Activation-induced cytidine deaminase (AID) is necessary for immunoglobulin somatic hypermutation (SHM) and class switch recombination (CSR) in T-dependent immune response in germinal centers (GCs). The structural similarity of AID with RNA-editing enzymes and its largely cytoplasmic location have fueled controversial views of its mode of interaction with DNA. We show that AID, a mature B-cell–restricted cytoplasmic antigen, is relocated into the nucleus in 2.5% of CDKN1B–, CCNB1– GC cells. The GC dark zone and the outer zone (OZ), but not the light zone, contain nuclear and cytoplasmic AID+ blasts. AID+ cells in the OZ are in contact with T cells and CD23– follicular dendritic cells. In addition, AID is expressed in extrafollicular large proliferating B cells, 14% of which have nuclear AID. GC and extrafollicular AID+ cells express E47 but not the inhibiting BHLH protein Id2. Outside the GC, AID+ B cells are in contact with T cells and show partial evidence of CD40 plus bcr stimulation-dependent signature (CCL22, JunB, cMYC, CD30) but lack early and late plasma cell markers. The distribution of nuclear AID is consistent with the topography of SHM and CSR inside the GC and in extrafollicular activated B cells.
Introduction
The host immune response to foreign antigens in higher vertebrates has evolved via separate cell lineages, such as immunoglobulin-producing B cells, effectors T cells, and a variety of accessory cells specialized in antigen processing and presentation (antigen-presenting cells, APCs). B cells can produce antigen-specific antibodies without (T independent, TI) or with (T dependent, TD) T-cell help.1 The latter response is characterized by a process of progressive refinement of the antibody by selecting the type of immunoglobulin heavy chain (class switch recombination, CSR) and improving the affinity of the antigen-binding site (affinity maturation). This process entails genetic alterations of the DNA sequences of the immunoglobulin genes, excision of intervening heavy-chain sequences in CSR, and mutation of nucleotides in the antigen-binding region (somatic hypermutation, SHM), a process followed by selection and resulting in affinity maturation. SHM and CSR take place in a lymphoid structure called the germinal center (GC). Typically, cognate T-B interactions with APC and antigen occur at the border of primary lymphoid follicles.2 Then, antigen-stimulated B cells enter the GC and experience a proliferative burst, mostly with unmutated Ig genes,3-5 followed by the onset of SHM and CSR, resulting in clonal selection of class-switched B cells carrying multiple mutations of their high-affinity antibodies.3,4,6-8 Transit of GC centroblasts from the dark zone to the light zone of the GC may signal 2 different phases of the generation and selection of high-affinity Ig-carrying B cells.9,10
The TI response in mice is typically extrafollicular,11 unmutated,12 and with no or short transit through a GC reaction.13-15 Recently, evidence of hypermutated memory B cells without GC transit has been identified in humans16,17 and mice.14
The gene expression changes that occur from the initial activation up to the GC exit have been extensively described.18-20
Genetic experiments have shown that activation-induced cytidine deaminase (AID) is essential for both CSR and SHM.21 AID is a 24-kDa molecule of the APOBEC family,22 restricted in its expression to lymphoid organs22 and embryonic stem cells.23 Two separate domains of the AID molecule are required for CSR and SHM, in cooperation with the DNA repair machinery.24
The mechanism by which AID performs SHM and CSR has been a matter of controversy and is not yet resolved. The structural similarity with the APOBEC family, an RNA-editing enzyme group of proteins, and other experimental evidence25 suggest an indirect mechanism by which nuclear AID binds and edits the message of an as-yet-unidentified endonuclease and exports the mRNA into the cytoplasm for translation, following which the endonuclease returns to the nucleus where it binds to the DNA sequences.25,26 Other experiments suggest a direct DNA-binding activity of AID, together with cofactors,27 resulting in a direct DNA deamination and consequent introduction of mutations upon activation of the DNA repair machinery.25,28 Consistent with both theories are experiments showing GFP-tagged transfected AID localizing to the nucleus in the presence of appropriate cobinding molecules.29,30
In situ hybridization and cDNA analysis of B-cell fractions suggest that AID is expressed in the GC.22,31 In addition, AID protein was detected in purified centroblast preparations by Western blot.32 Cellular fractionation shows AID to be largely in the cytoplasm,32 with less than 10% possibly localized in the nucleus.25
AID expression is induced by antigen- and T-cell interactions with B-cell surface receptors for interleukins (IL-4), bcr-associated costimulatory molecules (CD40), and Toll-like receptors (TLR-4).33-37 E2A activity is necessary for AID expression, which is in turn negatively influenced by BHLH proteins such as Id2 and Id3.35,36
Recently, a rat monoclonal antibody specifically recognizing AID in human-fixed tissue has been described.31 Here, we present a detailed analysis of the distribution of AID in normal human lymphoid tissues and its environmental context within and outside the GC and a description of the subcellular localization of this protein in vivo.
Patients, materials, and methods
Specimens
Paraffin-embedded normal lymphoid tissues (14 lymph nodes, 10 tonsils, 3 spleens) were obtained from the files of the Department of Pathology, Columbia University Medical Center, New York, according to Columbia University and federal provisions.
Tissue microarray (TMA) containing multiple normal tonsil samples was constructed using a Beecher Instruments microarrayer (Silver Spring, MD) as published in detail.38 At least 3 samples were used in each staining experiment, and results were duplicated in 2 independent institutions (in the United States and Germany).
Cell lines and in vitro stimulation
Cell lines were grown in vitro and stimulated with CD40L or mock transfectants, or anti-IgM (10 μg/mL for 12 or 21 hours) as previously published.39 EBNA2 conditional cell line ER-EB was grown with or without estrogen as published.40 Hydrogen peroxide treatment at 0, 100, 500, and 1000 μM41 was applied for 6 hours in culture and monitored for annexin V binding and γH2AX foci formation. Cells were fixed, embedded in molten agar, and processed similar to fresh surgical specimens.
Immunohistochemistry
Immunostaining with EK2-5G9 and other antibodies (previously fully validated and described, Table 1) was done essentially as previously published42 on 1 mM EDTA (pH 8) antigen-retrieved slides.
Primary antibodies
Antibody . | Source . | Species . | Clone or serum . | Isotype . | Dilution . |
|---|---|---|---|---|---|
| AID | E.K. (www.ascenion.de) | Rat | EK2-5G9 | G | 1:200 |
| BLIMP-1/PRDM1 | K. L. Calame, Columbia University | Mouse | 3H2E8 | G1 | 1:30 |
| MUM 1 | B. Falini, Perugia University | Mouse | 2H9 | G1 | 1:50 |
| MUM1/IRF4 | Santa Cruz Biotechnology (SCBT; Santa Cruz, CA) | Goat | sc-6059 | G | 1 μg/mL |
| c-Rel (NF-κB) | Oncogene Research (Cambridge, MA) | Rabbit | Ab-1 | G | 1:2000 |
| cMYC | SCBT | Rabbit | sc-764 | G | 0.5 μg/mL |
| Ki-67 | J. Gerdes, MIB Germany | Mouse | MIB 1 | G1 | 1:50 |
| Ki-67 | LabVision (Fremont, CA) | Rabbit | serum | G | 1:100 |
| CCNB1 (cyclin B1) | SCBT | Rabbit | sc-752 | G | 0.5 μg/mL |
| CDKN1B (p27) | BD PharMingen (San Diego, CA) | Mouse | 57 | G1 | 1 μg/mL |
| BCL6 | SCBT | Rabbit | sc-858 | G | 0.1 μg/mL |
| MTA3 | Calbiochem (San Diego, CA) | Rabbit | IM1012 | G | 0.5 μg/mL |
| PAX5 | SCBT | Goat | C-20 sc1974 | G | 1 μg/mL |
| PAX5 | BD PharMingen | Mouse | 24 | G1 | 1 μg/mL |
| IgD | Dako USA (Carpinteria, CA) | Rabbit | serum | G | 1:4000 |
| CD3e | Dako USA | Rabbit | serum | G | 0.5 μg/mL |
| CD20 | LabVision | Mouse | L26 | G2a | 0.5 μg/mL |
| CD23 | Novocastra (Newcastle-upon-Tyne, United Kingdom) | Mouse | 1B12 | G1 | 1:50 |
| CD30 | LabVision | Mouse | Ber-H2 | G1 | 1 μg/mL |
| CD138 | Serotec (Raleigh, NC) | Mouse | B-B4 | G1 | 1:200 |
| CD79a | LabVision | Rabbit | Sp18 | G | 1:200 |
| CCL22 (MDC) | PeproTech (Rocky Hill, NJ) | Rabbit | serum | G | 0.5 μg/mL |
| JunB | SCBT | Mouse | sc-8051 | G1 | 1 μg/mL |
| E47 | SCBT | Rabbit | sc-763 | G | 0.2 μg/mL |
| Id2 | Zymed/Invitrogen (South San Francisco, CA) | Rabbit | serum | G | 0.5 μg/mL |
| γH2AX | Upstate Biotechnology (Lake Placid, NY) | Mouse | JBW301 | G1 | 1 μg/mL |
| PhosphoH2B | Upstate Biotechnology | Rabbit | serum | G | 1:10 000 |
| TP53 | J. Bartek, Denmark | Mouse | BP53.11.1 | G2a | 1:100 |
| TP53 | J. Bartek, Denmark | Mouse | DO.1 | G2a | 1:100 |
| Negative control | Sigma (St Louis, MO) | Rabbit | serum | G | 1 μg/mL |
| Negative control | Sigma | Mouse | serum | G | 1 μg/mL |
Antibody . | Source . | Species . | Clone or serum . | Isotype . | Dilution . |
|---|---|---|---|---|---|
| AID | E.K. (www.ascenion.de) | Rat | EK2-5G9 | G | 1:200 |
| BLIMP-1/PRDM1 | K. L. Calame, Columbia University | Mouse | 3H2E8 | G1 | 1:30 |
| MUM 1 | B. Falini, Perugia University | Mouse | 2H9 | G1 | 1:50 |
| MUM1/IRF4 | Santa Cruz Biotechnology (SCBT; Santa Cruz, CA) | Goat | sc-6059 | G | 1 μg/mL |
| c-Rel (NF-κB) | Oncogene Research (Cambridge, MA) | Rabbit | Ab-1 | G | 1:2000 |
| cMYC | SCBT | Rabbit | sc-764 | G | 0.5 μg/mL |
| Ki-67 | J. Gerdes, MIB Germany | Mouse | MIB 1 | G1 | 1:50 |
| Ki-67 | LabVision (Fremont, CA) | Rabbit | serum | G | 1:100 |
| CCNB1 (cyclin B1) | SCBT | Rabbit | sc-752 | G | 0.5 μg/mL |
| CDKN1B (p27) | BD PharMingen (San Diego, CA) | Mouse | 57 | G1 | 1 μg/mL |
| BCL6 | SCBT | Rabbit | sc-858 | G | 0.1 μg/mL |
| MTA3 | Calbiochem (San Diego, CA) | Rabbit | IM1012 | G | 0.5 μg/mL |
| PAX5 | SCBT | Goat | C-20 sc1974 | G | 1 μg/mL |
| PAX5 | BD PharMingen | Mouse | 24 | G1 | 1 μg/mL |
| IgD | Dako USA (Carpinteria, CA) | Rabbit | serum | G | 1:4000 |
| CD3e | Dako USA | Rabbit | serum | G | 0.5 μg/mL |
| CD20 | LabVision | Mouse | L26 | G2a | 0.5 μg/mL |
| CD23 | Novocastra (Newcastle-upon-Tyne, United Kingdom) | Mouse | 1B12 | G1 | 1:50 |
| CD30 | LabVision | Mouse | Ber-H2 | G1 | 1 μg/mL |
| CD138 | Serotec (Raleigh, NC) | Mouse | B-B4 | G1 | 1:200 |
| CD79a | LabVision | Rabbit | Sp18 | G | 1:200 |
| CCL22 (MDC) | PeproTech (Rocky Hill, NJ) | Rabbit | serum | G | 0.5 μg/mL |
| JunB | SCBT | Mouse | sc-8051 | G1 | 1 μg/mL |
| E47 | SCBT | Rabbit | sc-763 | G | 0.2 μg/mL |
| Id2 | Zymed/Invitrogen (South San Francisco, CA) | Rabbit | serum | G | 0.5 μg/mL |
| γH2AX | Upstate Biotechnology (Lake Placid, NY) | Mouse | JBW301 | G1 | 1 μg/mL |
| PhosphoH2B | Upstate Biotechnology | Rabbit | serum | G | 1:10 000 |
| TP53 | J. Bartek, Denmark | Mouse | BP53.11.1 | G2a | 1:100 |
| TP53 | J. Bartek, Denmark | Mouse | DO.1 | G2a | 1:100 |
| Negative control | Sigma (St Louis, MO) | Rabbit | serum | G | 1 μg/mL |
| Negative control | Sigma | Mouse | serum | G | 1 μg/mL |
For EK2-5G9 antibody, sections were cooled, blocked with 10% egg white (All White, Papetti Foods, NJ)43 in PBS for 30 minutes, washed in TBS–0.01% TritonX100 (TBS-T), blocked for 30 minutes with 5% defatted powdered milk in TBS-T,44 and, after several washes in TBS-T, incubated overnight with 1:200 dilution of the antibody in TBS-BSA-NaN3. After two 15-minute washes in TBS-T, sections were incubated with 1:500 diluted goat anti–rat Ig-AP (Southern Biotechnology, Birmingham, AL) for 45 minutes, washed for 60 minutes in 0.5 M NaCl, 0.005 M TBS, and developed in NBT-BCIP up to 24 hours.
For selective detection of nuclear AID, sections were stained first with CD20 antibody and fully developed in immunoperoxidase with AEC, thus preventing any further staining of the membrane or cytoplasm of the heavily stained CD20+ cells and leaving the nucleus unstained. Then, EK2-5G9 or a control antibody (rabbit CD79a or rat CD45RA) was applied and developed in alkaline phosphatase as described above.
In order to exclude artifactual, processing-dependent nuclear relocalization of AID, all specimens were costained in double IF and IHC for proteins of small molecular weight and known subcellular distribution (eg, CCL22: 2.9 kDa, CD20: 33-36 kDa, and PCNA: 36 kDa), and the correct localization was verified. Furthermore, we costained AID with cREL (75 kDa), previously shown to be translocated into the nucleus selectively and specifically dependent on CD40 signaling.39
Lastly, the isolated presence of nuclear staining amid abundant cytoplasmic-positive neighboring cells was considered as evidence of absence of diffusion and section artifacts.
Immunofluorescence
Double immunofluorescence was performed with primary antibodies raised in different species or of different isotypes, counterstained with FITC- or Cy3-conjugated, species- or isotype-specific secondary antibodies (Jackson ImmunoResearch Laboratories, West Grove, PA) or biotin-conjugated secondary antibodies, followed by conjugated avidin (Jackson ImmunoResearch). The sequence was repeated twice to enhance staining brightness.45 Tyramide signal amplification for single or double labeling46 was performed according to the manufacturer's instructions (Perkin-Elmer, Wellesley, MA): Briefly, avidin-HRP–counterstained immunostains were followed by tyramide-FITC or tyramid-biotin and avidin Cy-3 (Jackson ImmunoResearch) or avidin-Alexa350 (Molecular Probes, Eugene, OR).
EK2-5G9 was first stained with an FITC-conjugated goat anti–rat F(ab) monomer (1:1000; Jackson Immunoresearch), followed by goat anti-FITC peroxidase-conjugated antibody (1:1000; Roche, Indianapolis, IN), and then processed for tyramide-FITC amplification for fewer than 6 minutes. Following this, slides were briefly boiled in antigen retrieval medium, cooled, and counterstained with non–cross-reactive primary Abs and rat-absorbed secondary antibodies. The slides were counterstained with DAPI (Molecular Probes) when indicated and mounted.
For triple IF staining, AID was stained first as described above, followed by a mouse primary Ab, then Envision+ mouse HRP polymer (Dako USA, Carpinteria, CA), and tyramide-Cy3. Finally, a rabbit primary Ab was incubated and developed with Envision+ rabbit, tyramide-biotin, and avidin-Alexa350. Peroxidase quenching was performed after each step. No cross-reactivity among any layer was obtained.
γH2AX staining sensitivity was assessed on formalin-fixed, paraffin-embedded, etoposide-treated Ramos cells47 and was found more than adequate to detect DNA breaks induced by the lowest etoposide concentration (0.2 μM).
Image acquisition and analysis
Gray-scale or color images were observed using a Nikon E600 microscope (Nikon USA, Melville, NY), fitted with Planachromat 4 ×/0.10/30.0, 10 ×/0.25/10.5, or 20 ×/0.40/1.3; PlanApo 40 ×/.095/.12-.16 (light microscopy); or PlanFluor 10 ×/0.30/16.0, 40 ×/0.75/0.72, or 60 ×/0.80/0.3 (immunofluorescence) objectives, with a SPOT-2 CCD camera and software (Diagnostic Instruments, Sterling Heights, MI). A Zeiss LSM 510 NLO Multiphoton confocal microscope (Carl Zeiss, Thornwood, NJ) equipped with a 10 ×/0.3 and a 40 ×/1.3 Fluar objective, one 25-mW argon laser exciting at 458, 488, and 514 nm, and one 1-mW helium-neon laser exciting at 543 nm. Proprietary image acquisition software was used for confocal analysis of sections stained with AID, Ki-67, and nuclear dyes (see previous paragraph). All images were edited for optimal color contrast with Adobe Photoshop 7 and Adobe Illustrator 10 (Adobe Systems, San Jose, CA), on an Apple G4 computer (Apple Computers, Cupertino, CA).
Gray-scale images of AID-stained sections from 5 tonsils were acquired on an Agfa flat-bed scanner (Agfa, Ridgefield Park, NJ) at 1500-pixel resolution or with a microscope and analyzed with The ImageJ software (RSB; NIH, Bethesda, MD). Nuclear AID+ cells were counted manually on 16 GCs from 1 tonsil and on interfollicular areas from 5 tonsils.
Results
Specificity and distribution of AID in normal lymphoid tissues
The specificity of the staining pattern of the EK2-5G9 antibody (referred to as anti-AID hereafter), matching the AID RNA in situ hybridization distribution,31 was further confirmed by staining AID-transfected, AID-null cell lines but not mock-transfected ones (kind gift of Dr L. Pasqualucci) (Figure 1A). In addition, EK2-5G9 Ab staining correctly identified a large panel of Epstein-Barr virus–positive (EBV+) and EBV– B-cell lines of known AID RNA expression (G.C., manuscript in preparation). The terms cytoplasmic AID (cytAID) and nuclear AID (nAID) will be used hereafter when subcellular localization is specified, leaving the term AID for positive cells in general, regardless of the localization of the molecule.
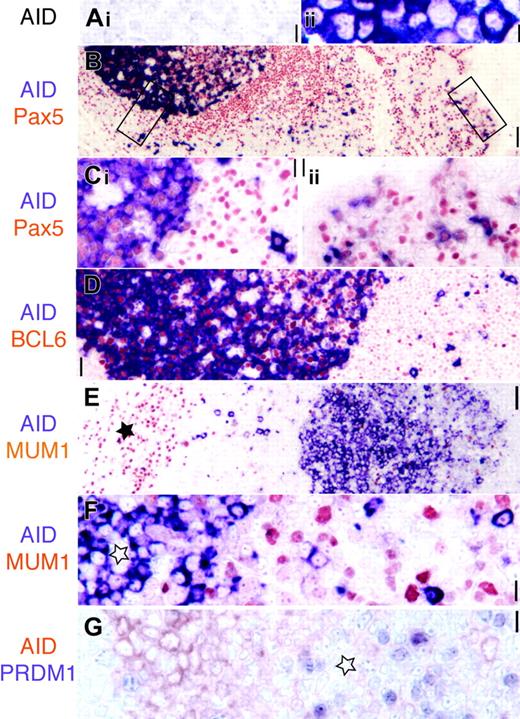
AID is expressed in Pax5+ GC and extrafollicular B cells. (A) AID transfectants (ii, purple) but not mock-transfected cell lines are stained by the EK2-5G9 antibody. Original magnification × 40. (B) Low-power image (original magnification × 10) of AID (purple) and panB cell Pax5 (red) in tonsil. The epithelial surface is on the right. Two rectangles indicate areas magnified in panel C. (Ci) AID stains Pax5+ GC and extrafollicular large blasts. (Cii) Subepithelial Pax5+ B cells are AID+. Original magnification × 40. (D) AID+ GC cells are BCL6+ (red), while extrafollicular AID+ are BCL6–. Original magnification × 20. (E) Low-power image (original magnification × 10) of AID (purple) and MUM1 (red). The star marks plasma cell collection in the septa. AID+ GC on the right is MUM1 negative. (F) High-power image of a MUM1–AID+ GC (star) and MUM1– extrafollicular blasts. Original magnification × 40. (G) AID+ centroblasts (red) are PRDM1– (purple). These latter are found in the AID– light zone (star). Original magnification × 40. Scale bar: 1 μmat × 40, 4 μmat × 10, and 10 μmat × 4.
AID is expressed in Pax5+ GC and extrafollicular B cells. (A) AID transfectants (ii, purple) but not mock-transfected cell lines are stained by the EK2-5G9 antibody. Original magnification × 40. (B) Low-power image (original magnification × 10) of AID (purple) and panB cell Pax5 (red) in tonsil. The epithelial surface is on the right. Two rectangles indicate areas magnified in panel C. (Ci) AID stains Pax5+ GC and extrafollicular large blasts. (Cii) Subepithelial Pax5+ B cells are AID+. Original magnification × 40. (D) AID+ GC cells are BCL6+ (red), while extrafollicular AID+ are BCL6–. Original magnification × 20. (E) Low-power image (original magnification × 10) of AID (purple) and MUM1 (red). The star marks plasma cell collection in the septa. AID+ GC on the right is MUM1 negative. (F) High-power image of a MUM1–AID+ GC (star) and MUM1– extrafollicular blasts. Original magnification × 40. (G) AID+ centroblasts (red) are PRDM1– (purple). These latter are found in the AID– light zone (star). Original magnification × 40. Scale bar: 1 μmat × 40, 4 μmat × 10, and 10 μmat × 4.
In tonsils and lymph nodes, AID was found to be expressed in the dark and outer zones of the GC and in extrafollicular large cells, often with dendritic morphology.31 AID+ cells of large and medium size were seen not only in the immediate vicinity of the follicular mantle, but also scattered in the interfollicular areas, and within the festooned tonsil epithelium, where marginal zone–type B cells normally reside48 (Figure 1B-D).
Comparison of lymph nodes with follicular versus paracortical hyperplasia shows a parallel and concomitant increase in extrafollicular AID staining together with an increase in AID+ GC size (data not shown).
Spleens with and without marginal zone hyperplasia contained AID+ GC and scattered single cells in the marginal zone (data not shown and Willenbrock et al49 ). The follicular mantle was invariably AID–.
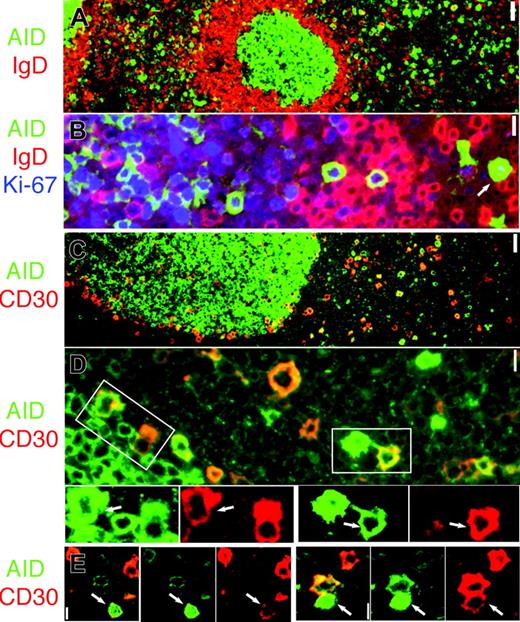
Proliferating IgD– AID+ cells and CD30+ B-cell partially overlap. (A) AID (green) and IgD are mutually exclusive. Original magnification ×4. (B) AID+ cells (green) are IgD– (red) and proliferate (Ki-67+, blue). Note nuclear AID+ extrafollicular cell (arrow). Original magnification ×40. (C) AID (green) and CD30 (red) are partially overlapping subsets. (D) Occasionally, AID+ (green) and CD30+ (red) double-positive cells can be seen outside and at the periphery of the GC. The areas marked by white rectangles are enlarged and colors are split below. Double-positive cells are indicated (arrows). Original magnification × 40. (E) Two representative nuclear AID+ (green) and CD30+ (red) extrafollicular cells are shown (arrows). Merged images show orange color where coexpressed. Original magnification × 40. Scale bar: 1 μm at ×40, 4 μmat ×10, and 10 μmat ×4.
Proliferating IgD– AID+ cells and CD30+ B-cell partially overlap. (A) AID (green) and IgD are mutually exclusive. Original magnification ×4. (B) AID+ cells (green) are IgD– (red) and proliferate (Ki-67+, blue). Note nuclear AID+ extrafollicular cell (arrow). Original magnification ×40. (C) AID (green) and CD30 (red) are partially overlapping subsets. (D) Occasionally, AID+ (green) and CD30+ (red) double-positive cells can be seen outside and at the periphery of the GC. The areas marked by white rectangles are enlarged and colors are split below. Double-positive cells are indicated (arrows). Original magnification × 40. (E) Two representative nuclear AID+ (green) and CD30+ (red) extrafollicular cells are shown (arrows). Merged images show orange color where coexpressed. Original magnification × 40. Scale bar: 1 μm at ×40, 4 μmat ×10, and 10 μmat ×4.
We found AID+ cells to be invariably PAX5+, CD3–, and Ki-67+. In the GC, positive cells in the dark zone (DZ) were CD20+, BCL6+, MTA3+, Oct-2+, bcl2–, IRF4–, PRDM1–, nuclear cREL–, and CDKN1B– (p27, Kip1) (Figures 1, 2B, and 3, and data not shown). The basal and apical light zone (LZ50 ; characterized morphologically, topographically, and by CD23 staining of follicular dendritic cells) had few weakly AID+ cells, mostly not in contact with CD3+ lymphocytes (data not shown) or CD23+ dendrites (Figure 4). AID+ cells were found in the outer zone (OZ), admixed with CD3+ T cells and plasmacytoid cells.
To further define the phenotype and boundaries of the OZ, 2 of 8 tonsils with a broad OZ in the majority of GCs were selected. Serial sections were double stained for IgD and BCL6 or Ki-67. The OZ uniquely contained a mixture of proliferating (Ki-67+) BCL6+ cells, ranging from weakly positive IgD and AID– to double IgD+ and AID+ to IgD– and AID+, reminiscent of the “GC founder cell” population previously defined by flow cytometry5 (Figure 4). In addition, numerous CD3+ T cells home in the OZ, where they come in contact with most, but not all, AID+ cells (Figure 4). The rare GCs that were totally occupied by IgD+ centroblasts and centrocytes were AID+ as well.
Outside the GC, AID+ cells were PAX5+, CD20+, Oct-2+, bcl-2–, BCL6–, PRDM1–, IRTA-1–, and Ki-67+. Smaller weakly AID+ cells were often CDKN1B+ (data not shown). One or more CD3+ T lymphocytes were in contact with more than 90% of extrafollicular AID+ cells (Figure 3A). IgD staining on AID+ extrafollicular cells was essentially absent (Figure 2A-B).
A reproducible minority of the extrafollicular AID+ cells was CD30+, as well as occasional large blasts at the border of the GC (Figure 2C-E).
To define the transcriptional activity in the GC and extrafollicular AID+ cells, tonsils were costained for AID and proteins known to be associated with activation and B-cell signaling.18,39 Extrafollicular AID+ cells were E47+ and Id2–, and occasionally cMYC+, JunB+, and CCL22+, and largely negative for IRF4 (Figure 5A-E). In the GC, AID+ cells were E47+ and Id2– (or borderline Id2+), and negative for cMYC, CCL22, IRF4, and JunB19,39,51 (and data not shown). Extrafollicular CD30+ cells uniquely shared some but not all of the AID+ cell phenotype, being virtually 100% cMYC+, JunB+, and Pax5+, and partially CD20+ and CCL22+ (in decreasing order of frequency). CD30+ cells were, however, IRF4+, Id2+, occasionally IgD+, and E47–. A transitional phenotype toward GC centroblasts was found in CD30+ GC cells (data not shown).
Distribution, phenotype, and cycle status of nuclear AID+ cells
Nuclear translocation of AID and subsequent mutation may occur in a small minority of cells, obscured by an overwhelming majority of nuclear-negative cells.52 In addition, previous experience with antibodies against nuclear antigens (eg, PRDM1) showed often inadequate detection with peroxidase complex–based biotintyramide amplification, compared with simpler methods that allow a linear IHC signal response42,53 (and G. C., unpublished observations, June 2000 through March 2004). Therefore, AID staining was modified as described in “Materials and methods.”
Nuclear AID is detected inside and outside the GC. (A) AID+ cells (green) express membrane CD20 (red; orange color when coexpressed) and are surrounded by CD3+ cells (blue) outside the GC. Several nuclear AID+ extrafollicular blasts (right, arrow) and small centroblasts (left) are shown. The areas marked by white rectangles are magnified below; the color split and phase contrast image of the same field were added. Original magnification ×40. (B) AID+ GC cells are strongly cytoplasmic cREL+ (red) in the GC (left field, orange color when coexpressed). Nuclei are counterstained with DAPI. The areas marked by white rectangles (GC on the left; extrafollicular on the right) are magnified below; the color split and nuclear AID+ cells are marked by a white arrow. Note the total absence of nuclear cREL in the nAID+ cells, while nuclear cREL+ nAID– cells (yellow arrowhead) are shown nearby. Original magnification ×40. (C) Confocal microscopy of AID (green) and Ki-67 (red) costained tonsil shows nuclear coexpression (yellow; arrows) in small centroblasts (left) and extrafollicular blasts (right). (D) Prior staining for CD20 (brown) prevents cytoplasmic AID staining, except in 2 AID+ CD20 ± extrafollicular blasts (red arrows). Nuclear AID (purple) can be shown in extrafollicular cells (black arrows) and in centroblasts (blue arrows). On the right, nAID+ centroblasts (left column) and large extrafollicular cells (right column) are shown, enlarged. Original magnification ×40.(E) As a control, no nuclear staining is shown in CD20 (brown) and CD79a (purple) double-stained tonsil. Original magnification ×40. Scale bar: 1 μm at × 40, 4 μm at ×10, and 10 μmat ×4.
Nuclear AID is detected inside and outside the GC. (A) AID+ cells (green) express membrane CD20 (red; orange color when coexpressed) and are surrounded by CD3+ cells (blue) outside the GC. Several nuclear AID+ extrafollicular blasts (right, arrow) and small centroblasts (left) are shown. The areas marked by white rectangles are magnified below; the color split and phase contrast image of the same field were added. Original magnification ×40. (B) AID+ GC cells are strongly cytoplasmic cREL+ (red) in the GC (left field, orange color when coexpressed). Nuclei are counterstained with DAPI. The areas marked by white rectangles (GC on the left; extrafollicular on the right) are magnified below; the color split and nuclear AID+ cells are marked by a white arrow. Note the total absence of nuclear cREL in the nAID+ cells, while nuclear cREL+ nAID– cells (yellow arrowhead) are shown nearby. Original magnification ×40. (C) Confocal microscopy of AID (green) and Ki-67 (red) costained tonsil shows nuclear coexpression (yellow; arrows) in small centroblasts (left) and extrafollicular blasts (right). (D) Prior staining for CD20 (brown) prevents cytoplasmic AID staining, except in 2 AID+ CD20 ± extrafollicular blasts (red arrows). Nuclear AID (purple) can be shown in extrafollicular cells (black arrows) and in centroblasts (blue arrows). On the right, nAID+ centroblasts (left column) and large extrafollicular cells (right column) are shown, enlarged. Original magnification ×40.(E) As a control, no nuclear staining is shown in CD20 (brown) and CD79a (purple) double-stained tonsil. Original magnification ×40. Scale bar: 1 μm at × 40, 4 μm at ×10, and 10 μmat ×4.
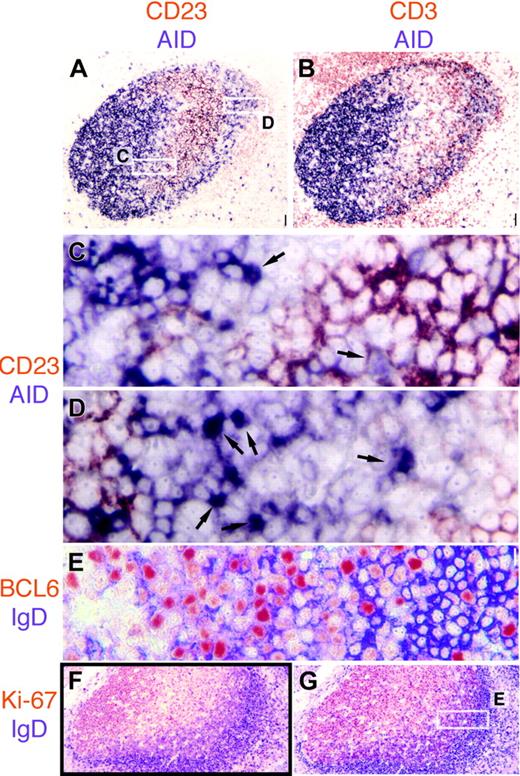
AID is expressed in the dark zone and the outer zone in GC. (A) AID (purple) and CD23 (red) are mutually exclusive in the GC. Note weakly CD23+ mantle zone. Original magnification ×4. (B) CD3+ T cells (red) are abundant in the AID+ outer zone at the right edge of the GC. Original magnification ×4. (C-D) The 2 white rectangles shown in panel A are magnified. In panel C, the basal light zone is empty, flanked by AID+ dark zone on top left, containing a single nAID+ cell (arrow) and the CD23+ apical light zone in the bottom right, in which a weakly AID+ nucleus contact CD23+ FDC (arrow). In panel D, the outer zone is magnified, containing numerous nAID+ cells (arrows). Original magnification ×40. Note the CD23+ mantle zone cells in the lower-right corner. (E) High-power detail of the area boxed in panel G, showing IgD strongly positive mantle zone cells on the right (purple), and double BCL6 (red) and IgD weakly positive cells of the outer zone, in the center. (F-G) Low-power images of serial sections stained for IgD (purple) and Ki-67 (F) or BCL6 (G). Note the double-positive cells under the IgD+ mantle zone. Scale bar: 1 μmat ×40, 4 μmat ×10, and 10 μmat ×4.
AID is expressed in the dark zone and the outer zone in GC. (A) AID (purple) and CD23 (red) are mutually exclusive in the GC. Note weakly CD23+ mantle zone. Original magnification ×4. (B) CD3+ T cells (red) are abundant in the AID+ outer zone at the right edge of the GC. Original magnification ×4. (C-D) The 2 white rectangles shown in panel A are magnified. In panel C, the basal light zone is empty, flanked by AID+ dark zone on top left, containing a single nAID+ cell (arrow) and the CD23+ apical light zone in the bottom right, in which a weakly AID+ nucleus contact CD23+ FDC (arrow). In panel D, the outer zone is magnified, containing numerous nAID+ cells (arrows). Original magnification ×40. Note the CD23+ mantle zone cells in the lower-right corner. (E) High-power detail of the area boxed in panel G, showing IgD strongly positive mantle zone cells on the right (purple), and double BCL6 (red) and IgD weakly positive cells of the outer zone, in the center. (F-G) Low-power images of serial sections stained for IgD (purple) and Ki-67 (F) or BCL6 (G). Note the double-positive cells under the IgD+ mantle zone. Scale bar: 1 μmat ×40, 4 μmat ×10, and 10 μmat ×4.
In the GC and in the extrafollicular areas, nuclear staining was demonstrated by both immunofluorescence and immunohistochemistry methods, mostly, but not always, associated with strong cytoplasmic AID (cytAID) staining (Figures 2B,E and 3). Nuclear AID (nAID) was prominent in larger extrafollicular cells, averaging 14.4% ± 3.9% (n = 5) of the total extrafollicular AID+ population, which represents up to 1% of the extrafollicular lymphocytes (Figure 3C). In the GC, nAID was weaker and expressed in smaller centroblasts, sometimes with irregular contours, often with faint cytoplasmic staining. Of the GC cells,2.5% ± 1.2% were nAID positive, irrespective of the size of the GC (n = 16; r = 0.92) (Figure 3C). The percentage of nAID in the OZ was 20% to 30% higher than in the DZ (data not shown).
Confocal microscopy confirmed nuclear deposition of nAID staining in a chromatin-like texture pattern (Figure 3C).
To gain insight into the cellular environment during AID nuclear translocation, we differentially examined nAID and cytAID cells.
Extrafollicular nAID+ cells were 50% weakly positive for CD30 (Figure 2E); 30% weakly positive for CCNB1+ (data not shown); negative for nuclear cREL (Figure 3A-B), BCL6, CDKN1B, and TP53; and did not contain foci of γH2AX or phosphorylated H2B histones (data not shown). Outside the GC, cytAID+ cells were also Ki-67+, CCNB1–, BCL6–, and CDKN1B– and rarely TP53+ (Figure 6). TP53 was also occasionally seen in CD30+ cells (data not shown) and in cytAID+ at the border of the GC (Figure 6).
Dark zone nAID+ cells were often BCL6 weak, but invariably Ki-67+, and mostly CDKN1B– (Figure 6) and TP53–. CCNB1 strongly stained the nucleus and cytoplasm of larger centroblasts, and did not stain smaller nAID+ cells (Figure 6). The surrounding cytAID+ cells were brightly BCL6+ and shared the same remaining phenotype, plus variable expression of membranous CCNB1 (Figure 6). γH2AX or phosphorylated H2B histones were negative in both.
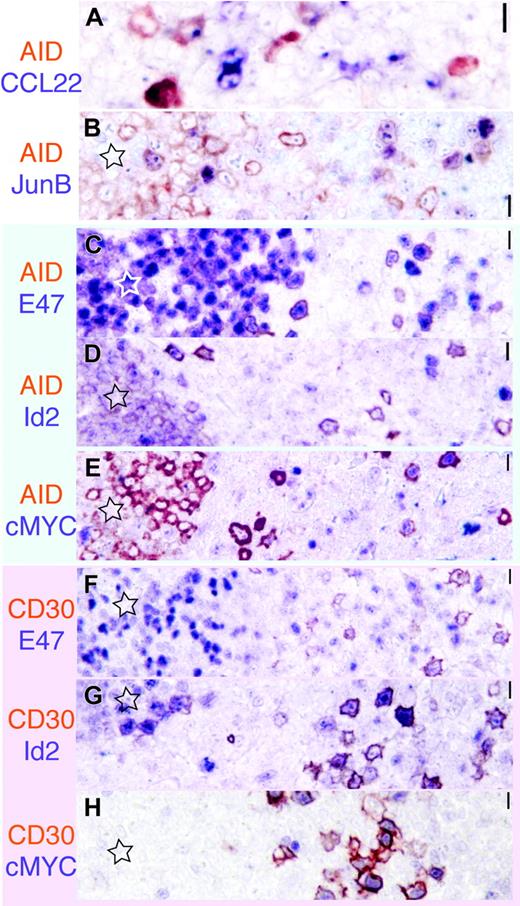
The transcription signature of AID+ and CD30+ B cells. AID is red-brown; the other marker is purplish in each image. Original magnification ×40.(A) A subset of AID+ cells display dotlike CCL22 positivity. (B) AID and JunB are largely nonoverlapping. (C-E) AID+ cells (red brown) inside and outside the GC (star) are E47+, Id2–, and largely cMYC–. (E-G) By comparison, CD30+ cells (red brown) show a reversed staining pattern, being E47– but Id2+ and cMYC+. Scale bar: 1 μm at ×40, 4 μmat ×10, and 10 μmat ×4.
The transcription signature of AID+ and CD30+ B cells. AID is red-brown; the other marker is purplish in each image. Original magnification ×40.(A) A subset of AID+ cells display dotlike CCL22 positivity. (B) AID and JunB are largely nonoverlapping. (C-E) AID+ cells (red brown) inside and outside the GC (star) are E47+, Id2–, and largely cMYC–. (E-G) By comparison, CD30+ cells (red brown) show a reversed staining pattern, being E47– but Id2+ and cMYC+. Scale bar: 1 μm at ×40, 4 μmat ×10, and 10 μmat ×4.
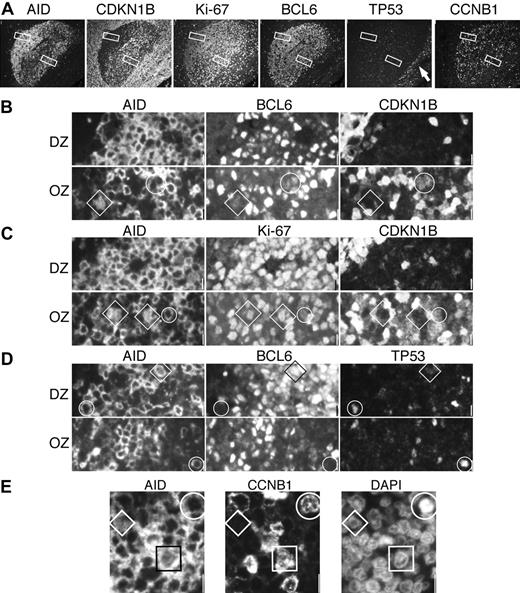
Proliferation and growth arrest in AID+ DZ and OZ GC cells. (A) A single GC was imaged with 4 sets of triple staining for AID, CDKN1B (p27), Ki-67, BCL6, TP53, and CCNB1. Each stain is shown as single color at low power. Original magnification ×10. Note at the right, TP53+ basal epithelial layer (arrow). The DZ and OZ areas enlarged below (original magnification ×40) are shown as white rectangles. (B) The DZ is homogeneously CDKN1B–; circled in the OZ is a cytAID+, BCL6+dim, CDKN1B+dim cell. A diamond highlights a nAID+, BCL6+dim, CDKN1B– cell. (C) DZ AID+ cells are Ki-67+ and CDKN1B–. In the OZ, a cytAID+, Ki-67+, CDKN1B+dim cell is circled. A diamond shows a nAID+, Ki67+, CDKN1B– cell. (D) At the outer border of the DZ and OZ, cytAID+ BCL6– cells (circles) shows TP53. A nAID+, BCL6+, TP53– cell is indicated by a diamond. (E) A portion of the DZ shows a CCNB1–nAID+ small centroblast (diamond), a rare CCNB1+ nAID+ centroblast (square), and a mitotic cell (circle) in which AID and CCNB1 are cytoplasmic. Scale bar: 1 μm at ×40 and 4 μmat ×10.
Proliferation and growth arrest in AID+ DZ and OZ GC cells. (A) A single GC was imaged with 4 sets of triple staining for AID, CDKN1B (p27), Ki-67, BCL6, TP53, and CCNB1. Each stain is shown as single color at low power. Original magnification ×10. Note at the right, TP53+ basal epithelial layer (arrow). The DZ and OZ areas enlarged below (original magnification ×40) are shown as white rectangles. (B) The DZ is homogeneously CDKN1B–; circled in the OZ is a cytAID+, BCL6+dim, CDKN1B+dim cell. A diamond highlights a nAID+, BCL6+dim, CDKN1B– cell. (C) DZ AID+ cells are Ki-67+ and CDKN1B–. In the OZ, a cytAID+, Ki-67+, CDKN1B+dim cell is circled. A diamond shows a nAID+, Ki67+, CDKN1B– cell. (D) At the outer border of the DZ and OZ, cytAID+ BCL6– cells (circles) shows TP53. A nAID+, BCL6+, TP53– cell is indicated by a diamond. (E) A portion of the DZ shows a CCNB1–nAID+ small centroblast (diamond), a rare CCNB1+ nAID+ centroblast (square), and a mitotic cell (circle) in which AID and CCNB1 are cytoplasmic. Scale bar: 1 μm at ×40 and 4 μmat ×10.
In the outer zone, AID+ cells had a more variegated phenotype. Similar to the DZ, OZ nAID+ cells were weakly BCL6+, Ki-67+, CDKN1B–, TP53–, and CCNB1–. CytAID+ cells were also variably BCL6+, and largely Ki-67+ and cytoplasmic CCNB1+; occasionally, the cells were double positive for CDKN1B and Ki-67, both weakly (Figure 6). No γH2AX or phosphorylated H2B histone foci were seen.
In vitro manipulation of AID nuclear translocation
Primary virgin B cells and GC blasts are both unsuitable for in vitro reproduction of GC physiology. To understand the signals that may induce nuclear AID translocation, we challenged 2 GC-derived ABC-type and 1 GC-type human B-cell lymphoma lines (OCY-Ly3: nAID, 0.55%; OCY-Ly10: nAID, 1.5%; Ramos: nAID, 0.5%) with CD40-bearing or mock-transfected fibroblasts.39 No increase of nAID was noticed, whereas a modest increase in cytoplasmic staining was seen in those and in 2 other DLBCL lines (data not shown). Complete nuclear relocation of the NFkB member c-REL was, however, specifically induced in the susceptible cell lines. Bcr stimulation via an anti-IgM antibody39 did not change the percentage of nAID in Ramos. In order to mimic the recruitment of cofactors to DNA lesions, hydrogen peroxide treatment54 was applied to the Ramos cell line, without effect on nAID (data not shown).
After identifying the presence of AID in 8 of 8 EBV-infected B-cell lines from cord or peripheral blood, with a range of nAID ranging from 2% to less than 0.01% (data not shown), we used one of them to further explore a surrogate of combined CD40 and bcr signaling. The ER-EB lymphoblastoid cell line (nAID, ≈ 0.01%) was conditionally induced into quiescence or EBNA2-dependent LMP1 and LMP2a signalling.40,55 No change in nAID was seen upon EBV activation or withdrawal.
Discussion
It is unclear whether AID edits DNA directly by binding to it or through editing the RNA coding for an unknown endonuclease22,26,56 ; either way, this requires a phase of AID localization in the nucleus.26 Indeed, tagged and transfected AID has been shown to transiently enter the nucleus, but then is actively prevented from lingering there.29,30 In addition, transfected AID increases the mutation rate of target sequences several fold30,57,58 and binds to actively transcribed DNA regions56 where it can be coprecipitated with members of the DNA transcription machinery. Therefore, it is not surprising that we were able to show in the GC a small subset (less than 3%) of small centroblasts and extrafollicular blasts (14%) displaying nuclear positivity.
The percentage of nAID-positive cells depends on several factors. The first factor is how long AID stays in the nucleus, how rapidly it moves out, and how often it shuttles between the nucleus and the cytoplasm in a given mutation cycle. In primary cells, this parameter is unknown. There seems to be a consensus that AID is safely kept in the cytoplasm most of the time; however, increasing its nuclear permanence does not seems to affect the mutation rate of physiologic targets.29 This is, possibly, because B-cell–specific cofactors shuttle AID into the nucleus and bring it to the correct target DNA sequence.27
The second factor is the mutation rate of the cell where AID nuclear relocation causes the DNA breaks. The mutation rate of the approximately 1-kb long heavy- and light-chain targets in the immune response to a specific antigen has been estimated to be up to 1 × 10–3 per base pair per generation.59 Hence, each mutating GC cell must have nuclear AID at some point during each cell cycle (lasting 6 hours in centroblasts60 ). A single nucleotide mutation is introduced per cell cycle; therefore, hypermutating cells must experience nuclear AID several times during their permanence in the GC.
The third factor is clonal representation in the population analyzed. Each clone is defined as multiple cells with identical mutation repertoire, belonging to a “lineage tree,” differing among each other by fewer than 2 nucleotides.3,4 Cells with an identical mutation repertoire are almost by definition not undergoing additional mutational activity. As exemplified in lymphomas, there is active hypermutation in the clone founder cell, which is shut off when the clone expands, bearing identical mutated sequences in the progeny.52 Quantification of clonal content gives an indirect estimate of how many founder cells must have had nuclear AID before that set of mutations occurred. The clonal progenitors in a given GC are approximately 1 in every 2000 cells (ie, 0.05%).4 In addition, the GC contains variable amounts of clones that do not undergo SHM3,4,61,62 (and D. Dunn-Walters, King's College School of Medicine, London; written personal communication, May 2005). Those clones may not have nuclear AID, in which case they may dilute the nAID percentage.
The fourth factor is the number of AID molecules loaded on each target together with the sensitivity threshold of the detection method. The AID-target ratio in vivo is unknown, and also the stoichiometry and the cofactors involved in the mechanism of mutation are largely unknown. Each Ig locus on both mutating chromosomes is approximately 500 bp, while the DNA segment targeted for CSR is several fold larger. Assuming a 1:1 AID molecule–base pair ratio, cells undergoing SHM are at the lower limit of our detection system, which we believe is in the range of 500 molecules per cell (eg, we detected IL-2Rβ or cMYC by IHC, known to average 500-1000 molecules/cell19,63,64 ).
Cells undergoing CSR may also contribute to the percentage of nuclear AID+ cells in the GC and, having a larger target, may load more AID molecules in the nucleus. RAG molecules, which analogously target a DNA segment of similar size, can indeed be detected in the nucleus of thymocytes and BM B cells.65
CSR has been localized in the tonsils in GC cells putatively identified as “centrocytes.”66 The excised, switched DNA circles characterizing those cells can be identified in at least 1 in 500 of the cells in the “bm4” fraction (light density, CD38+, CD77–, IgD± tonsil cells66 ), similar in frequency to the nAID+ cells detected. We did not find, however, evidence of DNA breaks in nAID+ cells by using phosphorylated histone staining.67,68 The absence of γH2AX and phosphorylated H2B foci in all viable AID+ GC cells may be due to the intrinsic limitation of sensitivity of the immunofluorescence technique69 in detecting in fixed tissue an unknown amount of histone phosphorylation in physiologic single-strand breaks.
GC and extrafollicular nAID+ cells were CDKN1B– and Ki-67+, and therefore fully proliferating and did not contain TP53, which was, however, found occasionally in cytAID+ cells at the border of the GC. This suggests that DNA damage sensors are rarely activated and when they get activated, AID is no longer in the nucleus. The absence of TP53 up-regulation in the GC may be explained by its repression by BCL6,47 or alternatively the genomic stress in vivo does not reach a threshold to activate the TP53 pathway and histone phosphorylation, as seen also in VDJ recombining cells.70 The absence of both CCNB1 and CDKN1B in nAID+ small centroblasts suggests that active replication is required for AID to reach its target, but its presence is not desirable when mutations are repaired or when the cell is ready to exit from an actively cycling state.
Finally, we sought to investigate the effect of known AID-inducing stimuli on cytoplasmic intensity versus nuclear relocation. None of these altered the percentage of nAID, suggesting that de novo AID induction and nuclear translocation respond to a separate and different set of stimuli. A more physiologic model is, however, needed to clarify these issues.
In summary, the GC nAID+ cells we have identified may represent cells undergoing CSR, SHM, or both, but it is not clear in which proportion or whether the actual nucleotide replacement occurs after one cell cycle in a nAID-negative cell.
The apical and basal light zone, where centrocytes encounter T cells and immunocomplex-bearing follicular dendritic cells (FDCs) and where affinity maturation may take place,50,66 are devoid of a significant amount of AID and its driving factors (eg, E47).71,72 Instead, AID is found in the DZ and OZ, 2 distinct but contiguous zones located at the opposite poles of the polarized GC.
DZ centroblasts comprise rather homogeneous and highly proliferating clones of IgD-negative B cells; on the contrary, the OZ is composed of a heterogeneous mixture of proliferating large and small AID+ and AID– B cells, T lymphocytes, and plasmacytoid cells.66 It is morphologically elusive unless CD23 immunohistochemistry is performed, and too close to the mantle zone to be sampled separately from the apical light zone.4 AID and optimal IgD staining reproducibly identifies the OZ and at the same time provides a topographic location for the so-called “germinal center founder cells,” coexpressing IgD, IgM, and GC markers.5 These cells have been shown to display a range of clonally unrelated IgV gene mutations, which are thought to be the start of the clonal expansion taking place in the DZ.5 As a note of caution, this unique population, identified by IHC and flow cytometry in the majority of the GC and expanded in a minority of tonsils, may define an uncommon IgD+ response to a mucosal antigen.
The most likely scenario would be continuous AID expression and transient nuclear relocation in the contiguous DZ and OZ66,73 in BCL6+ cells. Contact with T cells and CD23– FDCs in the OZ may be the site of SHM initiation and/or CSR as suggested previously.66 The DZ would then expand selected clones in a T-cell– and bcruntethered fashion.74 AID+ blasts, which need further mutation, would recycle back to the T-cell–rich OZ. Finally, in the LZ, deaminated DNA bases are repaired in slowly cycling, CDKN1B+ AID-negative cells (hence the detection of excised switch circles66 ) and where fully mutated prememory or preplasma cells are generated.
Of interest, none of the published GC B-cell clones undergoing ongoing SHM has an unmutated expanded progenitor clone in the same GC3,4,60 (and D. Dunn-Walters, written personal communication), suggesting that a B cell, destined to hypermutate in the GC, either disappears with time, leaving no detectable unmutated progenitor population behind, or starts the mutation process outside the GC and expands with diversification upon entering the GC. CD40-dependent and bcr signaling–dependent B-cell activation preceding GC entry has been localized at the border of the mouse lymphoid follicle.2
We describe here a population of extrafollicular large activated IgD– B cells, in which nuclear AID is frequently and strongly expressed. These cells were thought to undergo SHM or, more likely, CSR outside the GC.31 Extrafollicular AID+ cells are PRDM1 negative, a potent transcriptional repressor that is up-regulated starting 18 to 24 hours after LPS stimulation, which turns on CSR without SHM. In addition, these cells partially show evidence of CD40 and bcr signaling, which represses PRDM1-induced plasma cell differentiation.75,76 Thus, extrafollicular AID+ cells are unlikely to represent the early stages of plasma cell commitment (eg, PRDM1 or IRF4 expression), which is the “default” lineage of TI extrafollicular B-cell differentiation,77 unless they represent a very early uncommitted stage.
AID+ extrafollicular cells resemble antigen-activated B cells (Ki-67+ large immunoblasts with prominent nucleoli, partially cMYC, JunB, and CCL22 positive) and overlap with CD30+ large B cells, also considered activated B cells.78 The latter cells, carrying unmutated IgH genes,79 bear a more robust signature of T-cell–dependent, CD40-mediated, IL-4–mediated B-cell activation (cMYC+, CCL22+, IRF4+, JunB+),18 which has been shown necessary for AID induction.33,80 The relationship between the CD30+ cells and the extrafollicular AID+ cells is circumstantial but is suggested by 2 facts: the presence of a phenotypic continuum between the 2 populations, which is not shared by any other extrafollicular B-cell population, and the fact that the nAID population has the highest frequency of double CD30+AID+. Thus, we assume that the antigen- and T-dependent reaction generates a burst of CD30+ B cells, which in turn produces an AID+ progeny in which mutation may start. The destiny of such progeny would be to rapidly enter the GC reaction or to generate an extrafollicular response.
Of interest, another population of mutated large CD20+, IRF4+, CD30– B cells with dendritic morphology (LBC-DM) has been described.81 AID+ interfollicular cells are instead CD20+, IRF4–, highly proliferating, and occasionally CD30+. More studies are needed to define the relationship between these 2 populations.
The nuclear localization of AID described in this article will not solve the controversy of the mechanism by which AID acts. We found nucleolar exclusion of this protein, but further and more detailed studies are necessary. The largely cytoplasmic localization of AID remains to be explained; this may represent a reservoir, actively prevented from entering the nucleus except when needed, similar to NFkB complexes,82 or, alternatively, AID may have an independent cytoplasmic function that is yet to be discovered.
The established concepts of SHM and CSR location inside the GC are once more validated by our findings, although we suggest a more precise distribution within the GC zones. Whether AID extrafollicular expression is linked to the initiation of SHM or to a GC-independent CSR remains a question open for further investigation.
Prepublished online as Blood First Edition Paper, January 31, 2006; DOI 10.1182/blood-2005-10-4170.
G.C. participated in experimental analysis and writing, and provided cases; M.B. participated in experimental analysis; R.S. provided cases and participated in experimental analysis; E.K. participated in experimental analysis and provided vital reagents; B.A. provided cases, evaluated pathology, and contributed to writing; and G.N. evaluated pathology and contributed to writing.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We wish to thank Riccardo Dalla-Favera for scientific support, ample discussion, and continuous encouragement on the project, and Michael L. Shelanski for substantial support and advice. The suggestions and scientific insight of Ulf Klein, Ralf Küppers, Matthew Scharff, Ellen Hsu, Deborah Dunn-Walters, and Laura Pasqualucci are gratefully acknowledged. Deborah Dunn-Walters kindly shared unpublished observations. Maryellen Benito and Paula M. Smith provided outstanding technical help. Lin Yang and the Molecular Pathology Facility of the Herbert Irving Cancer Center of Columbia University Medical Center provided superb histology service. We thank the Optical Microscopy Facility, Herbert Irving Comprehensive Cancer Center, Columbia University, for the help. Johannes Gerdes (Molecular Immunology, Borstel, Germany), Jiri Bartek (Institute of Molecular Pathology, Copenhagen, Denmark), Brunangelo Falini (Perugia University, Perugia, Italy), and Anna Lasorella (ICG, Columbia University, NY) generously provided antibodies.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal