Abstract
We evaluated the expression of 2 members of the Syk family, ZAP-70 and Syk, in acute lymphoblastic leukemia (ALL) samples, using data derived from a series of 33 T-ALL and 95 B-lineage adult ALL patients analyzed by oligonucleotide arrays. Of the B-lineage ALL cases, 37 were BCR/ABL+, 10 were ALL1/AF4+, 5 were E2A/PBX1+, and 43 carried no known molecular abnormality. ZAP-70 was highly expressed in T-ALL. A high ZAP-70 expression was also found in a proportion of B-lineage ALL, the highest levels being associated with the E2A/PBX1+ group and the lowest with ALL1/AF4+ cases (P < .001). A higher ZAP-70 expression was also observed in the pre-B group (P < .001). Remarkably, Syk expression was always preserved, suggesting that ZAP-70 expression is not substitutive of Syk. At the protein level, ZAP-70 was evaluated on 39 newly diagnosed ALL patients (25 adults, 14 children) and was detected in 23 cases (59%). ZAP-70 expression was consistently found in Igμ+ cases. Evaluation of long-term outcome in cases without molecular abnormalities showed that the higher levels of ZAP-70 were coupled to a higher relapse rate. In ALL, ZAP-70 expression is associated with the E2A/PBX1 rearrangement and pre-B stage and may have a prognostic role and be a candidate molecule for targeted therapies.
Introduction
The zeta-associated protein (ZAP) is a 70-kDa molecule associated with the ζ chain of the CD3 receptor complex of the lymphocyte population1 and belongs to the Syk (spleen tyrosine kinase) family of tyrosine kinases. ZAP-70 plays an important role in signaling initiation, activation, and phosphorylation of multiple downstream targets. Briefly, after T-cell receptor (TCR) ligation, activation of Src kinase occurs and induces activation of the immune receptor tyrosine–based activation motifs (ITAMs).1,2 This process eventually leads to the recruitment and activation of ZAP-70, which in turn activates several downstream targets, such as phospholipase Cγ/Ca++ signaling pathway and the mitogen-activated protein kinase (MAPK) kinase pathway.3
A similar pathway is sustained in B cells by Syk: activation of the B-cell receptor (BCR) induces rapid phosphorylation of ITAMs of Igα and Igβ. Once phosphorylated, ITAMs recruit Syk, thus inducing its phosphorylation and the activation of several downstream targets.4
Historically, ZAP-70 has been considered a tyrosine kinase exclusively associated to the T and natural killer (NK)–cell compartments.1 The role of the ZAP-70 molecule in B-cell development has been recently established in a mouse model.5 This has shown that this protein is also expressed in B-lineage cells and that it appears to play a role in their early development and, in particular, during the pro-B to pre-B transition stage. This becomes evident in the absence of Syk. The study by Schweighoffer et al5 demonstrated functional redundancy between these 2 kinases and also showed that the absence of both kinases, and not only Syk, is necessary to block pre-BCR–induced events, such as differentiation into pre-B cells, cell proliferation, and heavy chain allelic exclusion.
In the present study, we sought to evaluate the expression of ZAP70 and SYK in acute lymphoblastic leukemia (ALL), using data derived from a series of 128 adult ALL patients evaluated by oligonucleotide arrays and by the quantification of the protein levels. Our results indicate that ZAP70 is, as expected, highly expressed in T-ALL but also in different amounts in B-lineage ALL and that the expression of this gene is strongly correlated with the E2A/PBX1 rearrangement. Moreover, our results indicate that in B-lineage ALL, the levels of ZAP70 expression increase along with the maturation process of the leukemic cell. In fact, evaluation of ZAP70 through oligonucleotide arrays showed that the lowest levels of expression are found in pro-B cells and the highest levels at the pre-B stage. Finally, stratification of ALL patients without known molecular aberrations on the basis of the degree of mRNA ZAP70 expression allowed for the definition of 2 groups with a different outcome, a shorter survival being associated with a higher expression of this molecule.
Patients, materials, and methods
Patient characteristics
One hundred twenty-eight adult patients with a diagnosis of ALL were studied at the onset of the disease. All patients were enrolled in the Italian multicenter clinical trial GIMEMA (Gruppo Italiano Malattie Ematologiche dell'Adulto) 0496 and gave their informed consent for biologic studies according to the Declaration of Helsinki. The study was approved by the Institutional Review Board of the Department of Cellular Biotechnologies and Hematology, University “La Sapienza” of Rome. After diagnosis, leukemic cells were collected and isolated by density gradient centrifugation over Ficoll-Hypaque (Pharmacia, Uppsala, Sweden). After isolation, samples were cryopreserved in liquid nitrogen at our Institution in Rome. All samples contained more than 90% leukemic cells. In agreement with the protocol guidelines, all samples were extensively characterized for immunophenotypic, cytogenetic, and molecular features. Follow-up data were collected at our Institution.
mRNA expression of ZAP70 and p72-SYK genes
For oligonucleotide array analysis, cryopreserved leukemia cells were rapidly thawed and total RNA was extracted using the TRIzol reagent (Gibco, Grand Island, NY) and further purified using the SV total RNA isolation system (Promega, Madison, WI), with minor modifications. Two microliters (μL) of total RNA was electrophoresed on agarose gels to assess RNA quality. HGU95aV2 gene chips (Affymetrix, Santa Clara, CA) were used to determine gene expression profiles. The detailed protocol for sample preparation and microarray processing is available on the manufacturer's website.6
Affymetrix U95Av2 gene expression data were processed and analyzed with dChip,7 which uses an invariant set normalization method where the array with median overall intensity was chosen as the baseline for normalization. Model-based expressions were computed for each array and probe set using the perfect match-mismatch (PM-MM) model.8
Statistical analysis
Statistical analyses on ZAP70 and SYK expression data were performed using a free software, StatCrunch.11 Group comparisons were performed using either a 2-sample test or analysis of variance (ANOVA), and correlation analysis was evaluated using the Pearson coefficient. Box plots were generated using the same program.
For comparison between high versus low ZAP70-expressing samples, an ANOVA method was applied. The ANOVA method is provided by R language12 and included in dChip. From this analysis, probe sets with a nominal P value less than .005 were retained.
Survival curves were estimated by the Kaplan-Meier method and the differences were compared by the log-rank test, using the package SAS (SAS Institute, Cary, NC). The cut point for categorizing ZAP70 was chosen on the basis of the quantiles of the observed distribution of the variable, considering “high expression” as the range defined by the 20% highest values of ZAP70.
PCR
Polymerase chain reaction (PCR) was performed on cDNA of 3 selected E2A/PBX1+ samples and 3 ALL1/AF4+ samples, as well as on 3 samples of purified B cells obtained from the peripheral blood of healthy donors. Positive and negative controls were represented by cDNA derived from the Jurkat and Raji cell lines, respectively.
cDNA was generated using the Advantage reverse transcriptase (RT) for PCR Kit (BD Biosciences, Palo Alto, CA), starting from 1 μg of total RNA, and resuspended in a final volume of 100 μL of water according to the manufacturer's instructions. PCR reactions were performed as follows: 1 cycle of denaturation at 94°C for 2 minutes, 35 cycles of amplification at 94°C for 1 minute, 60°C for 1 minute, 72°C for 1 minute, and a final extension at 72°C for 10 minutes. For evaluation, the following primers were used: ZAP70 forward (F), 5′-ATG TGC GCT TCC ACC ACT TTC-3′; ZAP70 reverse (R), 5′-AC GTC TGG CGC ACG TAG TCA-3′; GAPDH F, 5′-TGA TGA CAT CAA GAA GGT GGT GGT GAA G-3′; and GAPDH R, 5′-TCC TTG GAG GCC ATG TGG GCC AT-3′.
Leukemic samples were positively selected using the CD19 human microbeads (Miltenyi, Bergisch Gladbach, Germany), according to the manufacturer's instructions, thus achieving a purity over 99%. For healthy donors, a further separation step was performed: samples were first depleted for CD2 (Miltenyi) and, after an overnight incubation at 37°C and 5% CO2, cells were positively selected for CD19.
Flow cytometry
Mononuclear cells were stained with fluorochrome-conjugated monoclonal antibodies and analyzed by flow cytometry (FACSCalibur; Becton Dickinson, San Jose, CA). B-lineage leukemic cells were characterized for the surface expression of CD45, CD38, CD10, CD19, CD20, CD22, CD34, CD13, CD33, HLA-DR, CD133, and CD66c antigens (Becton Dickinson; Beckman Coulter, Fullerton, CA). After fixation and permeabilization (Fix and Perm; Caltag Laboratories, Burlingame, CA), leukemic cells were evaluated for the intracytoplasmic and nuclear expression of CD79a, IgM, and TdT.
ZAP-70 protein detection
ZAP-70 protein expression was evaluated on mononuclear cells, after fixation and permeabilization, using an immunocytochemical method. All B-lineage ALL patients evaluated expressed intracytoplasmic CD79a, surface CD19, and nuclear TdT antigens. Cytospins were prepared with a concentration of 5 × 104 cells per slide, air-dried overnight, wrapped in aluminum foil, and stored at -20°C until immunostaining. The unconjugated anti–ZAP-70 clone 2F3.2 (Upstate Biotechnology, Waltham, MA) monoclonal antibody used for the immunocytochemical detection was used at a concentration of 1:60. The immunocytochemical reaction was performed with the immunoperoxidase technique using Dako (Carpinteria, CA) reagents, as previously described.13 Again, positive and negative controls were represented by the Jurkat and Raji cell lines, respectively. The proportion of ZAP-70+ cells was evaluated by light microscopy with oil immersion (original magnification × 1000) examining 500 cells per sample. Cases were considered as ZAP-70+ if at least 20% of the mononucleated cells were positive.
Results
Patient characteristics
Of the 128 patients analyzed, 33 samples were of T-cell origin and 95 were represented by B-lineage ALL. Within the B-lineage group, further immunophenotypic characterization showed a pro-B stage in 21% of cases, whereas 54% were considered as common ALL (CD10+) and 25% were pre-B ALL. From a molecular standpoint, 37 samples were BCR/ABL+, 10 carried an ALL1/AF4 rearrangement, 5 harbored an E2A/PBX1 rearrangement, and 43 samples did not carry any known molecular abnormality.14,15
ZAP70 mRNA expression is associated with E2A/PBX1 rearrangements
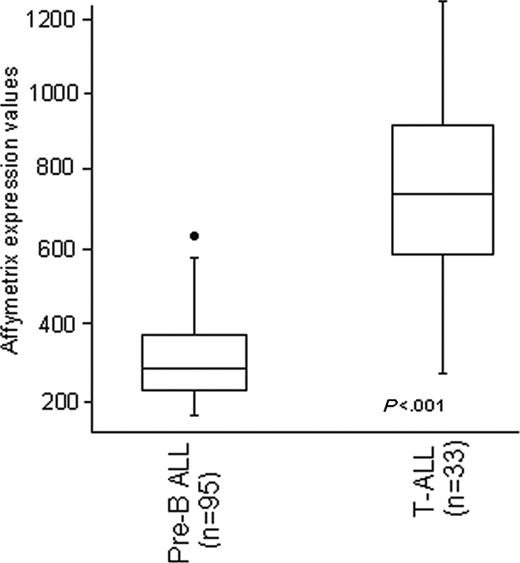
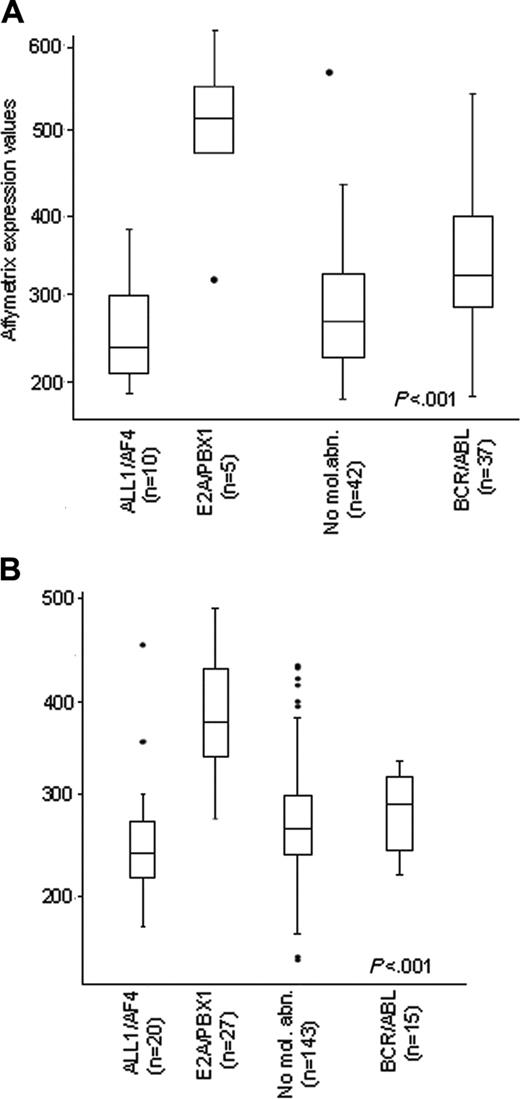
Our first approach aimed at evaluating ZAP70 expression in the 128 samples analyzed by oligonucleotide arrays. As shown in Figure 1, ZAP70 was found at significantly higher levels in T-ALL cases compared with B-lineage ALL (average values 752 ± 268 and 309 ± 105; P < .001). However, a proportion of B-lineage ALL also showed high levels of ZAP70. As shown in Figure 2A, the highest expression levels of ZAP70 were detected in samples carrying the E2A/PBX1 rearrangement, as opposed to samples positive for the ALL1/AF4 and BCR/ABL rearrangements and to cases without known molecular aberrations. In fact, all 5 E2A/PBX1+ samples had high levels of ZAP70 expression, whereas the lowest values of expression were found in the ALL1/AF4+ cases (P < .001). The percentage of T-cell contamination was not different in E2A/PBX1+ cases compared with non-E2A/PBX1+ B-lineage ALL cases (median CD2 expression 4.5% and 4%, respectively). The above findings were further validated by examining previously published data from Yeoh et al9 in a large pediatric ALL cohort (P < .001; Figure 2B).
Gene expression values evaluated by Affymetrix oligonucleotide arrays of ZAP70. The first box plot represents the 95 B-lineage cases and the second plot is comprehensive of the 33 T-ALL cases. Box plots define the median values, 25% to 75% of values around the median, and the range of values. Values were normalized using the dChip software. Values were significantly higher in T-ALL.
Gene expression values evaluated by Affymetrix oligonucleotide arrays of ZAP70. The first box plot represents the 95 B-lineage cases and the second plot is comprehensive of the 33 T-ALL cases. Box plots define the median values, 25% to 75% of values around the median, and the range of values. Values were normalized using the dChip software. Values were significantly higher in T-ALL.
ZAP70 is expressed in B cells of E2A/PBX cases
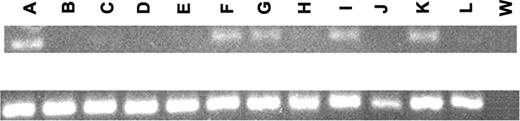
Since ZAP70 is strongly expressed in normal T cells, we validated, on an additional series of samples, the results obtained by oligonucleotide arrays by performing PCR analyses on 3 E2A/PBX1+ samples, 3 ALL1/AF4+ samples, as well as on purified CD19+ cells from 3 healthy donors and on the Jurkat and Raji cell lines. As expected, ZAP70 was found expressed in the Jurkat T-cell line, but not in the Raji B-cell line, and in the CD19+ cells from the peripheral blood of 3 healthy controls; conversely, ZAP70 was expressed in all the E2A/PBX1+ cases but in only 1 ALL1/AF4+ sample, confirming that this tyrosine kinase is preferentially expressed in cases carrying the E2A/PBX1 rearrangement. These findings are also consistent with a recent report that highlighted the absence of ZAP70 in peripheral blood B lymphocytes16 (Figure 3).
ZAP70 is expressed in cells that are undergoing maturation
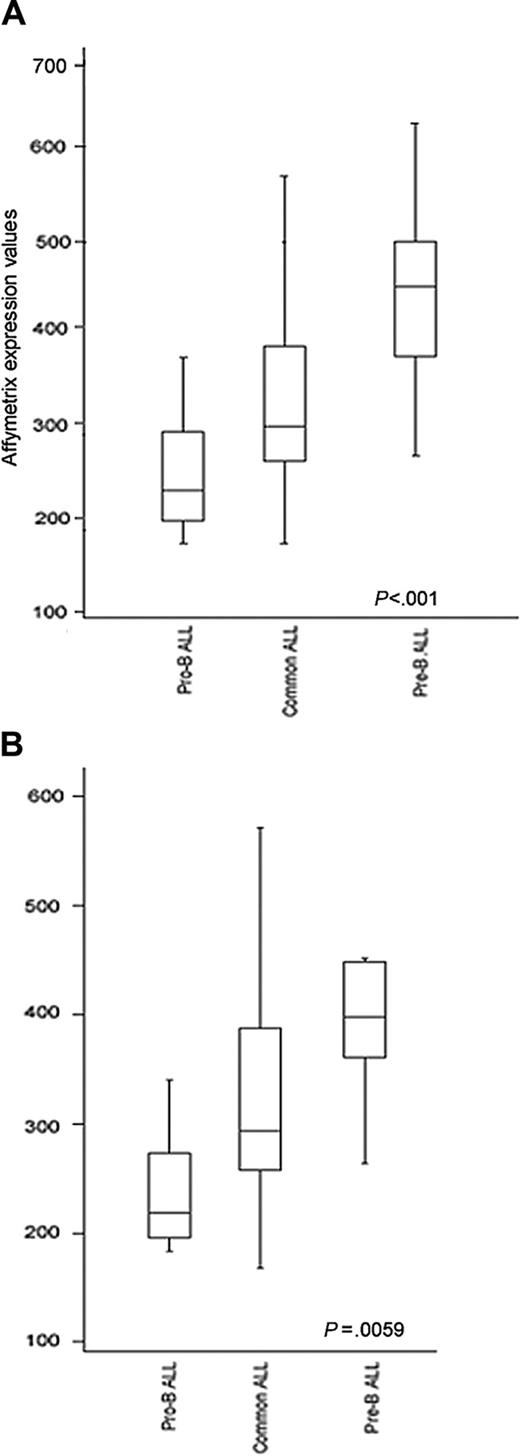
Since ZAP70 was found more highly expressed in E2A/PBX1+ cases and at low levels in the ALL1/AF4+ cases, this suggested that this protein may undergo changes in its expression during the maturation process, specifically increasing from the pro-B to pre-B stage. In fact, it is well established that ALL1/AF4+ cases are arrested at a pro-B stage,17 whereas the majority of E2A/PBX1+ cases have a pre-B phenotype.18 In order to validate this hypothesis, 2 approaches were used. First, samples were subdivided into 3 groups according to their degree of differentiation (pro-B, common, and pre-B ALL) and independently of molecular alterations. As shown in Figure 4A-B, the highest levels of expression were detected in the pre-B group (P < .001); this finding remained significant also when E2A/PBX1+ and ALL1/AF4+ cases were excluded (P = .005). Second, we evaluated the expression of ZAP70 in relation to 2 maturative antigens, CD20 and Igμ chain. The degree of correlation between CD20 mRNA and protein levels evaluated by the Pearson correlation coefficient was 0.67, showing a good concordance between these 2 techniques. Samples were divided according to CD20 antigen expression (20% cut-off on the cell surface): 42% were positive and 58% were negative. The average levels of ZAP70 mRNA expression were 339.53 ± 105.78 in CD20+ samples and 297.97 ± 104.93 in CD20- samples. However, this difference did not reach statistical significance (P = .095).
B-lineage ALL expression levels.ZAP70 RNA expression in 95 adult B-lineage ALL patients (A) subdivided according to molecular groups. E2A/PBX1 samples showed the highest levels of ZAP70 expression. Similar results were observed in the pediatric cohorts by Yeoh et al9 (B). No mol abn indicates no molecular abnormalities.
B-lineage ALL expression levels.ZAP70 RNA expression in 95 adult B-lineage ALL patients (A) subdivided according to molecular groups. E2A/PBX1 samples showed the highest levels of ZAP70 expression. Similar results were observed in the pediatric cohorts by Yeoh et al9 (B). No mol abn indicates no molecular abnormalities.
RT-PCR. RT-PCR of ZAP70 (top) and GAPDH (bottom). (A) Jurkat cell line; (B) Raji cell line; (C-E) CD19+ cells from the peripheral blood of 3 healthy donors; (F) primary T-ALL leukemic cells; (G-I) 3 E2A/PBX1+ leukemic cells; (J-L) 3 ALL1/AF4+ leukemic cells; and (W) negative control.
RT-PCR. RT-PCR of ZAP70 (top) and GAPDH (bottom). (A) Jurkat cell line; (B) Raji cell line; (C-E) CD19+ cells from the peripheral blood of 3 healthy donors; (F) primary T-ALL leukemic cells; (G-I) 3 E2A/PBX1+ leukemic cells; (J-L) 3 ALL1/AF4+ leukemic cells; and (W) negative control.
ZAP-70 RNA expression and phenotypic differentiation. ZAP-70 RNA expression in B-lineage ALL subdivided according to the degree of phenotypic differentiation with (A) or without E2A/PBX1+ and ALL1/AF4+ samples (B).
ZAP-70 RNA expression and phenotypic differentiation. ZAP-70 RNA expression in B-lineage ALL subdivided according to the degree of phenotypic differentiation with (A) or without E2A/PBX1+ and ALL1/AF4+ samples (B).
The degree of correlation between Igμ mRNA and protein expression was very poor, reflecting the fact that this transcript is always present at the RNA level in B-lineage ALL samples. Samples were stratified according to the Igμ protein expression (8% cut-off): 28% were positive and 78% were negative. The average levels of ZAP70 expression were significantly higher in Igμ+ samples (443.16 ± 105.21) than in Igμ- samples (294.05 ± 95.52; P = .002).
Comparative analysis between high versus low ZAP70 expression samples
Next, we examined if, within the samples that did not carry any known molecular abnormality, ZAP70 expression was associated with a different pattern of gene expression. Of the 43 samples that carried no known molecular abnormality, 2 samples were excluded because they showed by comparative genomic hybridization (CGH) analysis a gain of chromosome 1q and 1q23, respectively, and by gene expression analysis a profile that was strongly associated to that of E2A/PBX1+ samples.19 Of the 41 cases analyzed, 8 had high levels and 33 had low levels of ZAP70 expression.
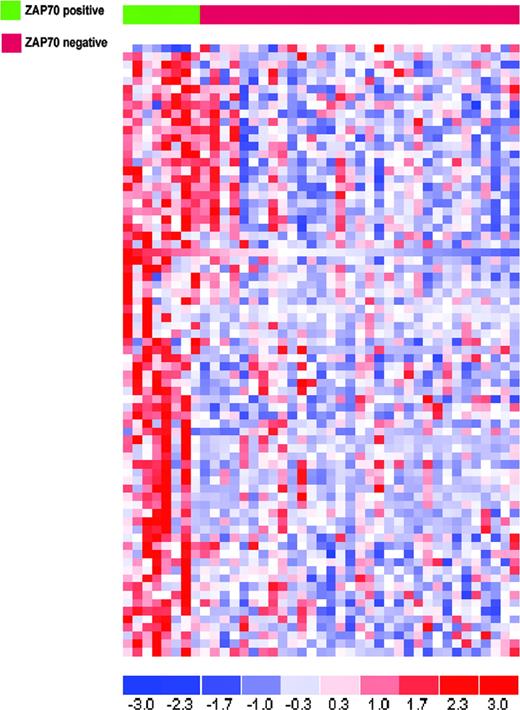
Seventy-five probe sets corresponding to 73 genes were selected by ANOVA with a P value cut-off less than .005. All but one gene were more highly expressed in the “ZAP70 high” group (Figure 5). The genes highly expressed in samples with high levels of ZAP70 can be functionally subdivided in the following categories: (1) immune response (TCRB, TNFRSF1B, IL27RA, LAT, ANXA11, CTSW, IGJ, and IL32); (2) apoptosis (CASP8, CASP10, CRADD, BBC3, and TNFRSF1B); (3) regulation of transcription (MYBL1, E2F4, FOXM1, TP53BP1, TCF20, BRF1, PBX3, and MLL4); and (4) DNA repair (XPC and ATM). A detailed list of the probe sets identified is summarized in Table 1.
ALL: differentially expressed genes between high versus low ZAP70 cases with no known molecular aberrations
Gene symbol . | Probe set . | Locuslink ID . | P . | GO process . | Chromosomal location . |
|---|---|---|---|---|---|
| ZAP70 | 1498_at | 7535 | <.001 | Immune response | 2q12 |
| AP3D1 | 36173_r_at | 8943 | <.001 | Intracellular protein transport | 19p13.3 |
| SEMA6A | 36275_at | 57556 | <.001 | Cytoskeleton organization and biogenesis | 5q23.1 |
| CSPG2 | 38112_g_at | 1462 | <.001 | Development | 5q14.3 |
| ITGA5 | 39753_at | 3678 | <.001 | Integrin-mediated signaling pathway | 12q11-q13 |
| CRADD | 1211_s_at | 8738 | <.001 | Induction of apoptosis via death domain receptors | 12q21.33-q23.1 |
| AGPS | 39225_at | 8540 | <.001 | Lipid metabolism | 2q31 |
| AP3D1 | 36172_s_at | 8943 | <.001 | Intracellular protein transport | 19p13.3 |
| LOC90410 | 41561_s_at | 55977 | <.001 | Unknown | 17q11.2 |
| LAT | 40688_at | 27040 | <.001 | Regulation of T-cell activation | 16p11.2 |
| ACVR2 | 35162_s_at | 92 | .001 | – | 2q22.2-q23.3 |
| SOCS1 | 41593_at | 8651 | .001 | Intracellular signaling cascade | 16p13.13 |
| CASP10 | 1326_at | 843 | .001 | Induction of apoptosis | 2q33-q34 |
| XPC | 35647_at | 7508 | .001 | Nucleotide-excision repair | 3p25 |
| TNFRSF1B | 33813_at | 7133 | .001 | Apoptosis | Ip36.3-p36.2 |
| TCRB | 1105_s_at | 6957 | .001 | Immune response | 7q34 |
| HPS1 | 38467_at | 3257 | .001 | – | 10q23.1-q23.3 |
| PTE1 | 36841_at | 10005 | .001 | Acyl-coA metabolism | 20q12-q13.1 |
| TNFRSF1B | 1583_at | 7133 | .001 | Apoptosis | 1p36.3-p36.2 |
| CASP8 | 33774_at | 841 | .001 | Apoptosis | 2q33-q34 |
| TR1P6 | 39341_at | 7205 | .001 | – | 7q22 |
| PCSK7 | 34361_at | 9159 | .001 | Proteolysis and peptidolysis | 11q23-q24 |
| LCK | 33238_at | 3932 | .001 | Intracellular signaling cascade | 1p34.3 |
| CASP8 | 33775_s_at | 841 | .001 | Induction of apoptosis | 2q33-q34 |
| KLRB1 | 35449_at | 3820 | .002 | Cell surface receptor linked signaling | 12p13.1 |
| LRRC15 | 34778_at | 131578 | .002 | – | 3q29 |
| PHC2 | 36960_at | 1912 | .002 | – | 1p34.3 |
| MYBL1 | 160043_at | 4603 | .002 | Regulation of transcription, DNA dependent | 8q22 |
| CKMT2 | 37592_at | 1160 | .002 | Muscle contraction | 5q13.3 |
| RQCD1 | 722_at | 9125 | .002 | Sex differentiation | 2q35 |
| RRAD | 1776_at | 6236 | .002 | GTPase activity | 16q22 |
| CNOT8 | 41361_at | 9337 | .002 | Negative regulation of cell proliferation | 5q31-33 |
| OR511 | 31428_at | 10798 | .002 | G-protein-coupled receptor protein signaling | 11q11 |
| GPR56 | 35769_at | 9289 | .002 | Cell-cell signaling | 16q13 |
| EPM2AIP1 | 35209_at | 9852 | .002 | – | 3p22.1 |
| GRIN1 | 37134_f_at | 2902 | .002 | Glutamate-signaling pathway | 9q34.3 |
| CTGF | 36638_at | 1490 | .002 | Epidermis development | 6q23.1 |
| PDE9A | 33709_at | 5152 | .003 | Signal transduction | 21q22.3 |
| IL27RA | 37844_at | 9466 | .003 | – | 19p13.11 |
| KIAA0542 | 36545_s_at | 9814 | .003 | – | 22q12.2 |
| FGR | 1780_at | 2268 | .003 | Protein amino acid phosphorylation | 1p36.2-p36.1 |
| SPINK1 | 38582_at | 6690 | .003 | – | 5q32 |
| FLJ10803 | 37610_at | 55744 | .003 | – | 7p13 |
| E2F4 | 38706_at | 1874 | .003 | Regulation of cell cycle | 16q21-q22 |
| FOXM1 | 41324_g_at | 2305 | .003 | Transcription from RNA polymerase II promoter | 12p13 |
| CTSW | 40718_at | 1521 | .003 | Immune response | 11q13.1 |
| TP53BP1 | 1711_at | 7158 | .003 | Transcription | 15q15-q21 |
| TTN | 40795_at | 7273 | .003 | Striated muscle contraction | 2q31 |
| BAIAP2 | 37760_at | 10458 | .003 | Insulin receptor signaling pathway | 17q25 |
| ANXA11 | 36637_at | 311 | .003 | Immune response | 10q23 |
| IGJ | 37006_at | 10569 | .003 | Immune response | 4q21 |
| PFAAP5 | 1532_g_at | 10443 | .004 | – | 13q12-q13 |
| HPN | 37639_at | 3249 | .004 | Proteolysis and peptidolysis | 19q11-q13.2 |
| CD200 | 37716_at | 4345 | .004 | – | 3q12-q13 |
| TCF20 | 33795_at | 6942 | .004 | Regulation of transcription, DNA dependent | 22q13.3 |
| BRF1 | 141_s_at | 2972 | .004 | Transcription initiation from RNA polymerase III | 14q |
| BBC3 | 1700_at | 27113 | .004 | Apoptosis | 19q13.3-q13.4 |
| KIAA0194 | 34221_at | 22993 | .004 | Regulation of transcription, DNA dependent | 5q33.1 |
| CDK9 | 387_at | 1025 | .004 | Regulation of cell cycle | 9q34.1 |
| CTNND1 | 40444_s_at | 1500 | .004 | Cell-cell adhesion | 11q11 |
| IL32 | 39119_s_at | 9235 | .004 | Immune response | 16p13.3 |
| DGKA | 32716_at | 1606 | .004 | Intracellular signaling cascade | 12q13.3 |
| PBX3 | 32696_at | 5090 | .004 | Regulation of transcription, DNA dependent | 9q33-q34 |
| MLL4 | 38284_at | 9757 | .004 | Regulation of transcription, DNA dependent | 19q13.1 |
| SF3B3 | 32753_at | 23450 | .004 | RNA splicing | 16q22.1 |
| Unknown | 34204_at | unknown | .004 | Unknown | Unknown |
| STK39 | 40966_at | 27347 | .004 | Protein amino acid phosphorylation | 2q24.3 |
| PIK4CA | 40783_s_at | 5297 | .004 | Phosphatidylinositol biosynthesis | 22q11.21 |
| ATM | 1863_s_at | 472 | .005 | DNA repair | 11q22-q23 |
| Unknown | 38423_at | Unknown | .005 | Unknown | 17q21.2 |
| AIP | 36586_at | 9049 | .005 | Protein folding | 11q13.3 |
| GAB1 | 1249_at | 2549 | .005 | Cell proliferation | 4q31.21 |
| SLC2A1 | 40507_at | 6513 | .005 | Glucose transport | 1p35-p31.3 |
| Unknown | 32467_at | Unknown | .005 | Unknown | Unknown |
| PTPN18 | 33453_at | 537 | .005 | Protein amino acid dephosphorylation | 2q21.1 |
Gene symbol . | Probe set . | Locuslink ID . | P . | GO process . | Chromosomal location . |
|---|---|---|---|---|---|
| ZAP70 | 1498_at | 7535 | <.001 | Immune response | 2q12 |
| AP3D1 | 36173_r_at | 8943 | <.001 | Intracellular protein transport | 19p13.3 |
| SEMA6A | 36275_at | 57556 | <.001 | Cytoskeleton organization and biogenesis | 5q23.1 |
| CSPG2 | 38112_g_at | 1462 | <.001 | Development | 5q14.3 |
| ITGA5 | 39753_at | 3678 | <.001 | Integrin-mediated signaling pathway | 12q11-q13 |
| CRADD | 1211_s_at | 8738 | <.001 | Induction of apoptosis via death domain receptors | 12q21.33-q23.1 |
| AGPS | 39225_at | 8540 | <.001 | Lipid metabolism | 2q31 |
| AP3D1 | 36172_s_at | 8943 | <.001 | Intracellular protein transport | 19p13.3 |
| LOC90410 | 41561_s_at | 55977 | <.001 | Unknown | 17q11.2 |
| LAT | 40688_at | 27040 | <.001 | Regulation of T-cell activation | 16p11.2 |
| ACVR2 | 35162_s_at | 92 | .001 | – | 2q22.2-q23.3 |
| SOCS1 | 41593_at | 8651 | .001 | Intracellular signaling cascade | 16p13.13 |
| CASP10 | 1326_at | 843 | .001 | Induction of apoptosis | 2q33-q34 |
| XPC | 35647_at | 7508 | .001 | Nucleotide-excision repair | 3p25 |
| TNFRSF1B | 33813_at | 7133 | .001 | Apoptosis | Ip36.3-p36.2 |
| TCRB | 1105_s_at | 6957 | .001 | Immune response | 7q34 |
| HPS1 | 38467_at | 3257 | .001 | – | 10q23.1-q23.3 |
| PTE1 | 36841_at | 10005 | .001 | Acyl-coA metabolism | 20q12-q13.1 |
| TNFRSF1B | 1583_at | 7133 | .001 | Apoptosis | 1p36.3-p36.2 |
| CASP8 | 33774_at | 841 | .001 | Apoptosis | 2q33-q34 |
| TR1P6 | 39341_at | 7205 | .001 | – | 7q22 |
| PCSK7 | 34361_at | 9159 | .001 | Proteolysis and peptidolysis | 11q23-q24 |
| LCK | 33238_at | 3932 | .001 | Intracellular signaling cascade | 1p34.3 |
| CASP8 | 33775_s_at | 841 | .001 | Induction of apoptosis | 2q33-q34 |
| KLRB1 | 35449_at | 3820 | .002 | Cell surface receptor linked signaling | 12p13.1 |
| LRRC15 | 34778_at | 131578 | .002 | – | 3q29 |
| PHC2 | 36960_at | 1912 | .002 | – | 1p34.3 |
| MYBL1 | 160043_at | 4603 | .002 | Regulation of transcription, DNA dependent | 8q22 |
| CKMT2 | 37592_at | 1160 | .002 | Muscle contraction | 5q13.3 |
| RQCD1 | 722_at | 9125 | .002 | Sex differentiation | 2q35 |
| RRAD | 1776_at | 6236 | .002 | GTPase activity | 16q22 |
| CNOT8 | 41361_at | 9337 | .002 | Negative regulation of cell proliferation | 5q31-33 |
| OR511 | 31428_at | 10798 | .002 | G-protein-coupled receptor protein signaling | 11q11 |
| GPR56 | 35769_at | 9289 | .002 | Cell-cell signaling | 16q13 |
| EPM2AIP1 | 35209_at | 9852 | .002 | – | 3p22.1 |
| GRIN1 | 37134_f_at | 2902 | .002 | Glutamate-signaling pathway | 9q34.3 |
| CTGF | 36638_at | 1490 | .002 | Epidermis development | 6q23.1 |
| PDE9A | 33709_at | 5152 | .003 | Signal transduction | 21q22.3 |
| IL27RA | 37844_at | 9466 | .003 | – | 19p13.11 |
| KIAA0542 | 36545_s_at | 9814 | .003 | – | 22q12.2 |
| FGR | 1780_at | 2268 | .003 | Protein amino acid phosphorylation | 1p36.2-p36.1 |
| SPINK1 | 38582_at | 6690 | .003 | – | 5q32 |
| FLJ10803 | 37610_at | 55744 | .003 | – | 7p13 |
| E2F4 | 38706_at | 1874 | .003 | Regulation of cell cycle | 16q21-q22 |
| FOXM1 | 41324_g_at | 2305 | .003 | Transcription from RNA polymerase II promoter | 12p13 |
| CTSW | 40718_at | 1521 | .003 | Immune response | 11q13.1 |
| TP53BP1 | 1711_at | 7158 | .003 | Transcription | 15q15-q21 |
| TTN | 40795_at | 7273 | .003 | Striated muscle contraction | 2q31 |
| BAIAP2 | 37760_at | 10458 | .003 | Insulin receptor signaling pathway | 17q25 |
| ANXA11 | 36637_at | 311 | .003 | Immune response | 10q23 |
| IGJ | 37006_at | 10569 | .003 | Immune response | 4q21 |
| PFAAP5 | 1532_g_at | 10443 | .004 | – | 13q12-q13 |
| HPN | 37639_at | 3249 | .004 | Proteolysis and peptidolysis | 19q11-q13.2 |
| CD200 | 37716_at | 4345 | .004 | – | 3q12-q13 |
| TCF20 | 33795_at | 6942 | .004 | Regulation of transcription, DNA dependent | 22q13.3 |
| BRF1 | 141_s_at | 2972 | .004 | Transcription initiation from RNA polymerase III | 14q |
| BBC3 | 1700_at | 27113 | .004 | Apoptosis | 19q13.3-q13.4 |
| KIAA0194 | 34221_at | 22993 | .004 | Regulation of transcription, DNA dependent | 5q33.1 |
| CDK9 | 387_at | 1025 | .004 | Regulation of cell cycle | 9q34.1 |
| CTNND1 | 40444_s_at | 1500 | .004 | Cell-cell adhesion | 11q11 |
| IL32 | 39119_s_at | 9235 | .004 | Immune response | 16p13.3 |
| DGKA | 32716_at | 1606 | .004 | Intracellular signaling cascade | 12q13.3 |
| PBX3 | 32696_at | 5090 | .004 | Regulation of transcription, DNA dependent | 9q33-q34 |
| MLL4 | 38284_at | 9757 | .004 | Regulation of transcription, DNA dependent | 19q13.1 |
| SF3B3 | 32753_at | 23450 | .004 | RNA splicing | 16q22.1 |
| Unknown | 34204_at | unknown | .004 | Unknown | Unknown |
| STK39 | 40966_at | 27347 | .004 | Protein amino acid phosphorylation | 2q24.3 |
| PIK4CA | 40783_s_at | 5297 | .004 | Phosphatidylinositol biosynthesis | 22q11.21 |
| ATM | 1863_s_at | 472 | .005 | DNA repair | 11q22-q23 |
| Unknown | 38423_at | Unknown | .005 | Unknown | 17q21.2 |
| AIP | 36586_at | 9049 | .005 | Protein folding | 11q13.3 |
| GAB1 | 1249_at | 2549 | .005 | Cell proliferation | 4q31.21 |
| SLC2A1 | 40507_at | 6513 | .005 | Glucose transport | 1p35-p31.3 |
| Unknown | 32467_at | Unknown | .005 | Unknown | Unknown |
| PTPN18 | 33453_at | 537 | .005 | Protein amino acid dephosphorylation | 2q21.1 |
Genes are rank ordered according to nominal P values. All genes have high expression, with the exception of AIP. GO indicates gene ontology; –, not recorded.
ZAP70 expression is associated with a worse long-term outcome
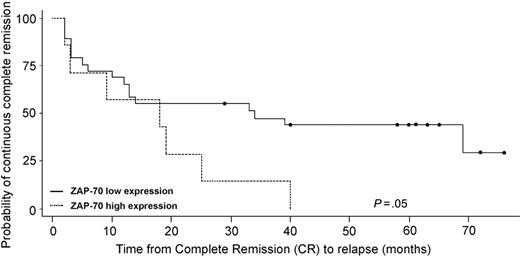
Finally, we sought to evaluate if ZAP70 mRNA expression in ALL patients was associated with a different clinical outcome. Considering the strict association of specific molecular aberrations (namely BCR/ABL, ALL1/AF4, and E2A/PBX1) with a very poor prognosis, this analysis was performed exclusively on the 43 samples that did not carry any molecular abnormality. As previously mentioned, all patients were enrolled in the Italian multicenter clinical trial GIMEMA 0496 and thus were uniformly treated. Of the 43 patients, 5 cases were not evaluable for long-term follow-up for the following reasons: 1 patient was refractory to induction chemotherapy, 2 patients died during induction chemotherapy, and 2 patients were lost to follow-up. Of the 2 patients who carried the gain of chromosome 1q, one died during induction chemotherapy and the other was lost to follow-up; both patients were therefore excluded from this analysis. The remaining 38 patients were evaluable for long-term outcome: 12 patients were in continuous complete remission (CCR) with a median follow-up of 61 months from achievement of complete remission (CR; range, 29-122 months), whereas the remaining 26 experienced a relapse or death within a median period of 12 months (range, 2-69 months). Samples were stratified into 2 groups according to ZAP70 mRNA expression. Using a cut-off level of normalized expression values based on the 80th percentile of the distribution (equal to 351), we could document an association between higher expression of ZAP70 and worse outcome (Figure 6; P = .045).
Comparative analysis between high-versus low-expressing ZAP70 samples that did not carry any known molecular abnormalities. The analysis was performed using an ANOVA and led to the selection of 75 probe sets. Blue indicates low expression; red, high expression.
Comparative analysis between high-versus low-expressing ZAP70 samples that did not carry any known molecular abnormalities. The analysis was performed using an ANOVA and led to the selection of 75 probe sets. Blue indicates low expression; red, high expression.
SYK mRNA expression in ALL
As stated in the “Introduction,” ZAP70 belongs to the family of spleen tyrosine kinases, of which SYK is one of the most important members. In order to establish if SYK was replaced by ZAP70, we evaluated the expression levels of Syk in this series of samples. As expected, Syk expression was significantly higher in B-lineage ALL cases as opposed to T-ALL (283.42 ± 106 and 210.03 ± 68, respectively; P < .001). Again, the highest levels of expression of SYK were found in the E2A/PBX1+ group as well as in pre-B ALL and the lowest in the samples with the ALL1/AF4 rearrangement and in the pro-B cases. However, this difference did not reach statistical significance when evaluated by ANOVA, strengthening the notion that this kinase is indeed expressed in all B cells, regardless of the molecular aberrations and maturative stage (data not shown).
Disease-free survival curves using the Kaplan-Meier method. Patients were stratified into 2 groups on the basis of ZAP70 mRNA expression levels. This analysis was performed exclusively on 38 samples that did not carry molecular abnormalities and had evaluable follow-up data. Patients who died during induction chemotherapy were excluded from the analysis.
Disease-free survival curves using the Kaplan-Meier method. Patients were stratified into 2 groups on the basis of ZAP70 mRNA expression levels. This analysis was performed exclusively on 38 samples that did not carry molecular abnormalities and had evaluable follow-up data. Patients who died during induction chemotherapy were excluded from the analysis.
ZAP-70 protein detection
Given the results obtained by gene expression analysis, we further validated our data on an independent set of 39 ALL cases, comprising 25 adults and 14 children. ZAP-70 protein–positive cells, evaluated using an immunocytochemical method, were found in 23 (59%) of 39 cases. Also at the protein level, we found that 7 of 8 pre-B ALL Igμ+ samples were ZAP-70+ (5/6 pre-B ALL and 2/2 mature B-ALL). In contrast, only 3 of 6 pro-B ALL samples were positive at the ZAP-70 protein level. Fifteen patients (47%) were CD20+, of these, 10 (66%) were also ZAP-70+. When samples were subdivided by molecular rearrangements, 0 of 3 ALL1/AF4+ samples analyzed expressed the ZAP-70 protein; the only sample harboring the E2A/PBX1 rearrangement was positive for ZAP-70, as well as 5 of 10 BCR/ABL+ cases and 14 of 22 cases without known molecular abnormalities (Table 2). Follow-up data were not available because of the short period of clinical observation.
ZAP-70 expression in ALL cases evaluated by immunocytochemistry
. | ZAP-70 positivity, no. positive/total (%) . |
|---|---|
| Immunophenoptypic results | |
| Pro-B ALL | 3/6 (50) |
| Common ALL | 13/25 (52) |
| Pre-B ALL | 5/6 (83) |
| Mature B-ALL | 2/2 (100) |
| Molecular abnormalities | |
| ALL1/AF4 | 0/3 (0) |
| BCR/ABL | 5/10 (50) |
| TEL/AML1 | 1/1 (100) |
| E2A/PBX1 | 1/1 (100) |
| No molecular abnormalities | 14/22 (64) |
. | ZAP-70 positivity, no. positive/total (%) . |
|---|---|
| Immunophenoptypic results | |
| Pro-B ALL | 3/6 (50) |
| Common ALL | 13/25 (52) |
| Pre-B ALL | 5/6 (83) |
| Mature B-ALL | 2/2 (100) |
| Molecular abnormalities | |
| ALL1/AF4 | 0/3 (0) |
| BCR/ABL | 5/10 (50) |
| TEL/AML1 | 1/1 (100) |
| E2A/PBX1 | 1/1 (100) |
| No molecular abnormalities | 14/22 (64) |
Samples are subdivided according to the immunophenotypic characteristics and molecular abnormalities.
Discussion
The role of ZAP-70 in T-cell function is well established1-3 and in the previous years this molecule was considered to be T-cell specific. More recent data have also documented a role of ZAP-70 in B cells.5 In the present study, we assessed the expression of ZAP70 in ALL samples, taking advantage of the opportunity of analyzing data derived from a previous study on gene expression profiling in a large cohort of 128 adult ALL patients studied at the onset of the disease.19 The results obtained indicate that ZAP70 is more strongly expressed in T-ALL samples compared with B-lineage ALL samples. This is in line with the biologic role of ZAP-70, which is a tyrosine kinase previously known to be T-lymphocyte specific and involved in T-cell signaling and activation. Nevertheless, we could also document that ZAP70 is expressed in a set of samples with B-lineage ALL, which accounts for about 40% to 50% of the whole population. These results are in line with 2 recent reports indicating that ZAP-70 expression is not limited to chronic lymphocytic leukemia (CLL) but can also be found in other hematologic malignancies, such as precursor B-cell ALL, mantle-cell lymphoma, diffuse large B-cell lymphoma, and Burkitt lymphoma.20,21
In B-lineage ALL, our data show that ZAP70 is preferentially expressed in cases carrying the E2A/PBX1 rearrangement; the availability of previously published experimental data allowed us to validate this finding also in pediatric ALL cases.9 Furthermore, our data clearly indicate that ZAP70 expression increases along with the differentiation and maturation process of the B-cell population. In fact, ZAP-70 was always expressed in pre-B ALL cases, both at the mRNA and protein levels, and was also found at the protein level in the B-mature ALL cases analyzed. These results strengthen the data previously reported in mice, indicating that ZAP70 plays a role in the process of pro-B to pre-B transition; its absence, combined with the absence of SYK, arrests the cells at the pro-B stage and induces a blockage of several pre-BCR–induced events, such as differentiation into pre-B cells, cell proliferation, and heavy chain allelic exclusion.5
In our series, another set of patients expressed ZAP70: roughly 50% of cases with common ALL were in fact positive at the protein level. Similarly, ZAP70 was also expressed in a set of pro-B cases, suggesting a maturative impairment that is often detected in acute leukemia. Interestingly, a supervised analysis performed exclusively on the samples without molecular abnormalities, aimed at comparing cases expressing high versus low levels of ZAP70, identified 75 genes differentially expressed between these 2 groups. Among the genes highly expressed in samples with high levels of ZAP70, it is of interest to underline the presence of a subset of genes that are usually associated to T cells and to T-cell activation. We exclude a contamination with residual T cells because of the stringent selection criteria and because reevaluation of immunophenotypic analysis did not reveal the presence of contaminating T lymphocytes. Moreover, among the genes selected with available chromosomal annotation, 11.4% resided on chromosome 2q, where ZAP70 is also located. Further studies are required to better evaluate if this region may be involved in the leukemogenic process.
We also evaluated SYK expression and found that this tyrosine kinase is more highly expressed in B-lineage ALL than in T-ALL. Similarly to ZAP70, this kinase showed higher levels of expression in the E2A/PBX+ cases and a lower degree of expression in ALL1/AF4+ cases. This finding leads to 2 considerations. First, in leukemic cells these 2 kinases are both expressed and therefore are not mutually exclusive. Second, the concurrent expression of both kinases may predispose the leukemic cells to a proliferative advantage. This hypothesis is supported by recent data showing that in CLL, after BCR ligation, cells that express ZAP-70 have increased levels of phosphorylated p72Syk, BLNK, and PLCγ and an increase in intracellular Ca2+.22 Furthermore, evidence that this effect is dependent on ZAP-70 expression is provided by infection studies with an adenovirus vector encoding this protein.23
Evaluation on long-term clinical outcome was feasible only on data derived by oligonucleotide arrays, mostly because the follow-up for patients evaluated at the protein level is at present too short. As previously reported, this analysis was performed exclusively on samples without molecular aberrations because of the strong impact on prognosis sustained by the known molecular abnormalities. Our data indicate that ZAP70 expression correlates with a shorter relapse-free survival after achievement of CR, although a cut-off value with strong statistical significance could not be identified due to the limited number of cases evaluated. These findings are similar to the results obtained in CLL, where in fact high levels of ZAP-70 expression have been detected in patients with an unmutated status of the Ig variable region genes (IgVH), who are usually characterized by a more aggressive clinical course and often require therapeutic intervention.24-28 ZAP-70 has raised great interest in CLL patients because it represents a powerful prognostic marker.29-33 Our results provide preliminary indication on the potential use of this protein as a prognostic marker also in ALL. Evaluation of a larger series of patients is needed to further validate this finding.
Finally, the discovery of tyrosine kinases that can be used as therapeutic targets is raising great interest. Since the introduction of STI-571 (imatinib), the outcome of patients with chronic myeloid leukemia (CML) and, probably, also of those suffering from BCR/ABL+ ALL rearrangement, has improved.34 At present, in fact, roughly 90%35,36 of CML patients who receive imatinib as a single drug achieve major clinical and complete cytogenetic responses.37,38 In Philadelphia chromosome–positive (Ph+) ALL, the association of imatinib with polychemotherapeutic regimens has resulted in an improved CR rate, disease-free survival, and overall survival.39,40 Similarly, the use of epidermal growth factor receptor (EGFR) inhibitors in patients with lung cancer carrying a mutation in the sequence of this receptor has improved the outcome in these patients.41,42
These results clearly show that ZAP-70 may indeed be a therapeutic target. ZAP-70 expression in a subset of patients with ALL opens the perspective of investigating the use of an inhibitor also in such cases.
In conclusion, our results provide evidence that in B-lineage ALL ZAP70 expression increases along the maturative process of B lymphocytes and that particularly high levels are associated with the E2A/PBX1 rearrangement. Furthermore, a high mRNA expression of ZAP70 appears to correlate with an increased relapse rate. Finally, these results raise the possibility of designing new compounds targeting this protein.
Prepublished online as Blood First Edition Paper, September 13, 2005; DOI 10.1182/blood-2005-04-1755.
Supported by Associazione Italiana per la Ricerca sul Cancro (AIRC); Ministero dell'Istruzione, Università e Ricerca (MIUR); Fondo per gli Investimenti della Ricerca di Base (FIRB); and Fondazione Internazionale di Ricerca in Medicina Sperimentale (FIRMS) project.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal