Natural antisense viral transcripts have been recognized in retroviruses, including human T-cell leukemia virus type 1 (HTLV-1), HIV-1, and feline immunodeficiency virus (FIV), and have been postulated to encode proteins important for the infection cycle and/or pathogenesis of the virus. The antisense strand of the HTLV-1 genome encodes HBZ, a novel nuclear basic region leucine zipper (b-ZIP) protein that in overexpression assays down-regulates Tax oncoprotein-induced viral transcription. Herein, we investigated the contribution of HBZ to HTLV-1–mediated immortalization of primary T lymphocytes in vitro and HTLV-1 infection in a rabbit animal model. HTLV-1 HBZ mutant viruses were generated and evaluated for viral gene expression, protein production, and immortalization capacity. Biologic properties of HBZ mutant viruses in vitro were indistinguishable from wild-type HTLV-1, providing the first direct evidence that HBZ is dispensable for viral replication and cellular immortalization. Rabbits inoculated with irradiated cells expressing HTLV-1 HBZ mutant viruses became persistently infected. However, these rabbits displayed a decreased antibody response to viral gene products and reduced proviral copies in peripheral blood mononuclear cells (PBMCs) as compared with wild-type HTLV-1–infected animals. Our findings indicated that HBZ was not required for in vitro cellular immortalization, but enhanced infectivity and persistence in inoculated rabbits. This study demonstrates that retroviruses use negative-strand–encoded proteins in the establishment of chronic viral infections.
Introduction
Human T-cell leukemia virus type 1 (HTLV-1) infection causes adult T-cell leukemia/lymphoma and is associated with a variety of lymphocyte-mediated diseases.1 Although infected subjects develop a vigorous and sustained immune response against the virus, infection is typically lifelong. The positive sense RNA genome of HTLV-1 encodes the typical retroviral structural and enzymatic genes Gag, Pol, and Env, the positive regulatory gene products Tax and Rex, and several accessory gene products, p12I, p27I, p13II, and p30II, which are important for viral infectivity and persistence in vivo.2-4 Tax, the transcriptional activator, is an important modulator of both viral and cellular gene expression,5 and its expression is essential for HTLV-mediated cellular transformation of T lymphocytes.6-8 Tax transactivates not only the viral gene promoter, but many host cellular promoters through cAMP response element-binding protein (CREB), nuclear factor κB (NF-κB), and serum response factor (SRF) pathways.9,10 The stimulation of such pathways by Tax leads to unregulated protein expression and heightened activation in signaling cascades, including Janus kinase/signal transducer and activator of transcription (JAK/STAT), phosphatidylinositol 3-kinase (PI3K), and c-Jun N-terminal kinase (JNK).11-14 It has also been shown that Tax can bind to cyclin-dependent kinase holoenzymes, inactivate tumor supressors such as p53 and discs large protein (DLG), and has the ability to silence cellular checkpoints.15-19 These pleiotropic effects of Tax on the cell have suggested that there may be differences between the initiation and maintenance of transformation.20
The majority of retroviral gene products are encoded by the sense strand of the genome. However, natural antisense viral transcripts have been recognized in retroviruses, including HTLV-1, HIV, and feline immunodeficiency virus (FIV).21-23 The novel HTLV-1 protein HBZ (HTLV-1 b-ZIP factor) is encoded by a minus-strand mRNA that is transcribed by a functional promoter present in the antisense strand of the HTLV-1 proviral genome (Larocca et al21 and Cavanagh et al24 ). Since exogenously overexpressed HBZ down-regulates Tax-induced HTLV-1 transcription and interacts with and disrupts the DNA binding activity of JunB and c-Jun (AP-1 components),25,26 it has been hypothesized that HBZ may play an important role in HTLV-1 biology by counteracting the effects of Tax at the transcriptional level and attenuating the activation of AP-1.
In this study, we evaluated the functional role of HBZ in the context of an infectious molecular clone and determined its contribution to viral-induced immortalization in vitro and viral replication kinetics and persistence in vivo. Our findings indicated that the reported repressive effects of HTLV-1 HBZ on Tax and AP-1 were not sufficient to disrupt the capacity of the virus to immortalize primary T lymphocytes in vitro, but rather, enhanced infectivity and modified virus persistence in vivo.
Materials and methods
Cells
293T cells and 729 B cells were maintained in Dulbecco modified Eagle and Iscove medium, respectively. The media were supplemented to contain 10% fetal bovine serum (FBS), 2 mM glutamine, penicillin (100 U/mL), and streptomycin (100 μg/mL). Human peripheral blood mononuclear cells (hPBMCs) were isolated using Ficoll Hypaque (Amersham, Piscataway, NJ) and cultured in RPMI 1640 medium supplemented with 20% fetal bovine serum (FBS), 2 mM glutamine, and antibiotics in the presence or absence of 10 U/mL recombinant human interleukin-2 (IL-2; Roche Applied Biosciences, Indianapolis, IN). The protocol to obtain human blood was approved by The Ohio State University human subjects internal review board.
Plasmids
The plasmid containing the wild-type HTLV-1 (wtHTLV-1) infectious proviral clone, pACHneo, was used in this study.27,28 HTLV-1HBZΔLZ was generated by changing the sequence 6760ACTCCA6765 to GCTAGC, which introduced a diagnostic NheI restriction enzyme site that truncates or terminates the HBZ reading frame at amino acid 175 of the published sequence29 (amino acid 172 of the HBZ major transcript24 ) by deleting the majority of the C-terminal leucine zipper (LZ) region. This mutation did not alter any other known HTLV-1 open reading frames (ORFs). HTLV-1ΔHBZ was generated by introducingaGtoA point mutation (nt 7258) that resulted in termination of the HBZ reading frame at amino acid 11 of the published sequence29 (amino acid 8 of the HBZ major transcript24 ). This single base mutation is 30 nucleotides 5′ to the p13II ATG on the sense strand and resulted in an Arg to Gln amino acid change in the accessory protein p30II. This p30II mutant displays a wild-type phenotype in both the transcriptional and posttranscriptional assays previously used to define p30II function in vitro30,31 (data not shown). HBZ and HBZΔLZ cDNA expression vectors (pME vector–based) were generated from the ACH proviral clone sequences (based on the HBZ major transcript24 ). The long terminal repeat 1 (LTR-1)–Luc and thymidine kinase (TK)–Renilla were described previously.30
Transfection, luciferase assay, and detection of viral p19 matrix antigen
To measure Tax function, 1.5 × 105 293T cells were transfected using Lipofectamine (Invitrogen, Carlsbad, CA). The amount of DNA was kept constant and was composed of 0.1 μg of LTR-1–Luc reporter and 10 ng TK-Renilla along with 2 μg of an empty plasmid or proviral clones. Cell supernatants (48 hours) were used for p19 enzyme-linked immunosorbent assay (ELISA; Zeptometrix Corporation, Buffalo, NY). Cell pellets were lysed and Tax activity was measured in light units as described previously.27 All experiments were performed independently 3 times in triplicate and the results were normalized for transfection efficiency using Renilla luciferase. Stable 729 transfectants containing proviral clones were isolated as described,27 and cell clones were screened by p19 Gag expression in the cell supernatant by ELISA.
DNA preparation and standard PCR
DNA was isolated from 729 producer cell lines, immortalized human PBMCs, or infected rabbit PBMCs using PURGENE DNA purification system (Gentra, Minneapolis, MN). DNA (0.5 μg) was subjected to 35-cycle polymerase chain reaction (PCR; 95° for 5 minutes, 95°C for 1 minute, 56°C for 1 minute, and 72°C for 1 minute, followed by 72°C for 10 minutes and held at 4°C). The HTLV-1–specific primer pair 7145GTCAAGCACAGCTTCCTCCTCCTC7168 and 7386GGGGCACCAGTCGCCTTGTACACA7363 was used to amplify a 241–base pair fragment for sequencing to confirm the ΔHBZ mutation. The HTLV-1–specific primer pair H1JA16659 TTATTGCAACCACATCGCCTCCAGCCTCCC6688 and H1JA2 6885AGGAGCGCCGTGAGCGCAAGT6865 was used to amplify a 226–base-pair fragment that was subjected to NheI diagnostic digestion and sequenced to confirm the HBZΔLZ mutation.
RT-PCR and quantitative Taqman real-time RT-PCR
RNA was extracted using the RNeasy kit (Quiagen, Valencia, CA) from SLB-1, 729.ACHneo, and 729 uninfected control cells. Total RNA was subjected to 3 consecutive DNase treatments followed by OLIGOTEX polyA+ mRNA isolation (Quiagen). PolyA+ mRNA (25 ng) was subjected to reverse transcriptase (RT)–PCR using the first-strand synthesis kit and the primer HMS6659 TTATTGCAACCACATCGCCTCCAGCCTCCC6688 (1 μM) to generate cDNA from the minus strand HBZ transcript RNA. Duplicate reactions were performed in the presence and absence of RT to control for DNA background/contamination. The cDNAs then were subjected to standard PCR using the HTLV-1–specific primer pair H1JA1 and H1JA2. Human GAPDH was amplified using primers previously described.32 PCR-amplified fragments were separated on a 1% agarose gel and visualized by ethidium bromide staining. Forty cycles of real-time Taqman PCR (Applied Biosystems, Foster City, CA) were conducted to quantitate proviral copy number per cell in infected rabbit PBMCs. Rabbit PBMC DNA was amplified in duplicate using the primers TaxS 7335CGGATACCCAGTCTACGTGTTT7356 and TaxAS 7495CTGAGCCGATAACGCGTCCA7476 and probe (5′-FAM-7456ATCACCTGGGACCCCATCGATGGA7476-TAMARA-3′) and final values were averaged. The 25-μL reactions contained 500 ng rabbit PBMC DNA, 100 ng (25 ng/mL) of each primer and probe concentration of 100 pmol/μL. Copy number was determined based on a standard curve generated from duplicate samples of log10 dilutions of a plasmid containing the Tax sequences. The copy number per cell value for a sample was generated based on the estimation that 1 μg PBMC DNA is equivalent to 67 300 cells.
Western blot
Western blot was performed as described30 using rabbit anti-pHBZ polyclonal antisera (1:500) and goat antirabbit conjugated with horseradish peroxidase (Santa Cruz Biotechnology, Santa Cruz, CA). Proteins were visualized using the electrochemiluminescence (ECL) Western blot analysis system (Amersham Biosciences, Piscataway, NJ).
Short-term coculture microtiter proliferation and long-term immortalization assays
Short-term microtiter proliferation assays were performed as described previously.33 Briefly, 729 HTLV producer cells (200) were irradiated with 100 Gy (10 000 rad) and cocultured with 104 prestimulated PBMCs in the presence of human (h) IL-2 in 96-well round-bottom plates. Wells were enumerated for growth and split 1:4 at weekly intervals. For long-term immortalization assays, 106 irradiated 729 producer cells were cocultivated with 2 × 106 freshly isolated PBMCs with 10 U/mL hIL-2 in 24-well culture plates.34 The presence of HTLV expression was confirmed at weekly intervals by detection of p19 Gag protein in the culture supernatant using an ELISA. Viable cells also were counted by trypan blue exclusion. Cells inoculated with wtHTLV-1 and HTLV-1 HBZ mutant viruses that continued to produce p19 Gag antigen and proliferate 12 weeks after coculture in the presence of exogenous hIL-2 were identified as immortalized.
Rabbit inoculation procedure
Twelve-week-old specific pathogen–free female New Zealand White rabbits (Hazelton, Kalamazoo, MI) were inoculated via the lateral ear vein with approximately 1 × 107 gamma-irradiated 75 Gy (7 500 rad) 729 viral producer cells or uninfected control cells. At weeks 0, 2, 4, 6, and 8 after inoculation, 10 mL blood was drawn from the central ear artery of each animal. This study protocol was approved by the University Laboratory Animal Resources (ULAR) of The Ohio State University. Serum reactivity to specific viral antigenic determinants was detected using a commercial HTLV-1 Western blot assay (GeneLabs Diagnostics, Singapore). Serum (dilution of 1:400) showing reactivity to Gag (p24 or p19) and Env (gp21 or gp46) antigens was classified as positive for HTLV-1 seroreactivity (data not shown). A commercial ELISA kit (Vironostika HTLV-1 MicroELISA system; BioMerieux, Durham, NC) was used to quantitate HTLV-1 serum antibody. Plasma was diluted 1:12 000 to obtain values in the linear range of the assay and data were expressed as absorbance values.
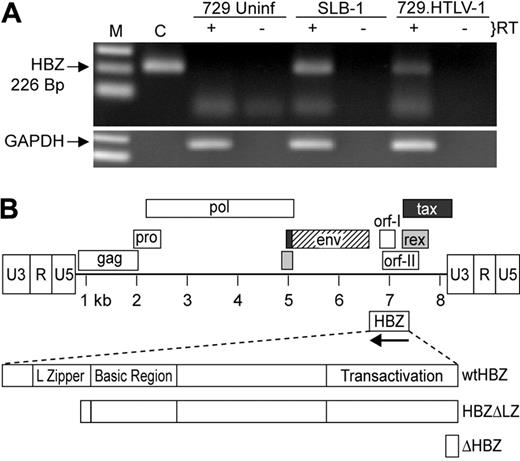
Detection of the HBZ RNA transcript. (A) HBZ transcript was detected in Poly A+ RNA isolated from SLB-1 and 729.HTLV-1, but not 729 uninfected (Uninf), using standard RT-PCR. First-strand synthesis with a specific oligo designed to copy only HTLV-1 antisense RNA containing the HBZ coding sequence was performed in the presence and absence of reverse transcriptase. The 226-bp PCR product was separated on a 2% agarose gel and visualized by ethidium bromide staining. (B) Schematic representation of the complete HTLV-1 proviral genome is shown. LTRs are depicted with their U3, R, and U5 regions. The location of the viral open reading frames and the opposite-strand HBZ are indicated. The reported HBZ coding sequence has been expanded showing the transactivation domain, basic region, and leucine-zipper region, as well as the 2 HBZ truncation mutants generated for this study (HBZΔLZ and ΔHBZ).
Detection of the HBZ RNA transcript. (A) HBZ transcript was detected in Poly A+ RNA isolated from SLB-1 and 729.HTLV-1, but not 729 uninfected (Uninf), using standard RT-PCR. First-strand synthesis with a specific oligo designed to copy only HTLV-1 antisense RNA containing the HBZ coding sequence was performed in the presence and absence of reverse transcriptase. The 226-bp PCR product was separated on a 2% agarose gel and visualized by ethidium bromide staining. (B) Schematic representation of the complete HTLV-1 proviral genome is shown. LTRs are depicted with their U3, R, and U5 regions. The location of the viral open reading frames and the opposite-strand HBZ are indicated. The reported HBZ coding sequence has been expanded showing the transactivation domain, basic region, and leucine-zipper region, as well as the 2 HBZ truncation mutants generated for this study (HBZΔLZ and ΔHBZ).
Results
The HBZ transcript was detected in HTLV-1–transformed and stably transfected cell lines
It was reported previously that the HBZ transcript was present in HTLV-1–infected SLB-1 cells, but in a relatively low quantity compared with tax/rex mRNA.21 In addition, the HBZ protein was detected in the HTLV-1–infected cell line C8166, but could not be detected in MT-2 cells.29 Since our studies make use of the HTLV-1 molecular proviral clone ACH,28 initial experiments were designed to verify the expression of the HBZ mRNA transcript in the HTLV-1 producer cell line 729.ACHneo (referred to as 729.HTLV-1). This cell line produces HTLV-1 that has the capacity to infect and transform human PBMCs.35,36 Poly A+ RNA isolated from SLB-1, 729.HTLV-1, and uninfected 729 cells was subjected to standard RT-PCR in the presence and absence of reverse transcriptase. HBZ mRNA was clearly detected in SLB-1 and 729.HTLV-1, but absent in 729-uninfected cells (Figure 1A). In addition, reactions in which the reverse transcriptase was omitted indicated that no amplified product resulting from DNA contamination was detected. Therefore, the ACH molecular clone produced a minusstrand transcript reported to encode HBZ.
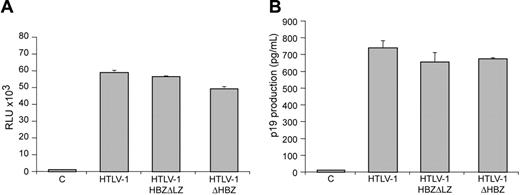
Characterization of proviral clones in vitro. 293 T cells (1.5 × 105) were cotransfected with 2 μg wtHTLV-1, HTLV-1ΔHBZ, HTLV-1HBZΔLZ proviral clones, or negative control DNA along with 0.1 μg LTR-1–Luc and 0.01 μg TK-Renilla. All transfections were performed in triplicate and normalized to TK-Renilla to control for transfection efficiency. Cell lysates or supernatants were harvested 48 hours after transfection. Histograms present the average values from 3 independent experiments; error bars denote (SD). (A) Measure of Tax activity presented as relative luciferase units. (B) Measure of p19 Gag in the cellular supernatants.
Characterization of proviral clones in vitro. 293 T cells (1.5 × 105) were cotransfected with 2 μg wtHTLV-1, HTLV-1ΔHBZ, HTLV-1HBZΔLZ proviral clones, or negative control DNA along with 0.1 μg LTR-1–Luc and 0.01 μg TK-Renilla. All transfections were performed in triplicate and normalized to TK-Renilla to control for transfection efficiency. Cell lysates or supernatants were harvested 48 hours after transfection. Histograms present the average values from 3 independent experiments; error bars denote (SD). (A) Measure of Tax activity presented as relative luciferase units. (B) Measure of p19 Gag in the cellular supernatants.
Construction and characterization of HTLV-1 HBZ mutant proviral clones
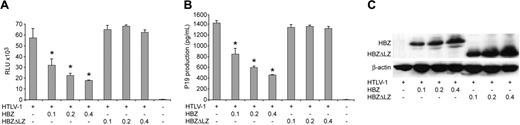
Although informative, studies to date investigating the function or activity of HBZ have been performed in overexpression systems outside the context of the entire provirus. In order to determine the role of HBZ in HTLV-1–mediated cellular immortalization in vitro and viral persistence in vivo, we generated 2 mutant proviral clones. HTLV-1HBZΔLZ has a deletion in the C-terminal leucine zipper that is a key functional domain for b-ZIP proteins and is required for the association of HBZ with CREB-2, JunB, and c-Jun.25,26,29 HTLV-1ΔHBZ has a severely truncated HBZ within the first 10 amino acids (Figure 1B). We first determined whether HBZ mutant proviruses had altered Tax-mediated LTR gene expression. Cotransfection of either HBZ wild-type or HBZ mutant HTLV-1 proviral clones, as a source of Tax, and the LTR-1–Luc reporter revealed no significant difference in LTR-directed gene expression (Figure 2A). Moreover, cells transfected with either HBZ mutant proviral clone produced levels of p19 Gag in the culture supernatant similar to wild-type HTLV-1 (Figure 2B). We next determined the effect of exogenously expressed HBZ on Tax-mediated transcription. Our results indicated that HBZ expressed from a cDNA vector significantly repressed Tax-mediated transcription in a dose-dependent manner (Figure 3A). Importantly, the HBZΔLZ cDNA expression vector failed to repress Tax-mediated transcription (Figure 3A). In direct correlation, the addition of HBZ, but not HBZΔLZ, resulted in a dose-dependent reduction of p19 Gag in the supernatant of transfected cells (Figure 3B). Western blot analysis confirmed that the amount of HBZ or HBZΔLZ protein expressed correlated directly with the amount of plasmid DNA transfected, whereas a control cellular protein, β-actin, remained unchanged (Figure 3C). Taken together, these data support previous published work that the leucine zipper is a key functional domain required for protein interaction and transcriptional repression,25,26,29 and also indicates that the repression is not the result of a significant RNA interference (RNAi) effect on Tax/Rex or Gag/Pol. These results are also consistent with the conclusion that either HBZ is not expressed from the proviral clone following transient transfection or that the levels of HBZ expressed from the proviral clone are below the threshold concentration required for a detectable repression of viral transcription.
Exogenously expressed HBZ results in dose-dependent repression of Tax-mediated transcription and p19 Gag production. 293T cells (1.5 × 105) were cotransfected with 1 μg wtHTLV-1 proviral clone or negative control DNA, 0.1 μg LTR-1–Luc, and 0.01 μg TK-Renilla, and varying concentrations (0.1-0.4 μg) of HBZ or HBZΔLZ expression vectors as indicated. (A) Tax function was measured as firefly luciferase activity (RLU indicates relative light units) from LTR-Luc normalized to Renilla luciferase activity. (B) Culture supernatant was collected from cells in panel A and assayed for p19 Gag production by ELISA. (C) Western blot analysis to confirm increasing concentrations of HBZ and HBZΔLZ used in panels A and B. β-actin levels were assessed as a loading control. *Statistically significant dose-dependent reduction of Tax transactivation activity or p19 Gag production. Statistical significance was determined by analysis of variance (ANOVA) followed by Tukey test. The histogram presents the average values from 3 independent experiments; error bars denote SDs.
Exogenously expressed HBZ results in dose-dependent repression of Tax-mediated transcription and p19 Gag production. 293T cells (1.5 × 105) were cotransfected with 1 μg wtHTLV-1 proviral clone or negative control DNA, 0.1 μg LTR-1–Luc, and 0.01 μg TK-Renilla, and varying concentrations (0.1-0.4 μg) of HBZ or HBZΔLZ expression vectors as indicated. (A) Tax function was measured as firefly luciferase activity (RLU indicates relative light units) from LTR-Luc normalized to Renilla luciferase activity. (B) Culture supernatant was collected from cells in panel A and assayed for p19 Gag production by ELISA. (C) Western blot analysis to confirm increasing concentrations of HBZ and HBZΔLZ used in panels A and B. β-actin levels were assessed as a loading control. *Statistically significant dose-dependent reduction of Tax transactivation activity or p19 Gag production. Statistical significance was determined by analysis of variance (ANOVA) followed by Tukey test. The histogram presents the average values from 3 independent experiments; error bars denote SDs.
HBZ repressed viral gene expression in stable viral producer cell clones
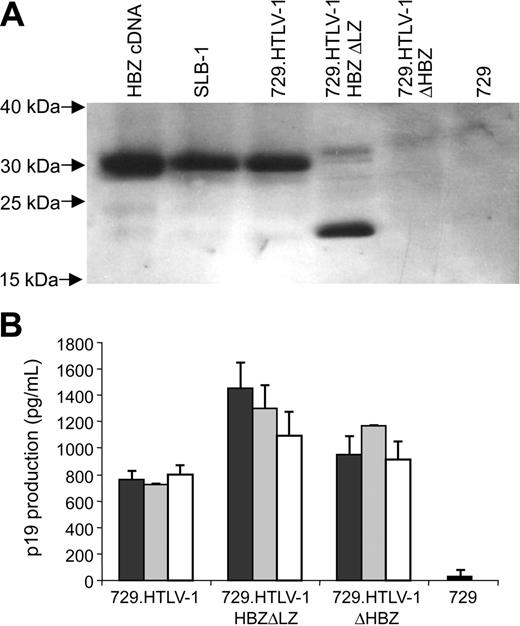
To determine the capacity of HBZ mutant proviral clones to synthesize viral proteins, direct viral replication, and induce cellular immortalization, stable 729 cell transfectants expressing the proviral clones were isolated and characterized. Each stable transfectant contained complete copies of the provirus with the expected mutations (data not shown). Using Western blot analyses, HBZ was detected in the HTLV-1–transformed cell line SLB-1 and the virus producer cell line 729.HTLV-1. A smaller form of HBZ was detected in 729.HTLV-1HBZΔLZ consistent with the truncation and as expected, HBZ was not detected in 729.HTLV-1ΔHBZ or in the negative control 729 (Figure 4A). To monitor the production of another viral protein in these mutant stable transfectants, the concentration of p19 Gag in the culture supernatant of several independently isolated cell clones was quantified by ELISA. p19 Gag expression can be variable from independent stable cell clones attributable to chromosomal location of proviral sequences and overall proviral copy number. However, we consistently observed a significant increase in p19 Gag production in cell clones expressing HBZ mutant proviral clones (Figure 4B), which is consistent with the conclusion that in stable cell lines the loss of HBZ function results in increased viral gene expression.
HBZ and p19 Gag protein expression in stably transfected cell lines. (A) HBZ protein was detected in stable provirus expressing cell lines by Western blot using a rabbit polyclonal antibody against HBZ. HBZ was not detected in 729 control and 729.HTLV-1ΔHBZ as expected. HBZ polypeptide of the expected molecular weight was detected in SLB-1, 729.HTLV-1, and 729.HTLV-1HBZΔLZ. (B) HTLV-1 p19 Gag was quantified by ELISA from 3 independently isolated stable 729 transfectants expressing wild-type HTLV-1, HTLV-1ΔHBZ, or HTLV-1HBZΔLZ. HBZ mutant virus producers expressed statistically greater amounts of p19 Gag than wild-type virus producers, which was consistent with a repressive role of HBZ on viral transcription. p19 Gag production of the 2 mutant virus producers (HTLV-1ΔHBZ and HTLV-1HBZΔLZ) were not statistically significant. Statistical significance was determined by ANOVA followed by Tukey test. The histogram presents the average values from 3 independent experiments; error bars denote SDs.
HBZ and p19 Gag protein expression in stably transfected cell lines. (A) HBZ protein was detected in stable provirus expressing cell lines by Western blot using a rabbit polyclonal antibody against HBZ. HBZ was not detected in 729 control and 729.HTLV-1ΔHBZ as expected. HBZ polypeptide of the expected molecular weight was detected in SLB-1, 729.HTLV-1, and 729.HTLV-1HBZΔLZ. (B) HTLV-1 p19 Gag was quantified by ELISA from 3 independently isolated stable 729 transfectants expressing wild-type HTLV-1, HTLV-1ΔHBZ, or HTLV-1HBZΔLZ. HBZ mutant virus producers expressed statistically greater amounts of p19 Gag than wild-type virus producers, which was consistent with a repressive role of HBZ on viral transcription. p19 Gag production of the 2 mutant virus producers (HTLV-1ΔHBZ and HTLV-1HBZΔLZ) were not statistically significant. Statistical significance was determined by ANOVA followed by Tukey test. The histogram presents the average values from 3 independent experiments; error bars denote SDs.
HBZ-deficient mutants promoted HTLV-1–induced proliferation and immortalization of PBMCs
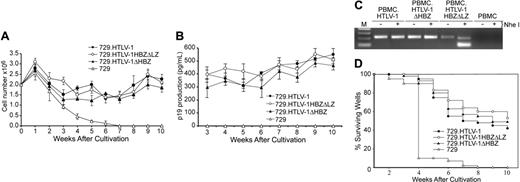
We next assessed the capacity of the HBZ mutant viruses to immortalize human PBMCs in coculture assays. Freshly isolated human PBMCs cocultured with lethally irradiated 729.HTLV-1, 729.HTLV-1ΔHBZ, or 729.HTLV-1HBZΔLZ in the presence of 10 U/mL hIL-2 showed very similar progressive growth patterns consistent with the HTLV-1 immortalization process (Figure 5A). We also detected continuous accumulation of p19 Gag in the culture supernatant, indicating viral replication and virion production (Figure 5B). Immortalized PBMCs harbored the expected HTLV-1 sequences, suggesting that viral transmission was responsible for the immortalization of PBMCs (Figure 5C and data not shown). In an effort to obtain a more quantitative measure of the ability of these viruses to infect and immortalize PBMCs, a fixed number of PBMCs were cocultured with different dilutions of virus-producing cells in a 96-well plate assay.33 Since this assay is very stringent, slowly growing or nondividing cells are eliminated very quickly and the percentage of surviving wells is an accurate measure of the immortalization efficiency of viruses. A Kaplan-Meier plot of HTLV-1–induced T-cell proliferation indicated that the percentage of wells containing proliferating lymphocytes was similar between HTLV-1 and HTLV-1 HBZ mutants (Figure 5D). Taken together, our results are consistent with the conclusion that HBZ is not required for efficient infectivity or HTLV-1–mediated immortalization of primary human T lymphocytes in vitro.
HTLV-1 HBZ enhanced infectivity and persistence in inoculated rabbits
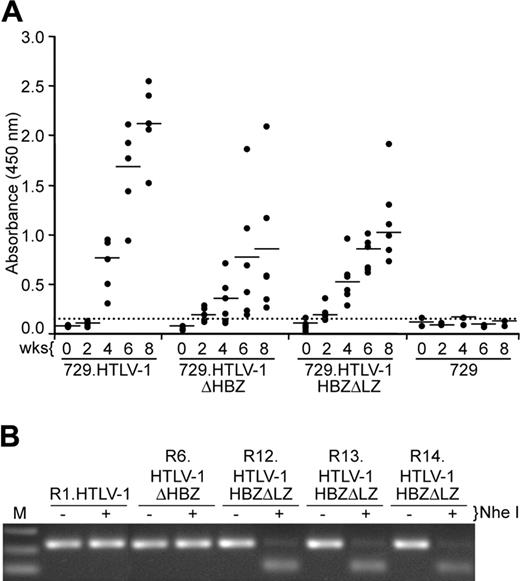
To evaluate the role of HBZ in vivo, we compared the abilities of 729, 729.HTLV-1, 729.HTLV-1ΔHBZ, or 729.HTLV-1HBZΔLZ cell lines to transmit virus to rabbits, which is an established model of infection and persistence.3,37 Rabbits were inoculated with lethally irradiated cell lines (cell inoculums were equilibrated based on their p19 Gag production) and blood was drawn at biweekly intervals. Antibody response to viral antigens was detected by Western blot in all rabbits inoculated with cells expressing either wild-type HTLV-1 or HBZ mutant viruses, and the antibody titers in the majority of the rabbits increased over the time course of the study (data not shown). Moreover, quantitative comparison of antibody responses between each rabbit was performed using a HTLV-specific ELISA (Figure 6A). Statistical analysis of titers at both 4 and 8 weeks after inoculation revealed a significantly lower antibody response to HTLV-1 antigens in the 729.HTLV-1HBZΔLZ– and 729.HTLV-1ΔHBZ–inoculated rabbits compared with the wild-type control group. Consistent with our antibody data, HTLV-1 DNA sequences were detected in all wild-type HTLV-1 and HBZ mutant virus–infected rabbits (Table 1). However, quantitative real-time Taqman PCR revealed that as early as 2 weeks after inoculation, and later at 8 weeks after inoculation, proviral loads in rabbits infected with either HBZ mutant virus were lower than rabbits inoculated with wild-type HTLV-1 (Table 1). Proviral loads in both wild-type and HBZ mutant virus–infected rabbits correlated with the observed antibody responses (Figure 6A). Diagnostic DNA PCR analyses and or nucleotide sequencing performed on PBMCs from rabbits 8 weeks after infection indicated that the infected cells contained the expected viral sequences (Figure 6B and data not shown). Taken together, our results indicated that HBZ, while dispensable for HTLV-1 infection, attenuated parameters of virus replication such as antibody response to viral antigens and proviral loads, which were decreased compared with wild-type HTLV-1–infected rabbits. This attenuation is apparent within 2 weeks after inoculation, suggesting that HBZ function is required early for efficient replication and survival in the host. In addition, since ΔHBZ and HBZΔLZ mutants displayed a similar phenotype in vivo, we concluded that the leucine zipper domain was critical for HBZ functional activity.
Detection and quantification of HTLV-1 DNA in rabbit PBMCs
Inoculum and rabbit . | Weeks after inoculation . | . | . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|
| . | 0 . | 2 . | 4 . | 6 . | 8* . | ||||
| 729.HTLV-1 | |||||||||
| R1 | - | + (0.001) | + | + | + (0.112) | ||||
| R2 | - | + (0.001) | + | + | + (0.108) | ||||
| R3 | - | + (0.004) | + | + | + (0.122) | ||||
| R4 | - | + (0.013) | + | + | + (0.035) | ||||
| R5 | - | + (0.002) | + | + | + (0.053) | ||||
| 729.HTLV-1ΔHBZ | |||||||||
| R6 | - | + (0.0003) | + | + | + (0.002) | ||||
| R7 | - | + (0.0003) | + | + | + (0.004) | ||||
| R8 | - | + (0.0008) | + | + | + (0.007) | ||||
| R9 | - | + (0.0003) | + | + | + (0.001) | ||||
| R10 | - | + (0.0002) | + | + | + (0.001) | ||||
| R11 | - | + (0.0005) | + | + | + (0.013) | ||||
| 729.HTLV-1HBZΔLZ | |||||||||
| R12 | - | + (0.0003) | + | + | + (0.001) | ||||
| R13 | - | + (0.0002) | + | + | + (0.005) | ||||
| R14 | - | + (0.0002) | + | + | + (0.014) | ||||
| R15 | - | + (0.0002) | + | + | + (0.006) | ||||
| R16 | - | + (0.0003) | + | + | + (0.004) | ||||
| R17 | - | + (0.0001) | + | + | + (0.003) | ||||
| 729 | |||||||||
| R18 | - | - (0.0001) | - | - | - (0.0001) | ||||
| Media | |||||||||
| R19 | - | - (0.0001) | - | - | - (0.0001) | ||||
Inoculum and rabbit . | Weeks after inoculation . | . | . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|
| . | 0 . | 2 . | 4 . | 6 . | 8* . | ||||
| 729.HTLV-1 | |||||||||
| R1 | - | + (0.001) | + | + | + (0.112) | ||||
| R2 | - | + (0.001) | + | + | + (0.108) | ||||
| R3 | - | + (0.004) | + | + | + (0.122) | ||||
| R4 | - | + (0.013) | + | + | + (0.035) | ||||
| R5 | - | + (0.002) | + | + | + (0.053) | ||||
| 729.HTLV-1ΔHBZ | |||||||||
| R6 | - | + (0.0003) | + | + | + (0.002) | ||||
| R7 | - | + (0.0003) | + | + | + (0.004) | ||||
| R8 | - | + (0.0008) | + | + | + (0.007) | ||||
| R9 | - | + (0.0003) | + | + | + (0.001) | ||||
| R10 | - | + (0.0002) | + | + | + (0.001) | ||||
| R11 | - | + (0.0005) | + | + | + (0.013) | ||||
| 729.HTLV-1HBZΔLZ | |||||||||
| R12 | - | + (0.0003) | + | + | + (0.001) | ||||
| R13 | - | + (0.0002) | + | + | + (0.005) | ||||
| R14 | - | + (0.0002) | + | + | + (0.014) | ||||
| R15 | - | + (0.0002) | + | + | + (0.006) | ||||
| R16 | - | + (0.0003) | + | + | + (0.004) | ||||
| R17 | - | + (0.0001) | + | + | + (0.003) | ||||
| 729 | |||||||||
| R18 | - | - (0.0001) | - | - | - (0.0001) | ||||
| Media | |||||||||
| R19 | - | - (0.0001) | - | - | - (0.0001) | ||||
Genomic DNA was isolated from rabbit PBMCs (weeks 2 and 8 after inoculation) and subjected to real-time Taqman PCR using HTLV-1-specific primers (670/671). Numbers in parentheses at weeks 2 and 8 denote copy number per cell in rabbit PBMCs.
- indicates no amplified PCR fragment; +, amplified PCR fragment.
Copy numbers in rabbits inoculated with the mutant proviruses at week 8 were significantly different from wild type, as determined by ANOVA followed by Tukey test (P < .001).
HTLV-1 T-lymphocyte proliferation and immortalization assays. PBMC (2 × 106) donor cells were cultured with (106) irradiated producer cells as indicated in 24-well plates. (A) Representative growth curve is presented showing cell viability at weekly intervals. The mean and standard deviation of each time point was determined from 3 random independent samples. (B) HTLV-1 gene expression was quantified by detection of Gag protein in the culture supernatant using ELISA. (C) The HTLV-1 genome fragment containing the HBZ coding region was amplified by PCR from DNA of immortalized PBMCs as indicated (HTLV-1HBZΔLZ DNA was cut by NheI). (D) Prestimulated PBMCs (104) were cocultured with 2000 irradiated 729 stable producer cells in 96-well plates. The Kaplan-Meier plot shows the percentages of proliferating wells as a function of time (weeks). Results indicated that the percentage of wells containing proliferating lymphocytes was similar between wtHTLV-1 and HBZ mutant viruses.
HTLV-1 T-lymphocyte proliferation and immortalization assays. PBMC (2 × 106) donor cells were cultured with (106) irradiated producer cells as indicated in 24-well plates. (A) Representative growth curve is presented showing cell viability at weekly intervals. The mean and standard deviation of each time point was determined from 3 random independent samples. (B) HTLV-1 gene expression was quantified by detection of Gag protein in the culture supernatant using ELISA. (C) The HTLV-1 genome fragment containing the HBZ coding region was amplified by PCR from DNA of immortalized PBMCs as indicated (HTLV-1HBZΔLZ DNA was cut by NheI). (D) Prestimulated PBMCs (104) were cocultured with 2000 irradiated 729 stable producer cells in 96-well plates. The Kaplan-Meier plot shows the percentages of proliferating wells as a function of time (weeks). Results indicated that the percentage of wells containing proliferating lymphocytes was similar between wtHTLV-1 and HBZ mutant viruses.
Assessment of HTLV-1 infection in rabbits. (A) Antibody response against HTLV-1 from each rabbit was measured by anti–HTLV-1 ELISA assay, using both HTLV-1 Gag and envelope proteins as antigens. Each dot represents the absorbance value of a single inoculated rabbit at 0, 2, 4, 6, and 8 weeks after inoculation within each group. Inoculum as indicated at bottom includes 729.HTLV-1 (n = 5), 729.HTLV-1ΔHBZ (n = 6), 729.HTLV-1HBZΔLZ (n = 6), or 729/media (n = 2). The horizontal line represents the average of the rabbit group at each weekly time point and the dotted line represents 3 times the standard deviation of uninfected control values. (B) The HTLV-1 genome fragment containing the HBZ coding region was amplified by PCR from DNA of PBMCs from a representative of at least 1 rabbit of each group (week 8 after inoculation). The expected HBZ mutations in rabbit PBMCs were confirmed using the diagnostic restriction enzyme NheI for HTLV-1HBZΔLZ and further confirmed by nucleotide sequencing (data not shown).
Assessment of HTLV-1 infection in rabbits. (A) Antibody response against HTLV-1 from each rabbit was measured by anti–HTLV-1 ELISA assay, using both HTLV-1 Gag and envelope proteins as antigens. Each dot represents the absorbance value of a single inoculated rabbit at 0, 2, 4, 6, and 8 weeks after inoculation within each group. Inoculum as indicated at bottom includes 729.HTLV-1 (n = 5), 729.HTLV-1ΔHBZ (n = 6), 729.HTLV-1HBZΔLZ (n = 6), or 729/media (n = 2). The horizontal line represents the average of the rabbit group at each weekly time point and the dotted line represents 3 times the standard deviation of uninfected control values. (B) The HTLV-1 genome fragment containing the HBZ coding region was amplified by PCR from DNA of PBMCs from a representative of at least 1 rabbit of each group (week 8 after inoculation). The expected HBZ mutations in rabbit PBMCs were confirmed using the diagnostic restriction enzyme NheI for HTLV-1HBZΔLZ and further confirmed by nucleotide sequencing (data not shown).
Discussion
The role of the novel HTLV-1–negative-strand gene product, HBZ, in viral replication and/or pathogenesis remains to be defined. Exogenously overexpressed HBZ has been shown to interact with several cellular transcriptional factors such as the CREB-2,25 JunD,38 JunB, and c-Jun25,26 and is a negative regulator of Tax-mediated HTLV-1 transcription.29 In the present study, site-directed mutations were introduced in an infectious molecular clone of HTLV-1 to severely truncate HBZ or delete the carboxy terminal leucine-zipper domain, while maintaining the ability to express other viral gene products. We examined the expression of HBZ and determined its biologic significance for the immortalization of primary human T lymphocytes in vitro and viral persistence in vivo. We showed that the HBZ antisense viral transcript (Figure 1A) and protein (Figure 4A) were expressed from the ACH HTLV-1 molecular proviral clone. Consistent with a previous report,21 quantitative real-time RT-PCR revealed that, although variable with each individual cell line, the level of HBZ mRNA transcription was approximately 20- to 50-fold lower than the abundant tax/rex mRNA (data not shown). We observed that in the context of a proviral clone, the repressive effects of HBZ on Tax transcription were not apparent following transient transfection (Figure 2A), but confirmed that overexpression of HBZ from a cDNA plasmid down-regulated Tax-mediated viral transcription (Figure 3A). We further demonstrated that the repressive effects of HBZ on Tax-mediated transcription were detectable in cell lines stably harboring the proviral clone (Figure 4B). Therefore, we speculate that HBZ gene expression is temporally regulated and not expressed following transient proviral DNA plasmid delivery or that a threshold level of HBZ is required for the repressive activity.
Data from our short-term proliferation and immortalization assays indicated that the reported repressive effects of the HTLV-1 HBZ on Tax and AP-125,26 were not sufficient to disrupt the capacity of the virus to infect, induce proliferation, and/or immortalize primary T lymphocytes in vitro (Figure 5A,D). Therefore, similar to the HTLV-1 pX open reading frame (ORF) I and II encoded accessory proteins,39,40 HBZ appears to be dispensable for efficient viral infectivity, replication and primary T-lymphocyte immortalization capacity in vitro.
Based on the efficient infectivity and immortalization of cells in vitro and our findings that 729.HTLV-1ΔHBZ– and 729.HTLV-1HBZΔLZ–inoculated rabbits became infected with HTLV-1, we hypothesized that the biologic function of HBZ and its role in HTLV-1 biology is not likely during the early phase of viral infection. However, future experiments designed to quantitatively assess viral infectivity of rabbits at 1 to 2 days after inoculation will ultimately be required to definitively rule out an early block in infection in vivo. It is clear that soon after inoculation, some parameters of viral replication are attenuated in HTLV-1 HBZ mutant virus–infected animals. We begin to observe a reduction in proviral load as early as 2 weeks after inoculation compared with the wild-type virus (Table 1). By 8 weeks HBZ mutant–inoculated rabbits show a significant reduction in antibody response to viral gene products and a 15- to 20-fold drop in proviral load compared with the wild-type virus (Figure 6A, Table 1). Combined with the data that HTLV-1 HBZ repressed Tax-mediated transcription and attenuated AP-1 activity, we speculated that HBZ might function in concert with other viral gene products to tightly regulate viral replication and/or influence the infected lymphocyte to ultimately promote infected cell survival, viral spread, and assist in establishment of persistent infection. Interestingly, HBZ's repressive function and potential contribution to HTLV-1 replication or leukemogenesis may be somewhat analogous to what has been reported for the HTLV-1 p30II accessory protein.41 p30II selectively increases key regulatory genes, influences T-cell signaling, apoptosis, and the cell cycle, but overall represses cellular gene expression. Many of these affects appear to counteract the HTLV-1 oncoprotein Tax. It remains a possibility that HBZ and p30II work synergistically to ultimately modulate viral and cellular gene expression during different stages of the infection to promote virus survival.
Since HBZ is expressed from an antisense transcript it might be predicted to result in a short interfering RNA (siRNA) or double-strand RNA (dsRNA) effect, ultimately resulting in a nucleic acid–based adaptive immunity. Recent evidence indicates that HIV-1 encodes viral siRNA precursors in its genome and provokes an RNA-silencing response.42 However, a domain of the HIV-1 Tat protein has been identified which suppresses the production of siRNAs and the cell's RNA-silencing defense.42 Thus far, there is no evidence to suggest that the HBZ antisense transcript induces RNAi. On the contrary, data presented in Figure 4A and 4B indicates that the HBZΔLZ expression vector, which makes the same RNA transcript as the HBZ vector with the exception of 4 nucleotide changes, fails to repress Tax activity and p19 Gag protein expression. However, it still remains a possibility that the HBZ antisense transcript may have an RNAi effect on the ORF I– or II–encoded proteins, but this is currently difficult to test since these proteins are not expressed at detectable levels from the provirus. It is exciting to speculate that HTLV-1, like HIV-1, may also have a suppressor of RNA silencing, but further studies are required to determine if the virus encodes such an activity.
In summary, our work provides the first demonstration that retroviruses use novel negative-strand gene products to enhance virus replication in vivo. Further studies are warranted to explore the effects of HTLV-1 HBZ on repression of viral transcription and its potential role in innate, cytotoxic T-cell, and natural killer (NK) cell immune surveillance. More broadly, our data elucidate future directions for studies to understand the role of negative-strand–encoded proteins in retroviral-mediated disease and offer new targets for therapy to disrupt virus replication in infected subjects.
Prepublished online as Blood First Edition Paper, January 19, 2006; DOI 10.1182/blood-2005-11-4551.
Supported by a grant from the National Institutes of Health (AI064440).
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Li Xie for technical assistance, Kate Hayes for editorial comments on the manuscript, and Tim Vogt for figure preparations.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal