Abstract
Hematopoietic effects of interferon-γ (IFN-γ) may be responsible for certain aspects of the pathology seen in bone marrow failure syndromes, including aplastic anemia (AA), paroxysmal nocturnal hemoglobinuria (PNH), and some forms of myelodysplasia (MDS). Overexpression of and hematopoietic inhibition by IFN-γ has been observed in all of these conditions. In vitro, IFN-γ exhibits strong inhibitory effects on hematopoietic progenitor and stem cells. Previously, we have studied the transcriptome of CD34 cells derived from patients with bone marrow failure syndromes and identified characteristic molecular signatures common to some of these conditions. In this report, we have investigated genome-wide expression patterns after exposure of CD34 and bone marrow stroma cells derived from normal bone marrow to IFN-γ in vitro and have detected profound changes in the transcription profile. Some of these changes were concordant in both stroma and CD34 cells, whereas others were specific to CD34 cells. In general, our results were in agreement with the previously described function of IFN-γ in CD34 cells involving activation of apoptotic pathways and immune response genes. Comparison between the IFN-γ transcriptome in normal CD34 cells and changes previously detected in CD34 cells from AA and PNH patients reveals the presence of many similarities that may reflect molecular signature of in vivo IFN-γ exposure.
Introduction
Bone marrow failure syndromes, including some forms of myelodysplastic syndromes (MDS), paroxysmal nocturnal hemoglobinuria (PNH), and aplastic anemia (AA) are characterized by a profound defect in the stem- and progenitor-cell compartments.1 Clinically, this defect is reflected by cytopenias and has been quantitated using flow cytometry,2,3 hematopoietic colony cultures,3,4 and long-term culture-initiating cells (LTCICs).5-9
Many clinical associations exist between bone marrow failure syndromes. For example, MDS (especially involving monosomy 7 and trisomy 8) can evolve from AA.10 PNH is closely related to AA; PNH clones are present in a high proportion of AA at presentation, and clinical PNH is a common long-term clonal complication of AA, similar to MDS.5,10-15 In the search for molecular defects responsible for the stem-cell dysfunction in these diseases, we have previously performed microarray analyses to identify characteristic gene-expression patterns of CD34 cells derived from patients with MDS, AA, or PNH.16-18
In PNH, both wild-type (WT) and glycosyl phosphatidyl inositol (GPI)-deficient CD34 cells were studied; PIG-A-deficient progenitors showed a molecular signature similar to that seen in CD34 cells derived from healthy individuals, whereas “normal” (WT) CD34 cells derived from PNH patients showed a pronounced expression of genes associated with apoptosis. In MDS, the gene-expression pattern in monosomy 7 progenitor cells showed functional characteristics of high proliferation and malignant potential with upmodulation of genes related to leukemia transformation, tumorigenesis, and apoptosis.17,18 By comparison, trisomy 8 CD34 cells showed a profile consistent with overexpression of immune and inflammatory gene groups.17 In AA, CD34 cells showed a downmodulation of the genes associated with cell cycling and increased gene expression accompanying apoptotic pathways.16
Various effector pathways have been hypothesized to mediate the dysfunction of hematopoietic stem cells in AA, ultimately leading to their depletion through cycling blockade and apoptosis.19,20 Cell-mediated mechanisms involving perforin and Fas pathways as well as the effects of cytokines such as IFN-γ have been shown to play a role in stem-cell destruction.21-24 IFN-γ involvement in the pathophysiology of AA has been well documented. IFN-γ is overexpressed in marrow and blood of patients with AA16,22,24-27 and detection of this cytokine in lymphocytes derived from patients with AA constitutes a marker for the responsiveness to immunosuppressive therapy.27 In vitro, IFN-γ broadly inhibits the hematopoietic activity of more committed as well as primitive progenitor and stem cells.28,29
Gene-expression patterns available from previous studies performed under standardized or comparable conditions may facilitate data mining to establish pathophysiologic links and explain clinical differences between the diseases especially if comparable target populations (CD34 cells) were investigated. At the outset of this study, we theorized that expression patterns induced in vitro under defined experimental conditions can be used to identify gene-expression signatures observed in vivo in patients. For example, expression patterns of CD34 cells in AA can be in part a result of effects of IFN-γ, and our experiments were designed to detect whether similar expression patterns can be generated by stimulation of normal CD34 cells with IFN-γ in vitro. Conversely, a signature profile of IFN-γ generated in CD34 cells in vitro may be detected in CD34 cells derived from patients with bone marrow failure.
Materials and methods
Bone marrow cells
Bone marrow cells were obtained through iliac crest aspiration. Informed consent from healthy volunteers was obtained according to a protocol approved by the Institutional Review Board of the National Heart, Lung, and Blood Institute. Bone marrow mononuclear cells (BMMNCs) were isolated by density gradient centrifugation using lymphocyte separation medium (Cappel, Aurora, OH), and were washed twice using phosphate-buffered saline (PBS; Cellgro, Herndon, VA).
Isolation of CD34
CD34 cells were positively selected using mini-MACS (magnetic-activated cell separation) immunomagnetic separation system (Miltenyi Biotec, Auburn, CA) according to the manufacturer's instructions. In brief, to obtain normal CD34 cells, up to 108 BMMNCs were suspended in 300 μL sorting buffer composed of 1 × PBS, 2 mM EDTA (ethylenediaminetetraacetic acid), and 0.5% bovine serum albumin. Cells were incubated with 100 μL human immunoglobulin-Fc receptor (FcR) blocking antibody and 100 μL monoclonal hapten-conjugated CD34 antibody (clone QBEND/10; Miltenyi Biotec) for 15 minutes at 4°C. After washing, cells were resuspended in 400 μL sorting buffer, and 100 μL paramagnetic microbeads conjugated to antihapten antibody were added, followed by incubation for 15 minutes at 4°C. Following washing, cells were diluted in sorting buffer, passed through a 30-μm nylon mesh, and separated in a column exposed to the magnetic field of the MACS device. The column was washed twice with sorting buffer and removed from the separator. Retained cells were eluted with sorting buffer subjected to a second separation. Purity of CD34 cells was 90% to 97% by flow cytometry analysis.
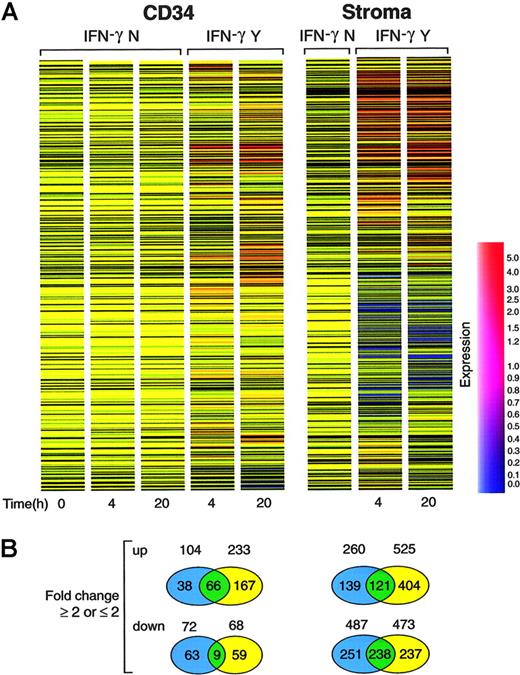
Gene-expression analysis of CD34 cells treated with IFN-γ. (A) Hierarchical clustering analysis of gene-expression patterns in CD34 and stromal cells over time. Each time point was averaged from 2 independent pools of CD34 or stromal cells. Genes were clustered on the horizontal axis and individual conditions on the vertical axis. IFN-γ N indicates control; IFN-γ Y, cells treated with IFN-γ. Blue-to-red scale indicates expression level from low to high. (B) The diagrams indicate genes that changed 2-fold or greater after 4 and 20 hours of culture. The top panel contains genes upregulated by IFN-γ; the bottom panel contains genes that were downregulated. Blue and yellow circles indicate genes changed in either 4 or 20 hours. Green circles indicate genes for which expression was changed at both time points.
Gene-expression analysis of CD34 cells treated with IFN-γ. (A) Hierarchical clustering analysis of gene-expression patterns in CD34 and stromal cells over time. Each time point was averaged from 2 independent pools of CD34 or stromal cells. Genes were clustered on the horizontal axis and individual conditions on the vertical axis. IFN-γ N indicates control; IFN-γ Y, cells treated with IFN-γ. Blue-to-red scale indicates expression level from low to high. (B) The diagrams indicate genes that changed 2-fold or greater after 4 and 20 hours of culture. The top panel contains genes upregulated by IFN-γ; the bottom panel contains genes that were downregulated. Blue and yellow circles indicate genes changed in either 4 or 20 hours. Green circles indicate genes for which expression was changed at both time points.
Stromal-cell preparation
BMMNCs were resuspended in 10 mL Myelocult 5100 media (Stem Cell Technologies, Vancouver, BC, Canada) supplemented with 1 × 10-6 M hydrocortisone sodium hemisuccinate (Sigma, St Louis, MO) at a concentration of 1 × 107 to 2 × 107 cells/mL. Cells were plated in a T75 flask. After 5-day culture at 37°C, media were replaced and nonadherent cells were removed. Subsequently, cultures were cultured at 33°C. Cultures were fed by replacing half of the media on a weekly basis. Through residual feeding cycles, all nonadherent cells were removed.
Sample preparation for microarray analysis
Due to the relatively high amount of mRNA required for microarray analysis, CD34 and stroma cells were pooled from 14 healthy donors (every pool composed of equal amounts of cells from each individual). Ten pools were obtained for both CD34 and stroma cells. Cells were cultured at time 0, 4, and 20 hours; time points 4 and 20 hours had 4 pools, 2 pools were cultured with 2000 U/mL IFN-γ (Boehringer Mannheim, Indianapolis, IN) and 2 pools without IFN-γ as control. In addition, 2 cell pools were obtained prior to culture. Cells were harvested and washed and total RNA was extracted with an RNeasy kit (Qiagen, Valencia, CA). The purity of RNA was checked with spectrophotometry. An additional 8 pools for CD34 and stroma cells at 4- and 20-hour time points with or without IFN-γ were used for real-time polymerase chain reaction (PCR; TaqMan; PE Applied Biosystems, Foster City, CA) analysis.
Overview of differential gene-expression patterns in CD34 and stromal cells following IFN-γ treatment. Genes selected for this analysis showed at least a 2.0-fold change. The right portion indicates upmodulation; the left portion indicates downmodulation. (A) CD34 cells. (B) Stromal cells. □ indicates change following 4 hours of culture; ▨, expression changes after 20 hours; ▪, concordant changes after 4 and 20 hours.
Overview of differential gene-expression patterns in CD34 and stromal cells following IFN-γ treatment. Genes selected for this analysis showed at least a 2.0-fold change. The right portion indicates upmodulation; the left portion indicates downmodulation. (A) CD34 cells. (B) Stromal cells. □ indicates change following 4 hours of culture; ▨, expression changes after 20 hours; ▪, concordant changes after 4 and 20 hours.
Affymetrix GeneChip assay and data analysis
Probes were prepared using standard Affymetrix protocols and hybridized to Affymetrix HG-U95Av2 arrays (Affymetrix, Santa Clara, CA). All of the microarrays were examined for surface defects, grid placement, background intensity, housekeeping gene expression, and 3′-5′ ratio of probe sets from genes of varying length (signal 3′-5′ ratio < 3). Expression analysis was conducted with standard Affymetrix analysis software algorithms (Microarray Suite 5.0). Comparative analysis between expression profiles for samples with or without IFN-γ was carried out on GeneSpring software version 7.1 (Silicongenetics, Redwood, CA). Gene-expression data were normalized in 2 ways: “per chip normalization” and “per gene normalization.” Each measurement was divided by the 50th percentile of all measurements in that sample. Specific samples were normalized to one another. Each measurement for each gene in those specific samples was divided by the mean of that gene's measurements in the corresponding control samples. Fold changes were compared between samples with or without IFN-γ at the same time point based on normalized data.
Real-time reverse transcriptase (RT)-PCR
Real-time PCR was performed to confirm expression levels of RNA transcripts with sequence-specific oligonucleotide primers (Table 1), according to the manufacturer's instruction (PE Applied Biosystems). For relative quantification, GAPDH mRNA served as an external control. All reactions were performed in the Model 7700 sequence detector (PE Applied Biosystems). The values obtained for the target gene expression were normalized to GAPDH and were expressed relative to the expression in control samples. For the calculation, the 2-ΔΔCT formula was used, where ΔΔCT = (CT, target - CT, GAPDH) experimental sample - (CT, target - CT, GAPDH) control samples.
Sequences for real-time PCR analysis
. | . | Sequence 5′-3′ . | . | PCR product length, bp . | |
|---|---|---|---|---|---|
| GenBank no. . | Gene symbol . | Forward . | Reverse . | . | |
| AF005775 | CFLAR | CTCTTTTTGTGCCGGGATGT | AAGTCCCCGACAGACAGCTTAC | 164 | |
| U37518 | TRAIL | CCAAAAGTGGCATTGCTTGTT | ACGGAGTTGCCACTTGACTTG | 107 | |
| X63717 | FAS | TTGGTGGACCCGCTCAGTA | AGCAATCCTCCGAAGTGAAAGA | 56 | |
| U11821 | FASL | TTCTGGTTGCCTTGGTAGGATT | TGTGTGCATCTGGCTGGTAGA | 101 | |
| X02910 | TNF | AGCCCCTCCCAGTTCTAGTTCT | CCTCAAAACCTATTGCCTCCAT | 110 | |
| L22474 | BAX-B | CGAGTGGCAGCTGACATGTT | CAGTTCCGGCACCTTGGT | 117 | |
| M31165 | TNF-AIP6 | AGAAGCACGGTCTGGCAAAT | GGCTGCCTCTAGCTGCTTGT | 103 | |
| M92357 | TNF-AIP2 | TATTGCCGTGACAGGTTTCCA | TGCCGACACCCAGATTTGA | 101 | |
| U64198 | IL12RB2 | TGAGTGGCCCCCACATTAAT | GGAGTCCCGTTCCTTCCAGTAT | 131 | |
| M21121 | RNTES | GCTTCCCCAACTAAAGCCTAGAA | TGTCACAGAGCCCTTGCAAA | 109 | |
| AF030514 | CXCL 11 | GATGCCTAAATCCCAAATCGAA | GGCAGTGGAATTCTGATTGTCA | 295 | |
| M27492 | IL1R | CAGAGCAAGACTCCGTCTCAAA | GTCCCCAGACAAGGCATAGC | 182 | |
| AF043129 | IL7R | AGCCCCAACTGCCCATCT | GGCGTCACATGCACTGACA | 102 | |
| U00672 | IL10RA | TGACAGTTGGCAGTGTGAACCT | CACCTTGCGAATGGCAATCT | 149 | |
| M13755 | IFI15 | ACAAATGCGACGAACCTCTGA | CCGCTCACTTGCTGCTTCA | 112 | |
| J04164 | IFITM1 | AAGCCAGAAGATGCACAAGGA | GGAGGTCTCGCTGTGGATGT | 106 | |
| L19871 | AIT3 | GGCTGTCACCACGTGCAGTA | GCTGCCCATGGATTTTGTGT | 107 | |
| M73255 | VCAM1 | TTCTCTCCCCACCCCCTTAA | TCCTCTCTCTGTCCTGGCAAA | 166 | |
| U66838 | CCNA1 | AGGTCCCGATGCTTGTCAGA | TCCTGTACTGCCCATTTGCA | 101 | |
| X04434 | IGF1R | CAGCCTTACGCCCACATGA | GTTTGCACAGATTCAGGATCCA | 101 | |
| M97935 | STAT1 | CCTGCTGCGGTTCAGTGA | GGTTCAACCGCATGGAAGTC | 102 | |
. | . | Sequence 5′-3′ . | . | PCR product length, bp . | |
|---|---|---|---|---|---|
| GenBank no. . | Gene symbol . | Forward . | Reverse . | . | |
| AF005775 | CFLAR | CTCTTTTTGTGCCGGGATGT | AAGTCCCCGACAGACAGCTTAC | 164 | |
| U37518 | TRAIL | CCAAAAGTGGCATTGCTTGTT | ACGGAGTTGCCACTTGACTTG | 107 | |
| X63717 | FAS | TTGGTGGACCCGCTCAGTA | AGCAATCCTCCGAAGTGAAAGA | 56 | |
| U11821 | FASL | TTCTGGTTGCCTTGGTAGGATT | TGTGTGCATCTGGCTGGTAGA | 101 | |
| X02910 | TNF | AGCCCCTCCCAGTTCTAGTTCT | CCTCAAAACCTATTGCCTCCAT | 110 | |
| L22474 | BAX-B | CGAGTGGCAGCTGACATGTT | CAGTTCCGGCACCTTGGT | 117 | |
| M31165 | TNF-AIP6 | AGAAGCACGGTCTGGCAAAT | GGCTGCCTCTAGCTGCTTGT | 103 | |
| M92357 | TNF-AIP2 | TATTGCCGTGACAGGTTTCCA | TGCCGACACCCAGATTTGA | 101 | |
| U64198 | IL12RB2 | TGAGTGGCCCCCACATTAAT | GGAGTCCCGTTCCTTCCAGTAT | 131 | |
| M21121 | RNTES | GCTTCCCCAACTAAAGCCTAGAA | TGTCACAGAGCCCTTGCAAA | 109 | |
| AF030514 | CXCL 11 | GATGCCTAAATCCCAAATCGAA | GGCAGTGGAATTCTGATTGTCA | 295 | |
| M27492 | IL1R | CAGAGCAAGACTCCGTCTCAAA | GTCCCCAGACAAGGCATAGC | 182 | |
| AF043129 | IL7R | AGCCCCAACTGCCCATCT | GGCGTCACATGCACTGACA | 102 | |
| U00672 | IL10RA | TGACAGTTGGCAGTGTGAACCT | CACCTTGCGAATGGCAATCT | 149 | |
| M13755 | IFI15 | ACAAATGCGACGAACCTCTGA | CCGCTCACTTGCTGCTTCA | 112 | |
| J04164 | IFITM1 | AAGCCAGAAGATGCACAAGGA | GGAGGTCTCGCTGTGGATGT | 106 | |
| L19871 | AIT3 | GGCTGTCACCACGTGCAGTA | GCTGCCCATGGATTTTGTGT | 107 | |
| M73255 | VCAM1 | TTCTCTCCCCACCCCCTTAA | TCCTCTCTCTGTCCTGGCAAA | 166 | |
| U66838 | CCNA1 | AGGTCCCGATGCTTGTCAGA | TCCTGTACTGCCCATTTGCA | 101 | |
| X04434 | IGF1R | CAGCCTTACGCCCACATGA | GTTTGCACAGATTCAGGATCCA | 101 | |
| M97935 | STAT1 | CCTGCTGCGGTTCAGTGA | GGTTCAACCGCATGGAAGTC | 102 | |
CFLAR indicates CASP8 and FADD-like apoptosis regulator; TRAIL, TNF-related apoptosis–inducing ligand; TNF, tumor necrosis factor; BAX-B, BCL2-associated X protein; TNF-AIP6, TNF alpha–induced protein 6; IL-12RB2, interleukin 12 receptor beta 2; RNTES, chemokine (C-C motif) ligand 5; CXCL 11, chemokine (C-X-C motif) ligand 11; IFI15, interferon alpha–inducible protein (clone IFI-15K); IFITM1, interferon-induced transmembrane protein 1 (9-27); AIT3, activating transcription factor 3; VCAM1, vascular cell adhesion molecule-1; CCNA1, cyclinA1; IGF1R, insulin-like growth factor 1 receptor; and STAT1, transcription factor interferon-stimulated gene factor-3.
Results
Global microarray analysis of gene expression in CD34 cells and stroma cells treated with IFN-γ
We devised an experimental microarray strategy that allowed for gene-expression analysis of rare cell populations. CD34 cells derived from 14 healthy individuals were cultured with and without IFN-γ. Following culture, RNA was combined into 2 pools that were independently analyzed using separate microarrays. Upmodulated and downmodulated genes that showed concordant changes in both arrays were selected for further analysis and their expression was averaged. Two separate dendrograms were generated for CD34 and stroma cells, respectively. Expression results were normalized to the baseline levels determined in the 2 cell pools not subjected to culture. Fold changes were compared for the same time point in samples cultured with and without IFN-γ (Figures 1, 2). Gene expression was analyzed using a hierarchical-clustering algorithm to identify the expression pattern as a gene or condition tree. Samples were subclustered in these trees based on the similarity of the expression pattern of all genes. CD34 and stroma cells clustered into 2 groups. Each group was further subclustered into IFN-γ-treated and untreated (control) groups, respectively. Globally, IFN-γ effects on gene expression were more pronounced by 20 hours in comparison to 4 hours after exposure (late vs early expression). In CD34 cells, 66 genes showed persistent upmodulation at 20 hours, whereas expression of only 38 genes was detected at 4 hours. For most of the genes, expression induction was delayed.
Similar analysis was performed in stroma cells in which 121 genes showed consistent (at least 2-fold) upregulation. Changes in the expression level occurred later in 139 genes (20 hours), whereas response was more immediate in 404 genes (4 hours). IFN-γ also decreased expression of a number of genes in stroma cells. These changes could be subdivided into a late and an early downmodulation. The expression downregulation was observed early for a majority of the genes (for CD34 cells, 63 genes vs 59 genes, at 4 and 20 hours, respectively; for stroma cells, 251 genes vs 237 genes, at 4 and 20 hours, respectively).
Gene-expression profiles in CD34 and stroma cells treated with IFN-γ
The genes that showed at least 2-fold altered expression levels were classified according to the Gene Ontology Groupings, and the nomenclature and functional descriptions were derived from public databases (CMAP30 and DAVID31 ). Among gene ontology groups, cell growth and maintenance, immune response, cell death, signal transduction, cytokine/chemokines and receptors, and IFN-induced genes showed the most pronounced modulation in CD34 cells, whereas the effects in stroma cells displayed a quite different pattern. In bone marrow stroma treated with IFN-γ, the gene groups showing a decreased expression included genes involving metabolism, cell growth and maintenance, signal transduction, biosynthesis, immune response, cell adhesion, and cell death (Figure 2). Subsequently, we have cross-referenced our results with those of other studies investigating interferon-stimulated genes (ISGs).49-54 In CD34 cells and stroma cells, the expression of a large number of previously reported ISGs was found to be increased by IFN-γ. These ISGs may constitute a pool of genes representing a signature of interferon response (Table 2). We have also found a large number of previously described ISGs that remained stable in CD34 cells or stromal cells; while in contrast, expression of certain ISGs may be restricted to CD34 cells.
Previously described ISGs confirmed in CD34 and stroma cells by gene-expression microarray
. | . | . | Fold change . | . | |
|---|---|---|---|---|---|
| GenBank no. . | Gene name . | Gene symbol . | Stroma . | CD34 . | |
| X67325 | Interferon alpha–inducible protein 27 | IFI27 (IFNa) | 59.6 | 5.0 | |
| U88964 | Interferon-stimulated gene, 20 kDa | ISG20 | 16.1 | 4.4 | |
| AF026939 | Interferon-induced protein with tetratricopeptide repeats 4 | IFIT4 (IFNa) | 57.1 | 6.3 | |
| M24594 | Interferon-induced protein with tetratricopeptide repeats 1 | IFIT1 (IFNb) | 17.5 | 3.7 | |
| U72882 | Interferon-induced protein 35 | IFI35 (IFNa) | 3.1 | 3.9 | |
| M13755 | Interferon alpha–inducible protein (clone IFI-15K) | G1P2 (IFNa,b) | 2.2 | 4.7 | |
| AA203213 | Interferon alpha–inducible protein (clone IFI-15K) | G1P2 (IFNa,b) | 8.1 | 6.2 | |
| M14660 | Interferon-induced protein with tetratricopeptide repeats 2 | IFIT2 | 78.6 | 13.1 | |
| L05072 | Interferon regulatory factor 1 | IRF1 | 9.4 | 3.6 | |
| M33882 | Myxovirus resistance 1, interferon-inducible protein p78 | MX1 | 43.6 | 7.1 | |
| M55542 | Guanylate binding protein 1, interferon-inducible, 67 kDa | GBP1 | 12.8 | 5.9 | |
| J04164 | Interferon-induced transmembrane protein 1 (9-27) | IFIM1 (IFNb) | 3.5 | 3.3 | |
| M63838 | Interferon gamma–inducible protein 16 | IFI16 | 4.8 | 2.3 | |
| D28915 | Interferon-induced protein 44 | IFI44 | 3.9 | 3.7 | |
| L78833 | Interferon-induced protein 35 | IFI35 | 11.4 | 3.5 | |
| U34605 | Interferon-induced protein with tetratricopeptide repeats 5 | IFIT5 | 3.7 | 2.2 | |
| J03909 | Interferon gamma–inducible protein 30* | IFI30 | – | 2.3 | |
| X57352 | Interferon-induced transmembrane protein 3 (1-8U)* | IFITM3 | – | 2.2 | |
| U50648 | Protein kinase interferon-inducible double-stranded RNA dependent* | PRKR | – | 2.2 | |
| J00219 | Interferon gamma* | IFNG | – | 5.4 | |
| U22970 | Interferon alpha–inducible protein (clone IFI-6-16)* | G1P3 (IFNb) | 3.0 | – | |
| M55543 | Guanylate binding protein 2, interferon-inducible* | GBP2 | – | 3.3 | |
. | . | . | Fold change . | . | |
|---|---|---|---|---|---|
| GenBank no. . | Gene name . | Gene symbol . | Stroma . | CD34 . | |
| X67325 | Interferon alpha–inducible protein 27 | IFI27 (IFNa) | 59.6 | 5.0 | |
| U88964 | Interferon-stimulated gene, 20 kDa | ISG20 | 16.1 | 4.4 | |
| AF026939 | Interferon-induced protein with tetratricopeptide repeats 4 | IFIT4 (IFNa) | 57.1 | 6.3 | |
| M24594 | Interferon-induced protein with tetratricopeptide repeats 1 | IFIT1 (IFNb) | 17.5 | 3.7 | |
| U72882 | Interferon-induced protein 35 | IFI35 (IFNa) | 3.1 | 3.9 | |
| M13755 | Interferon alpha–inducible protein (clone IFI-15K) | G1P2 (IFNa,b) | 2.2 | 4.7 | |
| AA203213 | Interferon alpha–inducible protein (clone IFI-15K) | G1P2 (IFNa,b) | 8.1 | 6.2 | |
| M14660 | Interferon-induced protein with tetratricopeptide repeats 2 | IFIT2 | 78.6 | 13.1 | |
| L05072 | Interferon regulatory factor 1 | IRF1 | 9.4 | 3.6 | |
| M33882 | Myxovirus resistance 1, interferon-inducible protein p78 | MX1 | 43.6 | 7.1 | |
| M55542 | Guanylate binding protein 1, interferon-inducible, 67 kDa | GBP1 | 12.8 | 5.9 | |
| J04164 | Interferon-induced transmembrane protein 1 (9-27) | IFIM1 (IFNb) | 3.5 | 3.3 | |
| M63838 | Interferon gamma–inducible protein 16 | IFI16 | 4.8 | 2.3 | |
| D28915 | Interferon-induced protein 44 | IFI44 | 3.9 | 3.7 | |
| L78833 | Interferon-induced protein 35 | IFI35 | 11.4 | 3.5 | |
| U34605 | Interferon-induced protein with tetratricopeptide repeats 5 | IFIT5 | 3.7 | 2.2 | |
| J03909 | Interferon gamma–inducible protein 30* | IFI30 | – | 2.3 | |
| X57352 | Interferon-induced transmembrane protein 3 (1-8U)* | IFITM3 | – | 2.2 | |
| U50648 | Protein kinase interferon-inducible double-stranded RNA dependent* | PRKR | – | 2.2 | |
| J00219 | Interferon gamma* | IFNG | – | 5.4 | |
| U22970 | Interferon alpha–inducible protein (clone IFI-6-16)* | G1P3 (IFNb) | 3.0 | – | |
| M55543 | Guanylate binding protein 2, interferon-inducible* | GBP2 | – | 3.3 | |
– indicates that the gene altered the expression level less than 2-fold, or that there was no change.
Altered gene expression detected in only one cell type
Detailed analysis of a transcriptome in CD34 and stromal cells exposed to IFN-γ
The IFN-γ transcriptome of CD34 cells was investigated at 2 different time points (Figure 3). After excluding known ISGs listed in Table 2, the newly identified IFN-γ-modulated genes were subdivided into gene categories that allow for determination of the global direction of the molecular response to IFN-γ. Although a large number of genes were affected, our analysis focused on those that may be involved in the pathophysiology of hematologic diseases. Examples of previously described ISGs that were found to be upmodulated in both CD34 and stroma cells include ASP4, VCAM1, CXCL10, CCL5, 2′-5′-oligoadenylate synthetase-2, and activating transcription factor-3. In contrast, some ISGs including CASP1, TRAIL, IL1 2RB1, and CCR3 were induced only in CD34 cells. Focused analysis performed with Ingenuity Pathways Analysis (Ingenuity Systems, Mountain View, CA) identified 36 upregulated genes previously associated with hematologic disease.
Confirmation of in vitro microarray expression by quantitative PCR and concordance with previous results
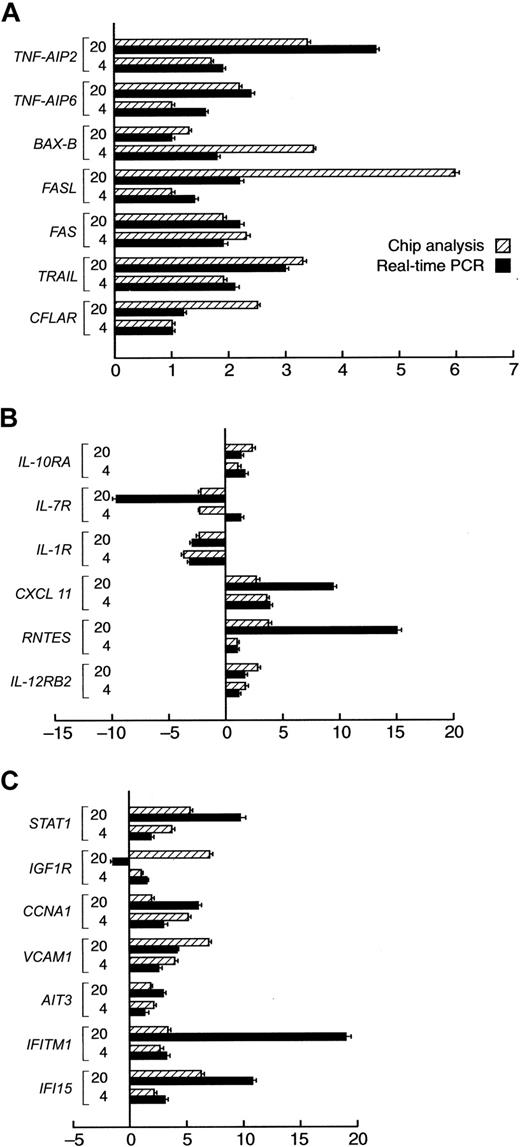
Subsequently, we validated the results of the array-based gene-expression analysis using quantitative RT-PCR. For that purpose, we selected a number of exemplary genes found to be characteristic of the IFN-γ effect in CD34 cells. Two additional sets of pooled cells that were harvested after 4 and 20 hours of culture with and without IFN-γ were analyzed. Our analysis involved 3 groups of genes found to be influenced by IFN-γ treatment by microarray analysis, including those involved in apoptosis, growth factor and chemokine receptors, as well as interferon-modulated transcription factors. We have found a good concordance between the array- and PCR-based analyses (Figure 4).
Gene-expression patterns of CD34 cells after exposure to IFN-γ. Gene-expression profiles of 2 independent pools of CD34 cells derived from healthy controls were generated using Affymetrix HG-U95Av2 oligonucleotide GeneChips. Differential expression was defined as at least a 2-fold change in expression level in cells treated with IFN-γ compared with controls (Y or N, respectively). Values on the right indicate degree of upregulation and values on the left show the degree of downmodulation. “Absolute” expression levels (in arbitrary fluorescence units [AFUs]) are indicated using a color scale; red bars indicate high level of expression (> 1000 AFU); yellow bars, medium level of expression (200-1000 AFU); green bars, low level (20-200). * indicates genes previously known to be regulated by IFN-γ. GenBank number and gene name in red indicate altered expression in both CD34 and stroma cells; black letters indicate that the genes changed their expression only in one of the cell types studied.
Gene-expression patterns of CD34 cells after exposure to IFN-γ. Gene-expression profiles of 2 independent pools of CD34 cells derived from healthy controls were generated using Affymetrix HG-U95Av2 oligonucleotide GeneChips. Differential expression was defined as at least a 2-fold change in expression level in cells treated with IFN-γ compared with controls (Y or N, respectively). Values on the right indicate degree of upregulation and values on the left show the degree of downmodulation. “Absolute” expression levels (in arbitrary fluorescence units [AFUs]) are indicated using a color scale; red bars indicate high level of expression (> 1000 AFU); yellow bars, medium level of expression (200-1000 AFU); green bars, low level (20-200). * indicates genes previously known to be regulated by IFN-γ. GenBank number and gene name in red indicate altered expression in both CD34 and stroma cells; black letters indicate that the genes changed their expression only in one of the cell types studied.
Comparison of IFN-γ-modulated expression profiles in CD34 cells with those seen in CD34 cells derived from marrow failure
IFN-γ has been implicated in the pathophysiology of hematopoietic suppression in bone marrow failure syndromes and mechanisms. Previously, we analyzed gene-expression profiles in CD34 cells derived from AA, PNH, and MDS using a methodology identical to that which was applied in this study.16,17 Similar to our current study, the changes in gene expression were calculated in relation to the baseline provided by the freshly isolated CD34 cells. The baseline expression profiles of CD34 cells did not vary between the studies. Therefore, we were able to globally compare the expression profile of CD34 cells treated with IFN-γ to that of CD34 cells derived from AA, GPI-deficient and phenotypically normal CD34 cells isolated from PNH patients, as well as CD34 cells from patients with MDS and trisomy 8 and monosomy 7 (Table 3).
Gene-expression profiles in CD34 cells treated with IFN-γ in vitro compared with expression profiles in bone marrow failure states
. | . | . | MDS . | . | . | . | PNH . | . | ||
|---|---|---|---|---|---|---|---|---|---|---|
| GenBank no. . | Gene name . | IFN-γ treated . | –7 . | +8 . | AA . | GPI-D . | GPI-N . | GPI-N vs GPI-D . | ||
| Similarly altered genes | ||||||||||
| Apoptosis genes | ||||||||||
| X63717† | FAS* | ↑4 | – | – | ↑ | – | ↑ | ↑ | ||
| U37518† | TRAIL* | ↑20 | ↑ | – | ↑ | – | ↑ | – | ||
| X98175 | CASP8* | ↑20 | ↑ | – | – | – | – | – | ||
| M92357†† | B94, TNFα-induced protein 2 | ↑20 | ↑ | ↑ | ↑ | – | – | – | ||
| Y16961 | Tumor protein, 63 kDa, with strong homology to p53 | ↑20 | ↑ | ↑ | – | – | – | – | ||
| U43746 | Breast cancer 2, early onset | ↑20 | ↑ | – | – | – | – | – | ||
| X04434 | Insulin-like growth factor 1 receptor | ↑20 | – | ↑ | – | – | – | – | ||
| M31165† | TNFα-induced protein 6 | ↑20 | – | – | ↑ | – | – | – | ||
| AF061034† | Optineurin | ↑20 | – | – | ↑ | – | – | – | ||
| AF005775 | CFLAR | ↑20 | – | – | ↑ | – | ↑ | – | ||
| Cytokines/chemokines and receptors | ||||||||||
| X02910 | TNF | ↑20 | – | – | ↑ | – | ↑ | ↑ | ||
| U00672 | IL10RA* | ↑20 | – | ↑ | ↑ | – | – | – | ||
| M21121 | CCL5* | ↑20 | – | – | ↑ | – | ↑ | ↑ | ||
| Y16645† | CCL8* | ↑20 | – | ↑ | – | – | ↑ | ↑ | ||
| AF030514† | CXCL11* | ↑20 | – | – | – | – | ↑ | ↑ | ||
| X72755† | CXCL9* | ↑4, ↑40 | – | – | – | – | ↑ | ↑ | ||
| X02530† | CXCL10* | ↑4, ↑40 | – | – | – | – | – | ↑ | ||
| Cell adhesion | ||||||||||
| M30257 | VCAM1 | ↑20 | – | ↑ | – | – | – | – | ||
| U88964† | IS320 | ↑20 | – | – | ↑ | – | ↑ | – | ||
| M63193† | EGF1 | ↑20 | – | – | ↑ | – | – | – | ||
| M73255† | VCAM1 | ↑20 | – | – | ↑ | – | – | – | ||
| Cell growth and maintenance | ||||||||||
| AF044309 | Syntaxin 11 | ↑20 | – | – | – | – | ↑ | – | ||
| X57522† | Transporter 1 (MDR/TAP)* | ↑20 | – | – | – | – | ↑ | – | ||
| M77016 | Tropomodulin 1 | ↑20 | – | – | – | ↑ | – | – | ||
| U20982 | Insulin-like growth factor-binding protein 4 | ↑20 | – | – | – | – | – | ↑ | ||
| J04164† | IFM1 (9-27)* | ↑20, ↑20 | – | – | ↑ | – | ↑ | – | ||
| X59892† | Tryptophanyl-tRNA synthetase* | ↑20, ↑20 | – | – | – | – | ↑ | ↑ | ||
| Signal transduction | ||||||||||
| U02020† | Pre–B-cell colony-enhancing factor 1 | ↑4 | – | – | ↑ | – | ↑ | – | ||
| D10923 | Putative chemokine receptor | ↑4 | – | – | – | – | ↑ | – | ||
| M97935† | STAT1* | ↑20 | – | – | ↑ | – | – | ↑ | ||
| Immune response | ||||||||||
| X16354† | Carcinoembryonic antigen-related cell-adhesion molecule 1* | ↑4 | – | – | ↑ | – | – | – | ||
| M55543† | GBP2 | ↑20 | – | – | ↑ | – | ↑ | ↑ | ||
| M55542† | GBP1 | ↑20 | – | – | – | – | ↑ | ↑ | ||
| M13755† | GIP2 | ↑4, ↑20 | – | – | – | – | ↑ | – | ||
| Transcription factor | ||||||||||
| L19871† | Activating transcription factor 3* | ↑4 | – | – | – | – | ↑ | – | ||
| Contradictory expression genes | ||||||||||
| U64198 | IL12RB2* | ↑4 | – | – | ↓ | ↓ | – | – | ||
| M36820† | CXCL2* | ↓4 | – | – | ↑ | – | ↑ | ↑ | ||
| M36821† | CXCL3* | ↓4 | – | – | ↑ | – | ↑ | ↑ | ||
| AF043129 | IL-7R | ↓4, ↓20 | – | ↑ | ↑ | – | ↑ | ↑ | ||
| M27492 | IL-1R1 | ↓4, ↓20 | – | – | ↑ | – | – | – | ||
. | . | . | MDS . | . | . | . | PNH . | . | ||
|---|---|---|---|---|---|---|---|---|---|---|
| GenBank no. . | Gene name . | IFN-γ treated . | –7 . | +8 . | AA . | GPI-D . | GPI-N . | GPI-N vs GPI-D . | ||
| Similarly altered genes | ||||||||||
| Apoptosis genes | ||||||||||
| X63717† | FAS* | ↑4 | – | – | ↑ | – | ↑ | ↑ | ||
| U37518† | TRAIL* | ↑20 | ↑ | – | ↑ | – | ↑ | – | ||
| X98175 | CASP8* | ↑20 | ↑ | – | – | – | – | – | ||
| M92357†† | B94, TNFα-induced protein 2 | ↑20 | ↑ | ↑ | ↑ | – | – | – | ||
| Y16961 | Tumor protein, 63 kDa, with strong homology to p53 | ↑20 | ↑ | ↑ | – | – | – | – | ||
| U43746 | Breast cancer 2, early onset | ↑20 | ↑ | – | – | – | – | – | ||
| X04434 | Insulin-like growth factor 1 receptor | ↑20 | – | ↑ | – | – | – | – | ||
| M31165† | TNFα-induced protein 6 | ↑20 | – | – | ↑ | – | – | – | ||
| AF061034† | Optineurin | ↑20 | – | – | ↑ | – | – | – | ||
| AF005775 | CFLAR | ↑20 | – | – | ↑ | – | ↑ | – | ||
| Cytokines/chemokines and receptors | ||||||||||
| X02910 | TNF | ↑20 | – | – | ↑ | – | ↑ | ↑ | ||
| U00672 | IL10RA* | ↑20 | – | ↑ | ↑ | – | – | – | ||
| M21121 | CCL5* | ↑20 | – | – | ↑ | – | ↑ | ↑ | ||
| Y16645† | CCL8* | ↑20 | – | ↑ | – | – | ↑ | ↑ | ||
| AF030514† | CXCL11* | ↑20 | – | – | – | – | ↑ | ↑ | ||
| X72755† | CXCL9* | ↑4, ↑40 | – | – | – | – | ↑ | ↑ | ||
| X02530† | CXCL10* | ↑4, ↑40 | – | – | – | – | – | ↑ | ||
| Cell adhesion | ||||||||||
| M30257 | VCAM1 | ↑20 | – | ↑ | – | – | – | – | ||
| U88964† | IS320 | ↑20 | – | – | ↑ | – | ↑ | – | ||
| M63193† | EGF1 | ↑20 | – | – | ↑ | – | – | – | ||
| M73255† | VCAM1 | ↑20 | – | – | ↑ | – | – | – | ||
| Cell growth and maintenance | ||||||||||
| AF044309 | Syntaxin 11 | ↑20 | – | – | – | – | ↑ | – | ||
| X57522† | Transporter 1 (MDR/TAP)* | ↑20 | – | – | – | – | ↑ | – | ||
| M77016 | Tropomodulin 1 | ↑20 | – | – | – | ↑ | – | – | ||
| U20982 | Insulin-like growth factor-binding protein 4 | ↑20 | – | – | – | – | – | ↑ | ||
| J04164† | IFM1 (9-27)* | ↑20, ↑20 | – | – | ↑ | – | ↑ | – | ||
| X59892† | Tryptophanyl-tRNA synthetase* | ↑20, ↑20 | – | – | – | – | ↑ | ↑ | ||
| Signal transduction | ||||||||||
| U02020† | Pre–B-cell colony-enhancing factor 1 | ↑4 | – | – | ↑ | – | ↑ | – | ||
| D10923 | Putative chemokine receptor | ↑4 | – | – | – | – | ↑ | – | ||
| M97935† | STAT1* | ↑20 | – | – | ↑ | – | – | ↑ | ||
| Immune response | ||||||||||
| X16354† | Carcinoembryonic antigen-related cell-adhesion molecule 1* | ↑4 | – | – | ↑ | – | – | – | ||
| M55543† | GBP2 | ↑20 | – | – | ↑ | – | ↑ | ↑ | ||
| M55542† | GBP1 | ↑20 | – | – | – | – | ↑ | ↑ | ||
| M13755† | GIP2 | ↑4, ↑20 | – | – | – | – | ↑ | – | ||
| Transcription factor | ||||||||||
| L19871† | Activating transcription factor 3* | ↑4 | – | – | – | – | ↑ | – | ||
| Contradictory expression genes | ||||||||||
| U64198 | IL12RB2* | ↑4 | – | – | ↓ | ↓ | – | – | ||
| M36820† | CXCL2* | ↓4 | – | – | ↑ | – | ↑ | ↑ | ||
| M36821† | CXCL3* | ↓4 | – | – | ↑ | – | ↑ | ↑ | ||
| AF043129 | IL-7R | ↓4, ↓20 | – | ↑ | ↑ | – | ↑ | ↑ | ||
| M27492 | IL-1R1 | ↓4, ↓20 | – | – | ↑ | – | – | – | ||
The number following ↑ or ↓ indicates the hours in which upregulation or downregulation occurs, respectively.
– indicates no change, or expression level altered less than 2-fold.
Known ISG
Similar change both in CD34 and stromal cells; rows without this symbol indicate a change only in CD34 cells
Validation of Genechip results by real-time PCR. For selected characteristic genes with altered expression by gene chip analysis an independent analysis using quantitative Taqman PCR was performed. Bars on the right of the axis represent upmodulated genes in CD34 cells after IFN-γ treatment. Bars on the left side of the axis represent downmodulated genes. ▨ indicates oligoarray results; and ▪, the corresponding real-time PCR results. Mean values of 2 independent experiments performed in duplicate. (A) Selected genes involved in apoptosis and cell death. (B) Selected genes coding for cytokines/chemokine receptors. (C) Miscellaneous genes. Error bars indicate SDs.
Validation of Genechip results by real-time PCR. For selected characteristic genes with altered expression by gene chip analysis an independent analysis using quantitative Taqman PCR was performed. Bars on the right of the axis represent upmodulated genes in CD34 cells after IFN-γ treatment. Bars on the left side of the axis represent downmodulated genes. ▨ indicates oligoarray results; and ▪, the corresponding real-time PCR results. Mean values of 2 independent experiments performed in duplicate. (A) Selected genes involved in apoptosis and cell death. (B) Selected genes coding for cytokines/chemokine receptors. (C) Miscellaneous genes. Error bars indicate SDs.
We identified 6 apoptotic genes in IFN-γ-treated CD34 cells that were also overexpressed in CD34 cells derived from AA; 3 of them were also upmodulated in normal CD34 cells derived from PNH. Concordant changes were also found for 2 cytokine receptor genes (TNF, IL10RA) in AA and 5 in normal CD34 cells derived from PNH (TNF, CCL5, CCL8, XCL11, and CXCL9). In addition, upmodulation IFN-γ-stimulated adhesion and transcription factors identified in cultured CD34 cells were also found in AA and WT CD34 cells derived from PNH patients. In contrast, little concordance with IFN-γ-induced changes was found in PIG-A-deficient CD34 cells, suggesting a relative resistance of these cells to the action of IFN-γ. In monosomy 7, a striking similarity was found with regard to the expression profile of apoptotic genes induced by IFN-γ. CD34 cells derived from patients with trisomy 8 showed increased expression of chemokine and cytokine genes also seen after IFN-γ treatment of CD34 cells in vitro.
Discussion
Microarray analysis of RNA-expression profiles is a powerful tool for hypothesis generation. Accumulation of microarray results in the form of an expression database derived from related conditions may facilitate a more complex analysis, provided that the conditions under which the results were generated are comparable. Data mining using existing sets of expression arrays may be an important tool of discovery.
In this study, we have performed an analysis of the in vitro-generated transcriptome of CD34 and bone marrow stroma cells treated with IFN-γ by using oligonucleotide arrays. Among the cytokines implicated in the pathogenesis of bone marrow failure syndromes, IFN-γ appears to be the most tightly regulated and shows the most stringent correlation with disease activity inAA.27,32 Consequently, our in vitro experiments dealt with the effects of IFN-γ on the hematopoietic progenitor and stem cells in vitro.
We have used RNA from CD34 cells derived from pooled sources; 2 independent pools have been investigated producing comparable results. The method of RNA-generating application and chip preparation has been established in previous studies and was shown to produce reliable results.16-18 These studies also showed excellent correlation between the pooled results and those obtained in individual donor RNA samples.17,18,33 Due to the application of the identical technology and protocols, we did not specifically repeat these control experiments. However, we did find good agreement between the array data and those obtained using Taqman PCR. Discordant results were found between the arrays and PCR for only 2 of 21 genes tested at 2 individual time points. In addition, our analysis confirmed induction of expression for many previously described ISGs. We were also able to reproduce the results of historic experiments. In these studies, we investigated various pathways by which IFN-γ can mediate its immunosuppressive effects. For example, we have shown that in CD34 cells, IFN-γ increased the expression of CASP8, TRAIL,15,34 and nitric oxide synthase,29 likely effects mediated by IRF2.35 Similarly, proapoptotic signals delivered by IFN-γ were shown to be mediated by IRF1 and 2 and activation of STAT1. In our current experiments, IFN-γ-treated CD34 and stroma cells showed high levels of both IRF1 and STAT1. Increased expression of CASP1 suggests that CD34 and stroma cells exposed to IFN-γ can be induced to undergo apoptosis, a conclusion that is consistent with previous findings.35 The historic studies were empiric and did not provide a comprehensive view of the effects of IFN-γ on the subcellular level. However, their concordance with historic results demonstrates the reliability of the microarray technology.
As a reference for the study of ISG transcription in CD34 cells, we focused our analyses on those genes that showed undetectable expression levels at the baseline but were detected in IFN-γ-treated cells. Known ISGs, such as IFI27, ISG20, and IRF1,36-38 were upregulated in both CD34 and stroma cells, whereas others such as IFI30 and PRKR39,40 were found only to be induced in CD34 cells but not in stroma cells by our cutoff criteria. Conversely, G1P3 gene expression was only detected in stroma and not in CD34 cells. While expression of some genes was found to be affected only at 4 hours, others were turned on after 20 hours or showed persistent upmodulation. Induction of some ISGs was shown to peak at 6 hours following stimulation41 but the exact kinetics may vary between the genes. Of note is that many previously known ISGs were not found to be expressed in CD34 cells and for some the induction levels were low (< 2-fold), suggesting that these genes may represent tissue-specific ISGs or their expression shows a tightly controlled time pattern.
Global analysis of the IFN-γ-induced transcriptome in CD34 cells showed profound effects on cell growth and/or maintenance genes such as tryptophanyl-tRNA synthetase and ECGF, in agreement with previously reported results. In addition to these expected results, IFN-γ induced a large number of new genes, including CCNA1, the oncogene RAB7,5 and IGF-BP4. These genes may play important roles in the regulation of hematopoiesis. Overexpression of murine cyclin A1 in transgenic mice leads to abnormal myelopoiesis42 and this gene is highly expressed in leukemias of myeloid origin.43 Our findings suggest that IFN-γ may be involved in modulation of myeloid differentiation.
As predicted from the previously described biologic effects of IFN-γ, this cytokine induced a variety of proapoptotic genes including cell-death-associated genes such as TRAIL, CASP8, and Fas. Moreover, profound changes were also observed in genes related to immune regulation, immune response, and signal transduction. In contrast, stromal cells treated with IFN-γ showed prominent expression of genes involved in metabolism, cell growth and/or maintenance, biosynthesis, and cell adhesion. This pattern is clearly different from that observed in CD34 cells, indicating again that IFN-γ exhibits distinct effects in various hematopoietic cell types.
Proinflammatory cytokines IFN-γ and tumor necrosis factor-alpha (TNF-α) have been shown to be overexpressed in bone marrow failure syndromes such as AA and MDS and were implicated in the mechanisms of hematopoietic suppression.25,29,44,45 Our previous microarray studies have shown that these genes are upregulated in both CD4 and CD8 marrow lymphocytes in AA.24 In concordance with this result, corresponding downstream response genes, including FAS, TNFR2, and TNFRSF1B, were also found to be increased in the marrow CD34 cells of the patients.16
IFN-γ can negatively affect human hematopoietic stem cell self-renewal.46 The balance between T helper 1 (Th1) and Th2 is shifted in AA, indicating a higher propensity for production of IFN-γ.47 In addition to its direct effects, IFN-γ may also alter stroma regulation of marrow homeostasis and produce an array of secondary effects (eg, by altering trophic and inhibitory factor expression or affecting adhesion molecule migration).48 In this study, we treated stromal cells with IFN-γ in vitro. Some adhesion molecules, such as ICAM1 and VCAM1, were found to be upregulated following treatment, whereas others (eg, integrin-α5 and integrin-β3) were downregulated. Similarly, in AA, in vivo IFN-γ produced by various bone marrow cell types may influence the physiologic adhesion between bone hematopoietic stem cells and the marrow microenvironment, resulting in their defective function. An increased level of IL-6 has been reported in stromal cells from MDS, consistent with our findings of upmodulation of IL-6 after exposure of stroma to IFN-γ.
Previously, we studied gene-expression profiles derived from AA, PNH, and MDS using an identical methodology and analogous analytic strategy.16,17 It is possible that the abnormal expression pattern seen in vivo in CD34 cells from AA, PNH, and perhaps some forms of MDS can be a result of the effects of IFN-γ secreted by the immune cells in these diseases. We have therefore taken advantage of these pre-existing data and compared the expression profile of CD34 cells exposed to IFN-γ in vitro to the transcriptome in bone marrow failure syndromes. The comparison was possible as the changes were expressed in relation to freshly isolated CD34 cells. Our comparisons included CD34 cells derived from AA, GPI-deficient and phenotypically normal CD34 cells from PNH, as well as CD34 cells from patients with MDS (trisomy 8 and monosomy 7). In addition to IRF1, Stat1, and CASP1 (previously shown to transduce apoptotic effects mediated by IFN-γ), 7 other classic apoptotic genes (FAS, TRAIL, B94, TFN-AIP6, optineurin, CASP8, and CFLAR) were found to be expressed by IFN-γ-treated CD34 cells; these genes were also overexpressed in CD34 cells derived from AA patients and 3 were also upmodulated in phenotypically normal CD34 cells derived from PNH. Concordant changes were also found for 3 cytokine receptor genes (TNF, IL10RA, and CCL5) in AA and 5 in “normal” CD34 cells derived from PNH (TNF, CCL5, CCL8, CXCL11, and CXCL9). In addition, upmodulation of IFN-γ-stimulated adhesion and transcription factors were found in IFN-γ-treated normal CD34 cells, AA CD34 cells, and WT CD34 cells derived from PNH patients. In contrast, little concordance with IFN-γ-induced changes was found in PIG-A-deficient CD34 cells, suggesting a relative resistance of these cells to the action of IFN-γ. In monosomy 7, a striking similarity was found with regard to the expression profile of apoptotic genes induced by IFN-γ, whereas CD34 cells derived from patients with trisomy 8 showed increased expression of chemokine and cytokine genes also seen after IFN-γ treatment of CD34 cells in vitro (such as IGF1R, VCAM1, CCL8, IL10RA, P63, TNF, and AIP2).17
For the expression analysis of CD34 cells in AA, pooling of a large number of patients was required due to the inherently low number of CD34 that can be recovered from aplastic bone marrow. As only a proportion of AA patients demonstrate upregulation of IFN-γ, targeted analysis of CD34 cells from those patients would be most appropriate. However, in order to obtain sufficient RNA for the expression arrays, marrow aspirates from 21 patients were used.16 Transcriptome results from AA-derived T cells previously reported by our group were obtained from lymphocytes pooled from the same cohort of AA patients and showed a net increase in IFN-γ expression.24
In summary, our results demonstrate the feasibility of combining the results of various gene-expression array experiments to query mechanisms of disease. The transcriptome of CD34 cells treated with IFN-γ reflects the pathophysiologic role of this cytokine in hematopoiesis and demonstrates the complexity of metabolic changes in bone marrow that result from the action of a singular cytokine.
Prepublished online as Blood First Edition Paper, August 30, 2005; DOI 10.1182/blood-2005-05-1884.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.


![Figure 3. Gene-expression patterns of CD34 cells after exposure to IFN-γ. Gene-expression profiles of 2 independent pools of CD34 cells derived from healthy controls were generated using Affymetrix HG-U95Av2 oligonucleotide GeneChips. Differential expression was defined as at least a 2-fold change in expression level in cells treated with IFN-γ compared with controls (Y or N, respectively). Values on the right indicate degree of upregulation and values on the left show the degree of downmodulation. “Absolute” expression levels (in arbitrary fluorescence units [AFUs]) are indicated using a color scale; red bars indicate high level of expression (> 1000 AFU); yellow bars, medium level of expression (200-1000 AFU); green bars, low level (20-200). * indicates genes previously known to be regulated by IFN-γ. GenBank number and gene name in red indicate altered expression in both CD34 and stroma cells; black letters indicate that the genes changed their expression only in one of the cell types studied.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/107/1/10.1182_blood-2005-05-1884/4/m_zh80010688860003.jpeg?Expires=1767706236&Signature=MrQXtG8E5h9Bp3PTZpoFy1hKlmKMUSuC0geITEb9ZDTWkm3~Emu4ZDlxK3DjfCjwYedK80ljIe0IwzwxY4PqmtR9J5wplzhy~SuBc21q8AyyQwvQDXNknrBBsh2EDeKcpwk-ZuKt~Aw0Uxr-cRcIUfpuZz6ERzSXUrSIasBeenyfCyvUKwuxmNzN8TuV9EzDlgYBqK0lYsJjWsUbKxkEodKJVeCgfz19jclZlL2~fse3PAEQhMxjyvNG36~SQ6Mg2JEOoPlpCWS8i9tXlqiwhmKz0m5ZqjXFMLIRZH83nrIGcaymJR4Ro3HPpJ68KiyYix~1cm5uj1Y-VKUGaNG5Fg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal