known enzymatic activity are FANCJ (helicase), FANCM (DNA translocase), and FANCL (E3 ubiquitin ligase). FANCD2 and FANCD1/BRCA2 have been shown to have direct DNA binding activity. dsDNA indicates double strand DNA; HD, helical domain; NES, nuclear export sequences; NLS, nuclear localization signals; OB, oligonucleotide/oligosaccharide-binding folds; ssDNA, single-strand DNA; TD, tower domain; TPR, tetratricopeptide repeat.
The hematologic complications of FA have also been extensively reviewed.2,3 Patients with FA develop BM failure, typically during the first decade of life. The actuarial risk of developing BM failure is 90% by 40 years of age.43 At least 20% of patients with FA also develop malignancies.43 The actuarial risks of developing hematologic and nonhematologic neoplasms are 33% and 28%, respectively, by 40 years of age.43 The median age of patients who develop cancer is 14 years for acute myelogenous leukemia (AML), 13 years for liver tumors, and 26 years for all solid tumors.44 In approximately 25% of patients with FA with cancer, the malignancy preceded the diagnosis of FA.44 Some patients with FA develop multiple cancers.44
The most common malignancies in patients with FA are AML and myelodysplastic syndrome. The most frequently reported chromosomal abnormalities in FA-associated leukemias are monosomy 7 and duplication of 1q.45 A study identified a high incidence of chromosome 3q abnormalities in BM cells of patients with FA. Gain of 3q is strongly associated with a poor prognosis.46
Patients with FA are also susceptible to solid tumors, such as head and neck squamous cell carcinoma (HNSCC), gynecologic squamous cell carcinoma (SCC), esophageal carcinoma, liver tumors, brain tumors, skin tumors, and renal tumors.43,44,47 Patients receiving androgen therapy for BM failure are prone to liver tumors, suggesting that androgens promote carcinogenesis in these patients.44 A high incidence (84%) of human papilloma virus (HPV) DNA and lack of p53 mutation in SCC (anogenital or head and neck) in patients with FA were reported,48 but another group reported conflicting results (lack of HPV positivity and high p53 mutation prevalence).49 The role of HPV should be further investigated, because HPV infection may be prevented by prophylactic HPV vaccination.
There is no clear evidence of increased cancer susceptibility in heterozygous carriers of mutation in FA genes, except for BRCA2, whose heterozygous carriers have increased susceptibility to breast/ovarian and other cancers.40,50 FA gene (FANCA, FANCC, FANCD2, FANCE, FANCF, and FANCG) mutations are rare in families negative for the BRCA1/2 mutation with inherited breast cancer.51 Two missense germ line mutations of BACH1/FANCJ associated with early-onset breast cancer were reported,52 but germ line mutations in FANCJ are rare in families with breast and ovarian cancers.53
Diagnosis and treatment of FA
Treatment of FA cells with DNA crosslinking agents, such as DEB, causes increased chromosome breakage and marked G2 accumulation. The DEB-induced chromosome breakage assay (DEB test) is widely used as a standard diagnostic test for FA.4 FA subtyping (determination of the complementation group of each patient with FA) has become increasingly important.54 Some patients with other rare chromosome breakage syndrome, such as NBS, can have a positive DEB test.17 Furthermore, a relation between genotype and phenotype has been reported. Patients in complementation group C (FA-C) had a significantly poorer survival than patients in groups A and G.43 FA-D1 subtype is associated with increased predisposition to medulloblastoma, Wilms tumor, and acute leukemia in early childhood and is clinically different from other subtypes.50,55,56 Family members of patients with FA-D1 may be carriers of mutation in BRCA2 and may be predisposed to cancers.50 Therefore, to confirm the diagnosis, to distinguish FA from other chromosome breakage disorders, and to manage each patient with FA and family better, FA subtyping should be performed routinely in the near future. For subtyping, cell fusion assays have been used, but a combination of retroviral gene transfer and G2 accumulation assay or a combination of retroviral gene transfer and FANCD2 immunoblotting are also useful.54,57,58
The treatment of FA has been extensively reviewed elsewhere.42 For BM failure, androgens and hematopoietic growth factors are effective in many cases, but most patients with FA become refractory to these treatments. For such patients, hematopoietic stem cell transplantation (SCT) is performed, providing a donor is available. The outcome of SCT has improved.42,59 Fludarabine-based regimens have proven to be a significant advance.42 Now that many patients with FA survive their hematopoietic problems, prevention, surveillance, and treatment of solid tumors are becoming more important. Because radiotherapy and chemotherapy may cause severe side effects, surgery is the primary therapy for solid tumors in FA. As a new modality of therapy, gene therapy will be an option in the future.60
Complementation groups and FA genes and proteins
The 12 FA complementation groups and the responsible genes and proteins are summarized in Table 1 and Figure 1. Eight FA proteins (FANCA, FANCB, FANCC, FANCE, FANCF, FANCG, FANCL, and FANCM) form a nuclear protein complex (the FA core complex), which is required for monoubiquitination of another FA protein, FANCD2 (Figure 2A).5,9,11,18 Monoubiquitination of FANCD2 is required for translocation of FANCD2 to chromatin and for nuclear foci formation of FANCD2, which may reflect accumulation of FANCD2 protein at the sites of DNA damage and repair.11,61
FANCA contains 2 overlapping bipartite nuclear localization signals (NLSs), 5 functional leucine-rich nuclear export sequences (NESs), and a partial leucine zipper sequence.62,64 The nuclear export of FANCA is regulated in a CRM1-dependent manner.64
FANCB (FAAP95) contains a putative bipartite NLS.65 The FANCB gene is located on the X chromosome, and all of the reported patients with FA-B are male.65
FANCC66 is a component of the nuclear FA core complex but localizes both in nucleus and cytoplasm.67 Some functions of FANCC outside of the FA core complex, such as involvement in Jak/STAT signaling and apoptotic signaling, have been also proposed.68
The gene for FA-D1 subtype, FANCD1, is identical to a breast/ovarian cancer susceptibility gene, BRCA2.13 Carriers of monoallelic germ line mutation of BRCA2 have breast/ovarian cancer susceptibility, but patients with biallelic germ line mutations develop FA-D1 subtype of FA. BRCA2 is not a component of the FA core complex nor is it required for monoubiquitination or nuclear foci formation of FANCD2.13 A principal function of BRCA2 is regulation of homologous recombination (HR) repair through control of RAD51 recombinase (eukaryotic homologue of bacterial RecA).69,70 BRCA2 also has other functions, such as stabilization of stalled replication forks, regulation of cytokinesis, and so forth.71 BRCA2 protein has 8 copies of 30 to 40 amino acid motif (BRC repeats). The BRC repeats directly bind to RAD51 protein,72 which can form a nucleoprotein filament with single-stranded DNA (ssDNA). The C-terminal region of BRCA2 is also implicated in RAD51 binding. The C-terminal domain has 5 domains; a helical domain (HD), 3 oligonucleotide/oligosaccharide-binding folds (OB1, OB2, and OB3), and a tower domain (TD) inserted in OB2.73 The OB folds bind to ssDNA. The TD interacts with double-stranded DNA (dsDNA).73 BRCA2 may bind to the ssDNA-dsDNA junction of resected DNA ends of double-strand DNA breaks and help RAD51 loading on ssDNA.74 DSS1 (deleted in split-hand/split foot syndrome) interacts with BRCA2 through the HD, OB1, and OB2 domains.73 DSS1 is required for stability of BRCA2.75 DSS1 is also a component of the lid subcomplex of the regulatory subunit of the 26S proteasome.76
FANCD2 does not have any known functional motifs.10 Normal cells express 2 isoforms of the FANCD2 protein, a nonubiquitinated form (155 kDa) and a monoubiquitinated form (162 kDa).11 Monoubiquitination occurs on a single residue, lysine 561. Monoubiquitinated FANCD2 protein is mainly located in the chromatin fraction.61 The C-terminal region of FANCD2 contains several acidic amino acid residues and is required for the function of FANCD2 in ICL resistance, although this region is not required for monoubiquitination or for chromatin localization of FANCD2.61
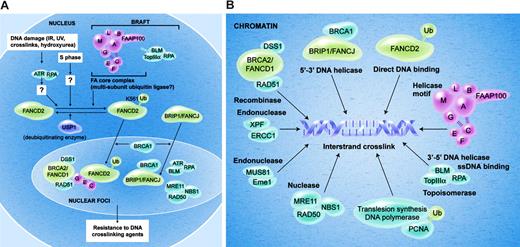
Current model of the Fanconi anemia pathway. (A) The FA proteins are depicted in the normal cell nucleus. In response to DNA damage, or during normal S-phase progression, the FANCD2 protein is monoubiquitinated on lysine 561. Efficient monoubiquitination requires several proteins, including ATR, RPA, and an intact FA core complex. Monoubiquitination of FANCD2 targets the protein into nuclear foci and chromatin fraction where it interacts with BRCA2. The function of monoubiquitinated FANCD2 (FANCD2-Ub) in chromatin remains unknown, and it may regulate HR repair, translesion DNA synthesis, or both. USP1 negatively regulates monoubiquitination of FANCD2. (B) This figure depicts several chromatin proteins that appear to interact in the repair of the DNA interstrand crosslink (see “Localization of FA proteins in chromatin”).
Current model of the Fanconi anemia pathway. (A) The FA proteins are depicted in the normal cell nucleus. In response to DNA damage, or during normal S-phase progression, the FANCD2 protein is monoubiquitinated on lysine 561. Efficient monoubiquitination requires several proteins, including ATR, RPA, and an intact FA core complex. Monoubiquitination of FANCD2 targets the protein into nuclear foci and chromatin fraction where it interacts with BRCA2. The function of monoubiquitinated FANCD2 (FANCD2-Ub) in chromatin remains unknown, and it may regulate HR repair, translesion DNA synthesis, or both. USP1 negatively regulates monoubiquitination of FANCD2. (B) This figure depicts several chromatin proteins that appear to interact in the repair of the DNA interstrand crosslink (see “Localization of FA proteins in chromatin”).
FANCE77 is a nuclear protein required for nuclear accumulation of FANCC.78,79 FANCE interacts with monoubiquitinated FANCD2 and BRCA2 in the chromatin fraction.80
FANCF has a region of homology with the prokaryotic RNA-binding protein ROM,81 but this homology appears to be nonsignificant.77 FANCF acts as a flexible adaptor protein, required for the assembly of the FA core complex.82
FANCG/XRCC9 was originally cloned as a cDNA that partially corrects the MMC sensitivity of a Chinese hamster mutant UV40 cell line.83,84 FANCG has 7 tetratricopeptide repeat motifs (TPRs).85 TPRs are degenerate 34–amino acid repeat motifs, mediating protein-protein interactions. Serines 383 and 387 on FANCG are phosphorylated in the M phase, presumably by cdc2.86 These 2 sites are important for exclusion of FANCG from chromatin in mitosis. Phosphorylation of serine 7 of FANCG is up-regulated after MMC treatment.87
FA-I cells lack monoubiqutination of FANCD2 but form the FA core complex normally.6 The still-unidentified FANCI protein is considered to be a factor required for monoubiquitination of FANCD2 but is not a subunit of the FA complex.6
FANCJ/BACH1/BRIP17,8,12 is a DNA-dependent ATPase and a 5′-to-3′ DNA helicase (DEAH helicase) that directly binds to the BRCT domain of BRCA1.52 BRCT domain is a phosphoprotein-binding domain, and phosphorylation of serine 990 of BACH1 is required for BRCA1-BACH1 interaction.88 FANCJ contains the 7 helicase-specific motifs and a C-terminal extension that has 39% homology with synaptonemal complex protein 1, a major component of the transverse filaments of developing meiotic chromosomes.52 FANCJ is not required for monoubiquitination of FANCD2.6,12,89
FANCL/PHF9/POG (FAAP43) has 3 WD40 repeats and a PHD finger motif.9 WD40 repeats are known to mediate protein-protein interactions. The PHD motif is a variant of RING finger and sometimes shows ubiquitin ligase activity. FANCL has an autoubiquitin ligase activity in vitro, is required for FANCD2 monoubiquitination in vivo, and is presumed to be the catalytic subunit of the FA core complex as an ubiquitin ligase for FANCD2. However, in vitro reconstitution of monoubiquitination of FANCD2 by FANCL has not been demonstrated. The mouse homolog of FANCL was identified as a gene responsible for the phenotype of gcd (germ cell-deficient) mice, which showed reduced fertility with defective proliferation of germ cells.90
FANCM (Hef) (FAAP250) protein is a component of the FA core complex and contains the 7 helicase-specific motifs and 1 degenerate endonuclease domain.5 FANCM has ssDNA and dsDNA-stimulated ATPase activity and DNA translocase activity. So far, the helicase activity or endonuclease activity of FANCM has not been shown. FANCM is closely related to the archeal protein Hef (helicase-associated endonuclease for fork-structured DNA),91 which has a functional helicase domain and a functional endonuclease domain and resolves stalled replication forks.91 Human XPF/ERCC4 endonuclease (the responsible protein for xeroderma pigmentosum complementation group F) and yeast MPH1 helicase also have high homology to archeal Hef. These proteins may have derived from a common ancestor and may have diverged.5 FANCM is phosphorylated in response to DNA damage.5
Interaction of the FA proteins: FA pathway
Because cells from all subtypes of FA are hypersensitive to ICLs, the FA proteins have been considered to cooperate in a common DNA damage response or DNA repair pathway (the FA pathway) (Figure 2A). The pathway is also referred to as the FA-BRCA pathway14 or the FA-BRCA network.15 The critical steps in the pathway are the monoubiquitination and nuclear foci formation of the FANCD2 protein.11
Eight FA proteins (A, B, C, E, F, G, L, and M) and 1 unidentified protein (FAAP100) form a nuclear complex (FA core complex).18 Because monoubiquitination of FANCD2 depends completely on the FA core complex and FANCL has ubiquitin ligase activity, the FA core complex is presumed to be a multi-subunit ubiquitin ligase complex responsible for monoubiquitination of FANCD2.9 FA core complex, BLM, RPA, and topoisomerase III α form a larger complex called BRAFT (for BLM, RPA, FA, and Topo III α). BLM is not required for monoubiquitination of FANCD2.18
Monoubiquitination of FANCD2 is required for nuclear foci formation, chromatin localization, and function of FANCD2 to restore resistance to ICLs in FA-D2 cells.11,80 In response to DNA damage, the FA pathway becomes activated. After treatment with DNA damaging agents, such as ionizing radiation (IR), ultraviolet radiation, DNA crosslinking agents, and hydroxyurea, FANCD2 is monoubiquitinated and targeted to BRCA1/BRCA2/RAD51-containing nuclear foci, presumably at the sites of DNA damage and repair.11,80,92 FANCD2 also colocalizes with a DNA damage–activated signaling kinase, ATR, and RPA in DNA damage–induced nuclear foci22 and partially colocalizes with FANCE,79 FANCC,93 NBS1,17 BLM,19,20 and FANCJ.12 FANCG is reported to colocalize with BRCA2 and RAD51,94 suggesting colocalization of FANCG and FANCD2. All of these factors are required for cellular resistance to ICLs.
ATR is required for the DNA damage–induced monoubiquitination and nuclear foci formation of FANCD2, although its specific mechanism in the process remains unclear.22 FANCD2 itself can be directly phosphorylated by ATR in vitro,22 and phosphorylation of FANCD2 in response to ICLs is reported to be ATR dependent.23 Therefore, ATR-mediated phosphorylation of FANCD2 may enhance monoubiquitination of FANCD2. However, ATR may phosphorylate some components of FA core complex and may enhance ubiquitin ligase activity of the complex. The recent finding of DNA damage–inducible FANCM phosphorylation is consistent with the latter hypothesis.5
BRCA1 is a RING finger protein with ubiquitin ligase activity. BRCA1 is required for efficient DNA damage–inducible increase of monoubiquitinated FANCD2 in a breast cancer cell line, HCC1937,11 but this phenomenon is not reproducible in HeLa cells exposed to BRCA1 siRNA or in BRCA1-deleted chicken DT40 cells.95 Therefore, the in vivo role of BRCA1 for monoubiquitination of FANCD2 is questionable, although BRCA1 together with BARD1 can monoubiquitinate FANCD2 in vitro.95 BRCA1 is required for DNA damage–inducible nuclear foci formation of FANCD2 in any of these cells.11,95 BRCA1 also facilitates FANCJ52 and RAD51 nuclear foci formation after DNA damage.
In the absence of DNA damage, FANCD2 is monoubiquitinated and forms nuclear foci with BRCA1 and RAD51 in the S phase of the cell cycle,96 and, for this activation in the S phase, ATR is not required.22 The functional significance and mechanism of activation of the pathway in the S phase remains unknown.
Monoubiquitinated FANCD2 is preferentially retained in the chromatin fraction.61,80 In the chromatin fraction, monoubiquitinated FANCD2 can be coimmunoprecipitated with BRCA2 and FANCE.80 Monoubiquitinated FANCD2 is required for the increase of nuclear foci of BRCA2 and RAD51 in response to DNA damage,80 suggesting a role for BRCA2 and RAD51 downstream of monoubiquitinated FANCD2. RAD51 foci formation is an important marker for the integrity of upstream HR machinery. BRCA2 is required for the increase of RAD51 foci after DNA damage. Whether FA proteins other than BRCA2 also increase the assembly of RAD51 foci is controversial. Some reports claim that FA core complex proteins are required for efficient RAD51 foci formation in response to certain types of DNA damage.21,80,97 Other reports claim that these FA proteins are not required for DNA damage–inducible RAD51 foci formation.12,98,101 This discrepancy may reflect the use of different anti-RAD51 antisera and different degrees of genotoxic stress.
A deubiquitinating enzyme, USP1, is a negative regulator of the FA pathway.102 Knockdown of USP1 causes increased monoubiquitination of FANCD2 and cellular resistance to MMC. Although direct deubiquitination of FANCD2 by USP1 has not been demonstrated, USP1 is likely to be the deubiquitinating enzyme for monoubiquitinated FANCD2 in vivo.
Monoubiquitination of PCNA is mediated by RAD6 and RAD18 and is implicated in DNA polymerase switch from a replicative polymerase to a translesion synthesis (TLS) polymerase, DNA polymerase eta (Rad30, XPV), at the site of blocked replication forks.103 These factors are part of the postreplication repair (PRR) pathway and are also required for cellular resistance to ICLs.104 Surprisingly, USP1 deubiquitinates monoubiquitinated PCNA,150 suggesting a common shut-off mechanism of 2 different DNA repair pathways required for cellular resistance to ICLs, the FA pathway and the PRR pathway.
Other proteins interacting with FA proteins include α-Spectrin II, FAZF, SNX5, NADPH cytochrome P450 reductase, a molecular chaperone GRP94, cdc2, GSTP1, CYP2E1, BRG1, STAT1, Hsp70, PKR, menin, and many other proteins identified in yeast 2 hybrid screens (summarized in Reuter et al105 ). The functional relevance of these protein-protein interactions remains largely unknown.
Localization of FA proteins in chromatin
Until recently, the FA core complex was believed to be a soluble nuclear complex whose primary function was to monoubiquitinate FANCD2. The identification of 2 new FA proteins with helicase motifs (FANCM and FANCJ) provide strong evidence that the FA complex proteins may in fact move in and out of the chromatin, depending on the state of DNA damage or cell-cycle phase.
The FA core complex does not simply function as the ubiquitin ligase for FANCD2. Increasing evidence suggests roles of components of FA core complex in the chromatin. First, FANCA, FANCC, and FANCG proteins are localized to chromatin in G1/S/G2 phases, and chromatin localization of these proteins increases after MMC treatment.106,108 Second, FANCE and FANCC are reported to form nuclear foci and colocalize with FANCD2 after DNA damage.79,93 FANCG also forms ICL-induced nuclear foci and colocalizes with BRCA2 and RAD51.94 Third, FANCE interacts with BRCA2 and FANCD2 in chromatin.80 Fourth, FANCM has a helicase motif and a degenerate nuclease motif, suggesting an important role for the FA core complex in moving along the DNA and sensing damage. FANCM also has DNA-stimulated ATPase activity and translocase activity, and it obviously interacts with DNA.5 Consistent with these findings, a recent report using a chicken FANCD2-monoubiquitin fusion protein suggests at least 2 more functions of the FA core complex other than monoubiquitination of FANCD2.93 According to that report, the monoubiquitination of FANCD2 is not sufficient for translocation of FANCD2 into chromatin. Even after FANCD2 is monoubiquitinated, the FA core complex is required for translocation of FANCD2 into chromatin. Furthermore, even after FANCD2 is localized in chromatin, the FA core complex is required for cellular resistance to ICLs.93 Those findings indicate that FA core complex is not merely the ubiquitin ligase for FANCD2. The FA core complex may ubiquitinate other substrates or may have functions other than ubiquitination.
Other FA proteins are also known to interact with DNA. FANCJ is a DNA-dependent ATPase and a 5′-to-3′ DNA helicase, which clearly interacts with DNA.52 Monoubiquitinated FANCD2 is mainly localized in the chromatin fraction, and purified FANCD2 protein has direct DNA-binding activity with specificity for dsDNA ends and Holliday junctions,109 suggesting a direct role of FANCD2 in DNA repair. BRCA2 interacts with DNA through OB folds and tower domain73 and recruits a recombinase, RAD51.
Some non-FA proteins interacting with FA proteins also interact with DNA. BLM is a part of the BRAFT complex,18 and ICL-induced nuclear foci formation of BLM depends on FANCC,19,20 FANCG,19 and FANCD2.20 ICL-induced phosphorylation of BLM depends on FANCC and FANCG but not on FANCD2.19 BLM colocalizes with FANCD2 in ICL-induced nuclear foci and can be coimmunoprecipitated with monoubiquitinated FANCD2.19,20 BLM-deficient cells are sensitive to ICLs to some extent.19 BLM has 3′-to-5′ DNA helicase activity and is associated with topoisomerase III α, which can break and rejoin DNA to alter its topology, and ssDNA binding protein, RPA.
The NBS1-MRE11-RAD50 complex also interacts with FA proteins.17,21 MRE11 has 3′-to-5′ dsDNA exonuclease and ssDNA and dsDNA endonuclease activity.25 NBS1-deficient cells and MRE11-deficient cells are sensitive to ICLs.17 FANCD2 and NBS1 partially colocalize in MMC-induced nuclear foci and are coimmunoprecipitated.17 FA core complex is required for phosphorylation of NBS1 and nuclear foci formation of MRE11 in response to ICLs specifically.21,110
XPF is an endonuclease required for incision of ICLs and forms a heterodimer with ERCC1. XPF-deficient cells and ERCC1-deficient cells are hypersensitive to ICLs. XPF colocalizes with FANCA in ICL-induced nuclear foci.111 A related endonuclease, MUS81-Eme1, which is required for cellular resistance to ICLs, may also have interactions with the FA pathway.
TLS DNA polymerases (REV1 and REV3) are also implicated in the FA pathway. Chicken DT40 cells deficient in REV3 or REV1 show crosslinker hypersensitivities that are epistatic to FANCC-deficient cells.112 Furthermore, REV1 and FANCD2 colocalize in nuclear foci on replication arrest.112 This raises the interesting possibility that FA proteins and error-prone TLS polymerases work in a common pathway, which is consistent with the reported hypomutability in FA cells.113
The important question is how these factors (helicases, nucleases, a topoisomerase, DNA polymerases, a recombinase, and other DNA-binding proteins) assemble in chromatin and repair damaged DNA lesions (especially ICLs) (Figure 2B). Step-by-step analysis of DNA repair intermediates from ICL lesions will be important. Investigation of the requirement of each factor for the formation of such intermediates will be required to envision the whole repair process.114
Cellular defects of DNA repair in FA
Increasing evidence suggests that FA cells are defective in HR, although there is some inconsistency among reports (Table 2). BRCA2-deficient cells show a severe decrease in overall HR efficiency.69,115,116 A mild HR defect in other FA gene–deficient cells (FA-A,116,117 FA-C,20,112 FA-D2,99,116 FA-G,100,116 and FA-J12 ) has been reported, although normal HR efficiency in FA-C,89 FA-D2,101 FA-J,89 and FA-M118 cells has also been reported. Some of these discrepancies may be explained by the difference in cell types used and systems used for measurement of HR. Overall, there seems to be at least a mild defect of HR in FA cells.
Defect of homologous recombination/single-strand annealing in FA cells
. | . | SCE . | . | . | . | |
|---|---|---|---|---|---|---|
| Subtype, cell type . | Efficiency of HR . | Spontaneous . | ICL induced . | SSA . | References . | |
| FA-A | ||||||
| Human FA-A fibroblast (SV40 transformed) | Decreased | NA | NA | Decreased | Nakanishi et al116 | |
| Fanca-/- mouse fibroblast | Decreased | NA | NA | Decreased | Yang et al117 | |
| FA-C | ||||||
| Chicken DT40 | Decreased | Increased | Poor response | NA | Niedzwiedz et al112 | |
| Chicken DT40 | Decreased | Increased | Poor response | NA | Hirano et al20 | |
| Chicken DT40 | Normal | Increased | NA | NA | Bridge et al89 | |
| FA-D1 | ||||||
| Human Capan1 pancreatic cancer (Brca2 mt) | Decreased | NA | NA | NA | Moynahan et al69 | |
| Mouse ES cells with a Brca2 exon 27 deletion | Decreased | NA | NA | Increased | Moynahan et al,69 Stark et al115 | |
| Chinese hamster cell (VC-8) | Decreased | NA | NA | Increased | Nakanishi et al,116 Yang et al117 | |
| FA-D2 | ||||||
| Human FA-D2 fibroblast (SV40 transformed) | Decreased | NA | NA | Decreased | Nakanishi et al116 | |
| Human FA-D2 fibroblast (SV40 transformed) | Normal | NA | NA | NA | Ohashi et al101 | |
| Chicken DT40 | Decreased | Increased | Poor response | NA | Yamamoto et al99 | |
| FA-G | ||||||
| Human FA-G fibroblast (SV40 transformed) | Decreased | NA | NA | Decreased | Nakanishi et al116 | |
| Chicken DT40 | Decreased | NA | NA | NA | Yamamoto et al99 | |
| FA-J | ||||||
| Human MCF7 breast cancer (FANCJ siRNA) | Decreased | NA | NA | NA | Litman et al12 | |
| Chicken DT40 | Normal | Increased | NA | NA | Bridge et al89 | |
| FA-M | ||||||
| Chicken DT40 | Normal | NA | NA | NA | Mosedale et al118 | |
. | . | SCE . | . | . | . | |
|---|---|---|---|---|---|---|
| Subtype, cell type . | Efficiency of HR . | Spontaneous . | ICL induced . | SSA . | References . | |
| FA-A | ||||||
| Human FA-A fibroblast (SV40 transformed) | Decreased | NA | NA | Decreased | Nakanishi et al116 | |
| Fanca-/- mouse fibroblast | Decreased | NA | NA | Decreased | Yang et al117 | |
| FA-C | ||||||
| Chicken DT40 | Decreased | Increased | Poor response | NA | Niedzwiedz et al112 | |
| Chicken DT40 | Decreased | Increased | Poor response | NA | Hirano et al20 | |
| Chicken DT40 | Normal | Increased | NA | NA | Bridge et al89 | |
| FA-D1 | ||||||
| Human Capan1 pancreatic cancer (Brca2 mt) | Decreased | NA | NA | NA | Moynahan et al69 | |
| Mouse ES cells with a Brca2 exon 27 deletion | Decreased | NA | NA | Increased | Moynahan et al,69 Stark et al115 | |
| Chinese hamster cell (VC-8) | Decreased | NA | NA | Increased | Nakanishi et al,116 Yang et al117 | |
| FA-D2 | ||||||
| Human FA-D2 fibroblast (SV40 transformed) | Decreased | NA | NA | Decreased | Nakanishi et al116 | |
| Human FA-D2 fibroblast (SV40 transformed) | Normal | NA | NA | NA | Ohashi et al101 | |
| Chicken DT40 | Decreased | Increased | Poor response | NA | Yamamoto et al99 | |
| FA-G | ||||||
| Human FA-G fibroblast (SV40 transformed) | Decreased | NA | NA | Decreased | Nakanishi et al116 | |
| Chicken DT40 | Decreased | NA | NA | NA | Yamamoto et al99 | |
| FA-J | ||||||
| Human MCF7 breast cancer (FANCJ siRNA) | Decreased | NA | NA | NA | Litman et al12 | |
| Chicken DT40 | Normal | Increased | NA | NA | Bridge et al89 | |
| FA-M | ||||||
| Chicken DT40 | Normal | NA | NA | NA | Mosedale et al118 | |
Efficiency of HR was not reported for the following subtypes: FA-B, FA-E, FA-F, FA-I, and FA-L.
HR indicates homologous recombination; SCE, sister chromatid exchange; SSA, single-strand annealing; NA, not available; ES, embryonic stem.
Spontaneous sister chromatid exchange (SCE) is elevated in chicken DT40 cells deficient in FANCC,20,112 FANCD2,99 or FANCJ.89 These are intriguing data, because (1) SCE is thought to be mediated by HR and (2) in human FA cells spontaneous SCE is not elevated. One speculative explanation is that in FA cells a subpathway of HR which is not required for spontaneous SCE may be affected99 and that something that masks increased SCE in human FA cells may be missing in chicken DT40 cells. Interestingly, in DT40 cells, increased SCE in FANCC-deficient cells is epistatic with BLM-deficient cells, indicating a functional linkage between FANCC and BLM in suppressing SCE.99
BRCA2-deficient cells show increased single-strand annealing (SSA).115,116 In contrast, cells deficient in BRCA1, FANCA, FANCG, or FANCD2 show reduced SSA,115,117 suggesting a role of FA proteins (other than BRCA2) in common steps for both SSA and HR.
HR activity is elevated in FA-C and FA-G cells in assays using extra-chromosomal plasmid-based templates,119 suggesting that regulation of HR by FA proteins outside of the chromosome may be different from regulation in the chromosome. In plasmid-based end-joining assays, the end-joining fidelity of DNA double-strand breaks is low in FA cells.120
FA cells are generally not hypersensitive to IR,121 but clinically some patients with FA show increased sensitivity to IR (unexpected toxicity as a result of radiation therapy).122 In the absence of DNA-PK, which is an essential component of nonhomologous end joining (NHEJ) repair and which phosphorylates many substrates in response to DNA damage,123 Fancd2 knockout sensitizes fibroblasts and mice to IR.124 These results suggest that normal NHEJ repair may mask the IR hypersensitivity of FA cells. This may explain inconsistent IR sensitivities among FA cells and patients with FA.
Common fragile sites are chromosomal loci that preferentially exhibit gaps and breaks when cells have been cultured under replicative stress. FA cells, ATR-deficient cells, and BRCA1-deficient cells show increased common fragile site instability, further supporting the idea of FA pathway interactions.125
Function of FA proteins in intra-S phase cell-cycle checkpoints
The FANCD2 protein has been implicated in IR-induced intra-S phase checkpoint.16 After IR exposure, FANCD2 is phosphorylated by an IR-activated signaling kinase called ATM on several residues, including serine 222. ATM kinase phosphorylates and activates proteins involved in cell-cycle checkpoint responses, including p53, CHK2, NBS1, SMC1, BRCA1, and FANCD2.24 Normal cells stop synthesizing DNA in response to exposure to IR; this response is called intra-S phase checkpoint. Cells deficient in ATM, or some substrates of ATM, such as CHK2, NBS1, SMC1, BRCA1, and FANCD2, do not stop synthesizing DNA after exposure to IR. IR-inducible ATM-dependent phosphorylation of FANCD2 on serine 222 is required for establishment of IR-induced intra-S phase checkpoint but is not required for ICL resistance.16 This phosphorylation also depends on NBS1.17 In contrast, monoubiquitination of FANCD2 on lysine 561 is required for ICL resistance, but it is not required for intra-S phase checkpoint. Therefore, FANCD2 has 2 independent functions, resulting from 2 independent posttranslational modifications.
FA cells fail to arrest DNA synthesis in response to ICLs, whereas normal cells arrest DNA replication. This means FA cells have a defect in the ICL-induced S phase checkpoint function, which is similar to a defect in IR-induced intra-S phase checkpoint function seen in ATM-, NBS1-, or FANCD2-deficient cells. The ICL-induced ATR-dependent phosphorylations of NBS1 and FANCD2 seem to be required for the establishment of ICL-induced intra-S phase checkpoint.23
Other functions of FA proteins
FA cells are hypersensitive to oxygen, and FA proteins have been implicated in the handling of oxidative stress.126 Interaction of FANCC protein and some cytoplasmic proteins involved in the handling of reactive oxygen species (for example, GSTP1) has been reported.127 FANCC has also been implicated in JAK/STAT signaling and apoptotic signaling.68,128,132 These functions of FA proteins, which appear to fall outside of DNA repair, are reviewed elsewhere133 and are beyond the scope of this article.
Cancer in murine models for FA
Generation of Fanca, Fancc, Fancg, Fancd2, Fanca-Fancc double, and Fancl/Pog knockout mice have been reported. Importantly, some of these FA mice develop tumors. Fancd2 knockout mice develop tumors, including adenocarcinoma and lymphoma.134 Fanca knockout mice develop lymphoma, sarcoma, and ovarian granulosa cell tumors.135 Fancc knockout mice develop mammary adenocarcinoma and histiocytic sarcoma.136 Fancc knockout reduces latency to tumor development of p53–/– and p53+/– mice.137 Heterozygosity for p53 (p53+/–) accelerates epithelial tumor formation in Fancd2 knockout mice.138 Many Brca1- or Brca2-deficient mice models with cancer susceptibility have been published (reviewed in Moynahan139 ). Those results demonstrate the importance of the FA pathway in tumor suppression.
Inactivation of the FA pathway in human cancer in the general population
Abnormalities of the FA pathway have been identified in a wide variety of human cancers, suggesting a molecular mechanism for their sensitivity to chemotherapy with DNA crosslinking agents (cisplatin, MMC, and melphalan). Table 3 summarizes recent reports describing abnormalities of the FA genes (except for BRCA2) in cancer in the general population.30,38,140,144 Abnormalities of BRCA1/2 in cancer have been reviewed elsewhere.40
Abnormalities of FA genes in human cancer in the general population
Gene, type of abnormality, tissue . | Frequency in clinical samples (%) . | Cell lines with FA abnormalities . | Reference . |
|---|---|---|---|
| FANCF | |||
| Methylation | |||
| Ovarian cancer | 4/19 (21) | TOV-21G, 2008, C13*, OAW42 | Taniguchi et al30 |
| Stage III/IV epithelial ovarian tumors | 0/? (0) | NA | Teodoridis et al140 |
| Granulosa cell tumors of the ovary | 6/25 (24) | NA | Dhillon et al35 |
| Breast cancer | 13/75 (17) | NA | Olopade and Wei32 |
| Non-small cell lung cancer | 22/158 (14) | NA | Marsit et al33 |
| Head and neck squamous cell carcinoma | 13/89 (15) | NA | Marsit et al33 |
| Cervical cancer | 27/91 (30) | SiHa, ME-180, SW756 | Narayan et al34 |
| Testicular germ cell tumor (nonseminoma) | 4/60 (6.7) | NA | Koul et al36 |
| Acute myeloid leukemia | 0/36 (0) | CHRF-288 | Tischkowitz et al31 |
| Decreased expression (unknown mechanism) | |||
| Breast cancer | NA | UACC812 | van der Heijden et al141 |
| FANCA | |||
| Reduced expression (heterozygous deletion) | |||
| Acute myeloid leukemia (adult) | 4/101 (4.0) | NA | Tischkowitz et al143 |
| Point mutation | |||
| Acute myeloid leukemia (adult) | 6/79 (7.6) | NA | Condie et al142 |
| Functional abnormality (unknown mechanism) | |||
| Acute myeloid leukemia (adult) | NA | UoC-M1 | Lensch et al144 |
| FANCC | |||
| Point mutation/frameshift + LOH | |||
| Pancreatic cancer | 2/22 (9)* | NA | van der Heijden et al37 |
| Large homozygous deletion | |||
| Pancreatic cancer | NA | PL11 | van der Heijden et al38 |
| FANCG | |||
| Point mutation (nonsense) + LOH | |||
| Pancreatic cancer | 0/22 (0)† | Hs766T | van der Heijden et al37 |
| Unknown | |||
| Lack of monoubiquitination of FANCD2 | |||
| Head and neck squamous cell carcinoma | NA | FaDu | van der Heijden et al141 |
Gene, type of abnormality, tissue . | Frequency in clinical samples (%) . | Cell lines with FA abnormalities . | Reference . |
|---|---|---|---|
| FANCF | |||
| Methylation | |||
| Ovarian cancer | 4/19 (21) | TOV-21G, 2008, C13*, OAW42 | Taniguchi et al30 |
| Stage III/IV epithelial ovarian tumors | 0/? (0) | NA | Teodoridis et al140 |
| Granulosa cell tumors of the ovary | 6/25 (24) | NA | Dhillon et al35 |
| Breast cancer | 13/75 (17) | NA | Olopade and Wei32 |
| Non-small cell lung cancer | 22/158 (14) | NA | Marsit et al33 |
| Head and neck squamous cell carcinoma | 13/89 (15) | NA | Marsit et al33 |
| Cervical cancer | 27/91 (30) | SiHa, ME-180, SW756 | Narayan et al34 |
| Testicular germ cell tumor (nonseminoma) | 4/60 (6.7) | NA | Koul et al36 |
| Acute myeloid leukemia | 0/36 (0) | CHRF-288 | Tischkowitz et al31 |
| Decreased expression (unknown mechanism) | |||
| Breast cancer | NA | UACC812 | van der Heijden et al141 |
| FANCA | |||
| Reduced expression (heterozygous deletion) | |||
| Acute myeloid leukemia (adult) | 4/101 (4.0) | NA | Tischkowitz et al143 |
| Point mutation | |||
| Acute myeloid leukemia (adult) | 6/79 (7.6) | NA | Condie et al142 |
| Functional abnormality (unknown mechanism) | |||
| Acute myeloid leukemia (adult) | NA | UoC-M1 | Lensch et al144 |
| FANCC | |||
| Point mutation/frameshift + LOH | |||
| Pancreatic cancer | 2/22 (9)* | NA | van der Heijden et al37 |
| Large homozygous deletion | |||
| Pancreatic cancer | NA | PL11 | van der Heijden et al38 |
| FANCG | |||
| Point mutation (nonsense) + LOH | |||
| Pancreatic cancer | 0/22 (0)† | Hs766T | van der Heijden et al37 |
| Unknown | |||
| Lack of monoubiquitination of FANCD2 | |||
| Head and neck squamous cell carcinoma | NA | FaDu | van der Heijden et al141 |
Values in column labeled “Frequency in clinical samples” indicates number of cases with the identified abnormality out of total number of cases.
? indicates number unknown; NA, not available.
LOH at 9q22.3.
LOH at 9p13.
Ovarian tumor cells are initially sensitive to cisplatin but become refractory to cisplatin over time. Understanding the molecular basis of cisplatin sensitivity and acquired resistance is critical to the management of this tumor. In 2 ovarian cancer cell lines (TOV-21G and 2008), methylation of the promoter region of the FANCF gene causes suppression of FANCF expression and increased sensitivity to MMC and cisplatin.30 FANCF was partially demethylated, and FANCF expression was restored in a cisplatin-resistant derivative of the 2008 cell line, suggesting that reactivation of the FA pathway is a mechanism for acquired resistance. At least in the cell lines, the integrity of the FA pathway is one of the critical determinants of cisplatin resistance.30 In contrast, the significance of FANCF methylation in clinical samples of ovarian cancer remains unclear. In the initial report, it was detected in 21% of a relatively small number of samples,30 but one subsequent report with a larger number of samples failed to detect FANCF methylation in stage III and IV epithelial ovarian tumors.140 The true prevalence in epithelial ovarian cancer may be lower than initially estimated. FANCF methylation was found in 24% of clinical samples of a relatively rare subtype of nonepithelial ovarian tumor, granulosa cell tumors.35
For lung adenocarcinoma, FANCF methylation is a significant predictor of poor survival, and for non–small cell lung cancer, it is associated with a shorter duration of tobacco use.33 In lung SCC, increased p16INK4a homozygous deletion occurred at a higher frequency in those with FANCF methylation.145 In HNSCC, FANCF methylation is associated with a greater number of years of alcohol drinking.33 Interpretation of these associations is difficult, but FANCF methylation seems to have some clinical significance in these types of cancers.
In cervical cancer, FANCF methylation was frequently observed (30%).34 Patients younger than 45 years showed a higher frequency of FANCF methylation, suggesting that FANCF methylation may play a role in initiation or progression of cervical cancer in younger patients.34
Germ cell testicular cancer (nonseminoma type) is usually highly responsive to cisplatin. FANCF methylation was found in a small subset (6.7%),36 indicating that the role of FANCF methylation in general cisplatin sensitivity of nonseminoma is limited.
FANCA abnormalities are involved in a small subset of AML. FANCA missense mutations were found in 7.6% of cases of adult AML, although functional relevance of most of these mutations is unknown.142 Heterozygous deletion of FANCA with reduced expression of FANCA was identified in 4% of adult patients with AML.143
Inherited and somatic mutations of FANCC and FANCG are present in a subset of young-onset pancreatic cancer.37 Two germ line–truncating FANCC mutations associated with loss of heterozygosity in tumor samples were identified in cases of young-onset pancreatic cancer,39 suggesting that inherited mutations in FANCC may predispose to pancreatic cancer.
Melphalan is widely used in the treatment of multiple myeloma. A comparison of a melphalan-sensitive myeloma cell line and its melphalan-resistant derivative identified FANCF as a gene expressed significantly higher in the resistant derivative.146 Modulation of FANCF expression in a myeloma cell line by siRNA or by overexpression of FANCF causes melphalan sensitivity or resistance, respectively.41 Whether the FA pathway is inactivated in clinical samples of myeloma remains to be investigated.
One possible therapeutic approach for DNA crosslinker-resistant cancers is sensitization of tumor to DNA crosslinkers using a specific small-molecule inhibitor of the FA pathway. As a proof of principle, it has been shown that suppression of the FA pathway by an adenovirus overexpressing dominant-negative FANCA sensitizes tumor cells to cisplatin.147
Concluding comments and unanswered questions
The 11 identified FA proteins appear to cooperate in a common pathway, regulating the repair of interstrand DNA crosslinks. The characterization of the cloned FA genes and proteins has allowed new diagnostic approaches to FA and suggests novel treatment options for patients with FA.
Critical questions remain for the FA pathway. First, although FANCD2 monoubiquitination is a required event in this pathway, little is known about the downstream function of this posttranslational modification. For instance, monoubiquitinated FANCD2 may activate crosslink repair, bind to a novel TLS polymerase, or bind other DNA repair subunits. Second, the role of the pathway is primarily in DNA damage response. Yet, the role of the new DNA binding components (FANCM and FANCJ) in damage recognition versus DNA repair is largely unknown. Third, it will be important to identify the FANCI protein and perhaps other proteins in the pathway.
Finally, further examination of the role of the FA pathway in the cause and treatment of cancers in the general population is warranted. As is often the case with the study of a rare pediatric disease, FA research has already led to general insights to other more prevalent human diseases, such as aplastic anemia, infertility, and cancer.
Prepublished online as Blood First Edition Paper, February 21, 2006; DOI 10.1182/blood-2005-10-4240.
An Inside Blood analysis of this article appears at the front of this issue.
We thank Hans Joenje, Weidong Wang, Kevin Hiom, K. J. Patel, Sharon Cantor, and Minoru Takata for sharing data prior to publication and Blanche Alter for helpful discussions. We thank Arleen D. Auerbach for permission to use unpublished data regarding the prevalence of the different FA complementation groups. T.T. is a Searle Scholar and a V Scholar. We apologize that we cannot cite some of the literature on FA with significance because of the limitation of space.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal