Abstract
Runx1 expression marks the putative hemogenic endothelium between embryonic days (E) 8.5 to 11.5 of mouse gestation and is required for the formation of intra-aortic hematopoietic clusters, leading to the hypothesis that Runx1 is required for the transition from endothelial to hematopoietic cell. To address this hypothesis, we ablated the Runx1 gene by Cre-recombinase-mediated excision, with Cre expression under the control of the Tek promoter and enhancer. Most embryos died between E12.5 and E13.5 with a phenotype almost identical to Runx1 deficiency. We conclude that Runx1 function in establishing definitive hematopoiesis is required in a Tek+ cell.
Introduction
Definitive hematopoietic cells emerge from the dorsal aorta within the aorta-gonad-mesonephros (AGM) region, the vitelline, and umbilical arteries, and the yolk sac during fetal development.1-5 The observation of intra-aortic clusters of hematopoietic cells in these sites led to the hypothesis that definitive hematopoietic progenitors and hematopoietic stem cells (HSCs) differentiate from a “hemogenic endothelium.”6,7 According to this concept, some of the endothelial cells lining specific blood vessels lose their endothelial-like properties and give rise to hematopoietic cells that are then released into the circulation and into the para-aortic mesenchyme.6,8
Before intra-aortic hematopoietic clusters are apparent, endothelial cells in the vitelline and umbilical arteries, in the ventral aspect of the dorsal aorta, and some endothelial cells in the yolk sac express Runx1.9 Para-aortic mesenchymal cells in the ventral aspect of the dorsal aorta also express Runx1, as do the intra-aortic clusters, and the formation of intra-aortic clusters requires Runx1 function.9,10 Runx1 encodes the DNA-binding subunit of a core-binding factor (CBF) and is required for the formation of HSCs during embryogenesis.11-13 Runx1 expression also marks all long-term repopulating HSCs in the midgestation mouse embryo and in the adult bone marrow.14,15 Although essential for generating fetal HSCs, Runx1 function is not required for maintaining HSCs in the adult.16,17
Here, we selectively disrupted Runx1 function in Tek+ cells. The Tek promoter and enhancer sequences used to drive Cre recombinase were previously shown to direct the expression of lacZ and gfp transgenes in all endothelial cells of the mouse embryo and also in a subset of definitive hematopoietic progenitors in the fetal liver.18-20 We show that by selectively ablating Runx1 function in Tek+ cells we reproduce essentially all of the phenotypes associated with Runx1 deficiency, demonstrating that Runx1 function is required in a Tek+ cell to establish definitive hematopoiesis.
Study design
Embryo and mouse generation
Runx1f/f mice (Runx1tm3Spe/tm3Spe) are described elsewhere.17 Tg(Tek-cre) [B6.Cg-Tg(Tek-cre)12Flv], ROSA26 [B6;129S-Gt(ROSA)26Sor/J], R26R [B6;129-Gt(ROSA)26Sortm1Sho/J], and Tg(Tek-lacZ) [FVB/N-Tg(TIE2-lacZ)182Sato/J] mice were purchased from Jackson Laboratories (Bar Harbor, ME). We crossed Tg(Tek-cre) mice with either Runx1rd/+ (Runx1tm1Spe/+)12 or Runx1lz/+ (Runx1tm2Spe/+)9 mice to generate Runx1rd/+Tg(Tek-cre) or Runx1lz/+Tg(Tek-cre) male mice, respectively, and then crossed these with Runx1f/f female mice.
Genotyping
Primers to detect the Tek-cre transgene are described elsewhere.21 We used RDint (GAGTCCCAGCTGTCAATTCC) and RDneo (TCGCAGCGCATCGCCTTCTA) for the Runx1rd allele and RDint and RDex5 (GGTGATGGTCAGAGTGAAGC) for the unexcised Runx1f allele. We detected the excised Runx1f allele with RDint and RDint3 (CACCATAGCTTCTGGGTGCAG) and the Runx1lz and R26R alleles with LacZ forward (ATCCTCTGCATGGTCAGGTC) and LacZ reverse (CGTGGCCTGATTCATTCC).
Histology
We detected β-galactosidase expression with X-gal (Sigma, St Louis, MO) as described previously9 and counterstained 8-μm sections with nuclear fast red (Vector Laboratories, Burlingame, CA). Whole mount staining of c-kit+ hematopoietic clusters was done as described by Takakura et al.22 Images of intact embryos in phosphate-buffered saline (Figure 1A,B) were visualized using a Leica MZFLIII stereomicroscope equipped with a Plan 1× 0.14 (.025-.125) objective lens (Leica, Heerbrugg, Switzerland). A Color Mosaic 11.2 camera and Spot Insight 4.0 acquisition software (Diagnostic Instruments, Sterling Heights, MI) were used to capture and acquire images. Embryos in Figure 1D were photographed in a 1:2 ratio of benzyl alcohol to benzyl benzoate. Images of histologic sections were visualized with an Optiphot-2 microscope equipped with 4×/0.10 and 40×/1.0 objective lenses (Nikon, Tokyo, Japan), and were captured and acquired with the same camera and acquisition software as described for Figure 1A,B. All images were processed with Adobe Photoshop 7.0 software (Adobe Systems, San Jose, CA).
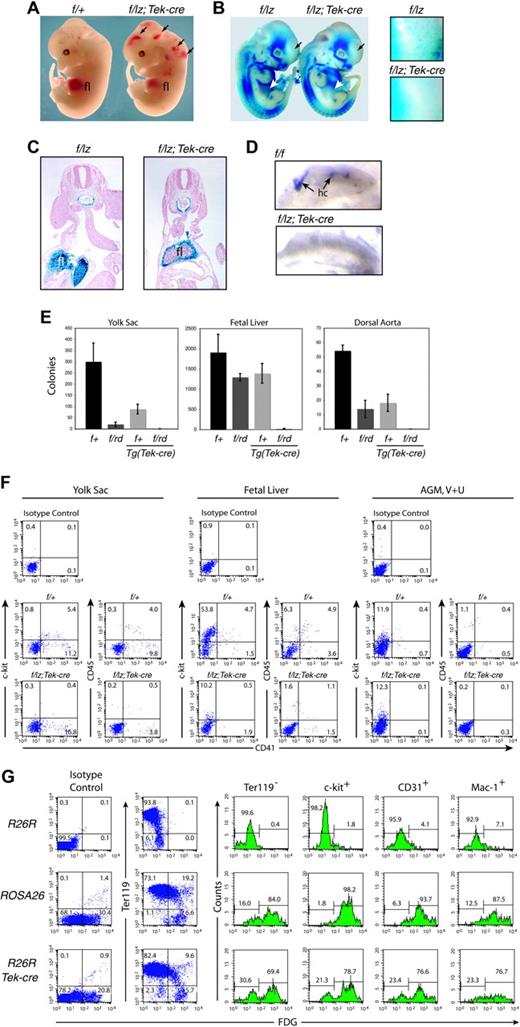
Hematopoietic analyses. (A) Gross appearance of E12.5 fetuses. Arrows indicate hemorrhages. (B) Expression from the Runx1lz allele in E11.5 Runx1f/lz and Runx1f/lz Tg(Tek-cre) littermates. The individual Runx1+ cells distributed throughout the Runx1f/lz fetus are absent in the Runx1f/lz Tg(Tek-cre) fetus (regions in head indicated by black arrows are enlarged in panels to the right). White arrows indicate fetal livers. (C) Transverse section through the AGM region and fetal liver of E10.5 fetuses illustrating the lack of Runx1+ progenitors in the center of the fetal liver (fl) in Runx1f/lz Tg(Tek-cre) animals. (D) C-kit+ hematopoietic clusters (hc's) in the umbilical artery, whole mount. (E) Colony-forming assays represented as total colonies per organ. Error bars represent 95% confidence intervals. Yolk sac (E9.5): nexperiments = 3, nf/+ = 6, nf/rd = 6, nf/+;Tek-cre = 5, nf/rd; Tek-cre = 5. Fetal liver (E11.5): nexperiments = 3, nf/+ = 5, nf/rd = 11, nf/+;Tek-cre = 3, nf/rd; Tek-cre = 8. Dorsal aorta and mesenchyme (E11.5): nexperiments = 4, nf/+ = 6, nf/rd = 11, nf/+;Tek-cre = 11, nf/rd;Tek-cre = 8. The difference between Runx1f/+ and all other genotypes is significant at P < .001. (F) FACS analysis of the yolk sac (E9.5), fetal liver (E11.5), and AGM region plus vitelline and umbilical (V + U) arteries (E11.5). The average percentage ± SD of c-kit+CD41+ cells in the yolk sac is 5.8 ± 1.1 Runx1f/+; 0.5 ± 0.5 Runx1f/lz Tg(Tek-cre), and of CD45+CD41+ cells is 4.4 ± 0.6 Runx1f/+; 0.5 ± 0.3 Runx1f/lz Tg(Tek-cre). The average percentage of c-kit+CD41+ cells in the fetal liver is 5.0 ± 0.8 Runx1f/+; 0.4 ± 0.4 Runx1f/lz Tg(Tek-cre), and CD45+CD41+ cells is 6.3 ± 2.0 Runx1f/+; 1.4 ± 1.4 Runx1f/lz Tg(Tek-cre). Average c-kit+CD41+ cells in the AGM, V + U is 1.1 ± 0.7 Runx1f/+; 0.0 ± 0.0 Runx1f/lz Tg(Tek-cre), and of CD45+CD41+ cells is 1.2 ± 1.0 Runx1f/+; 0.1 ± 0.1 Runx1f/lz Tg(Tek-cre). (G) Assessment of excision efficiency by Tek-cre, determined by comparing the percentage of β-gal+ fetal liver cells in ROSA26 and R26R Tg(Tek-cre) mice. Representative plots and histograms are shown. Histograms indicate the percentage of β-gal+ and β-gal- cells expressing (or not expressing, in the case of Ter119) the indicated cell-surface markers.
Hematopoietic analyses. (A) Gross appearance of E12.5 fetuses. Arrows indicate hemorrhages. (B) Expression from the Runx1lz allele in E11.5 Runx1f/lz and Runx1f/lz Tg(Tek-cre) littermates. The individual Runx1+ cells distributed throughout the Runx1f/lz fetus are absent in the Runx1f/lz Tg(Tek-cre) fetus (regions in head indicated by black arrows are enlarged in panels to the right). White arrows indicate fetal livers. (C) Transverse section through the AGM region and fetal liver of E10.5 fetuses illustrating the lack of Runx1+ progenitors in the center of the fetal liver (fl) in Runx1f/lz Tg(Tek-cre) animals. (D) C-kit+ hematopoietic clusters (hc's) in the umbilical artery, whole mount. (E) Colony-forming assays represented as total colonies per organ. Error bars represent 95% confidence intervals. Yolk sac (E9.5): nexperiments = 3, nf/+ = 6, nf/rd = 6, nf/+;Tek-cre = 5, nf/rd; Tek-cre = 5. Fetal liver (E11.5): nexperiments = 3, nf/+ = 5, nf/rd = 11, nf/+;Tek-cre = 3, nf/rd; Tek-cre = 8. Dorsal aorta and mesenchyme (E11.5): nexperiments = 4, nf/+ = 6, nf/rd = 11, nf/+;Tek-cre = 11, nf/rd;Tek-cre = 8. The difference between Runx1f/+ and all other genotypes is significant at P < .001. (F) FACS analysis of the yolk sac (E9.5), fetal liver (E11.5), and AGM region plus vitelline and umbilical (V + U) arteries (E11.5). The average percentage ± SD of c-kit+CD41+ cells in the yolk sac is 5.8 ± 1.1 Runx1f/+; 0.5 ± 0.5 Runx1f/lz Tg(Tek-cre), and of CD45+CD41+ cells is 4.4 ± 0.6 Runx1f/+; 0.5 ± 0.3 Runx1f/lz Tg(Tek-cre). The average percentage of c-kit+CD41+ cells in the fetal liver is 5.0 ± 0.8 Runx1f/+; 0.4 ± 0.4 Runx1f/lz Tg(Tek-cre), and CD45+CD41+ cells is 6.3 ± 2.0 Runx1f/+; 1.4 ± 1.4 Runx1f/lz Tg(Tek-cre). Average c-kit+CD41+ cells in the AGM, V + U is 1.1 ± 0.7 Runx1f/+; 0.0 ± 0.0 Runx1f/lz Tg(Tek-cre), and of CD45+CD41+ cells is 1.2 ± 1.0 Runx1f/+; 0.1 ± 0.1 Runx1f/lz Tg(Tek-cre). (G) Assessment of excision efficiency by Tek-cre, determined by comparing the percentage of β-gal+ fetal liver cells in ROSA26 and R26R Tg(Tek-cre) mice. Representative plots and histograms are shown. Histograms indicate the percentage of β-gal+ and β-gal- cells expressing (or not expressing, in the case of Ter119) the indicated cell-surface markers.
Methylcellulose colony-forming assays
Flow cytometry
Cells from yolk sac, liver, AGM, vitelline, and umbilical artery regions from E9.5 (yolk sac) or E11.5 fetuses were prepared as described previously.14 We performed immunostaining with fluorescein isothiocyanate (FITC)-conjugated antibodies to CD41 (MWReg30) and phycoerytherin (PE)-conjugated antibodies to c-kit2B8 CD45 (LCA), Sca-1 (Ly-6A/E, D7), CD-31 (MEC 13.3), B220 (RA3-6B2), CD4 (L3T4), CD8a (Ly-2), Gr-1 (RB6-8C5), CD11c (HL3), TER-119 (TER-119), or CD41 (MWReg30) (BD Pharmingen, San Diego, CA), and analyzed the cells on a Becton Dickinson FACSCalibur or FACScan flow cytometer. Dead cells were excluded with TO-PRO-3 (Molecular Probes, Eugene, OR). We detected Runx1-β-galactosidase expression with fluorescein-di-β-d-galactopyranoside (FDG; Molecular Probes).14 All procedures involving mice were approved by the Institutional Animal Care and Use Committee at Dartmouth College (Hanover, NH).
Results and discussion
Deletion of Runx1 in Tek+ cells mimics Runx1 deficiency
We crossed either Runx1rd/+Tg(Tek-cre) or Runxllz/+Tg(Tek-cre) mice to Runx1f/f mice to delete Runx1 in Tek+ cells. Runx1rd has a deletion of exon 4, which encodes part of the DNA binding domain, and is a nonfunctional Runx1 allele.12 Runx1lz has exons 5.3 and 6 replaced with lacZ sequences and is also nonfunctional.9 Of the approximately 200 mice weaned we identified only 2 animals with the genotype Runx1f/rd Tg(Tek-cre) (Table 1), indicating that most mice with this genotype died in utero. Peripheral blood from the 2 adult Runx1f/rd Tg(Tek-cre) mice contained primarily the nonexcised Runx1f allele (not shown), indicating that hematopoiesis originated from cells that had escaped Cre excision.
Genotype and phenotype of offspring derived from crossing Runx1rd/+ Tg(Tek-cre) with Runx1f/f mice
. | Genotype, no. mice . | . | . | . | Hemorrhage, no. mice . | |||
|---|---|---|---|---|---|---|---|---|
| Stage . | f/+ . | f/rd . | f/+;Tek-cre . | f/rd;Tek-cre . | f/rd;Tek-cre . | |||
| E10.5 | 2 | 5 | 4 | 3 | 1 | |||
| E11.5 | 9 | 21 | 9 | 16 | 8 | |||
| E12.5 | 20 | 14 | 17 | 24 | 16 | |||
| E13.5 | 7 | 8 | 11 | 10 | NS | |||
| Postnatal | 54 | 69 | 69 | 2 | NS | |||
. | Genotype, no. mice . | . | . | . | Hemorrhage, no. mice . | |||
|---|---|---|---|---|---|---|---|---|
| Stage . | f/+ . | f/rd . | f/+;Tek-cre . | f/rd;Tek-cre . | f/rd;Tek-cre . | |||
| E10.5 | 2 | 5 | 4 | 3 | 1 | |||
| E11.5 | 9 | 21 | 9 | 16 | 8 | |||
| E12.5 | 20 | 14 | 17 | 24 | 16 | |||
| E13.5 | 7 | 8 | 11 | 10 | NS | |||
| Postnatal | 54 | 69 | 69 | 2 | NS | |||
NS indicates not scored.
Most Runx1f/rd Tg(Tek-cre) or Runx1f/lz Tg(Tek-cre) fetuses died at E12.5 to E13.5 with hemorrhaging in the central nervous system and pale livers (Figure 1A). There was a dearth of Runx1+ hematopoietic progenitors in the interior of the liver, although the mesenchymal capsule was Runx1+ (Figure 1C). Most E12.5 Runx1f/rd Tg(Tek-cre) embryos hemorrhaged (Table 1; Figure 1A). Also, missing from Runx1f/lz Tg(Tek-cre) fetuses was a population of Runx1+ (Figure 1B) or F4/80+ (not shown) cells that are distributed throughout Runx1f/lz fetuses. We found no intra-aortic clusters in the AGM region, vitelline, or umbilical arteries in Runx1f/lz Tg(Tek-cre) animals (Figure 1D). The age of death and gross morphologic appearance were identical to that seen in animals homozygous for nonfunctional Runx1 or Cbfb alleles, or Runx1lz/rd animals.9,11,12,23,24
Colony-forming unit in culture (CFU-C) assays detected very few hematopoietic progenitors in the livers of Runx1f/rd Tg(Tek-cre) embryos and none in the yolk sac or dorsal aorta and its surrounding mesenchyme (Figure 1E). The reduction in fetal liver CFU-Cs was similar to that in CBFβ-deficient animals23 but less severe than in Runx1-deficient embryos in which CFU-Cs were totally absent.11,12 C-kit+CD41+ and CD45+CD41+ cells were reduced approximately 12.5- and 4.5-fold in the fetal liver, respectively; 11.6- and 8.8-fold in the yolk sac; and were below detection levels in the AGM region, vitelline, and umbilical arteries (Figure 1F). Mikkola et al25 showed that most definitive hematopoietic progenitors in the yolk sac are in the c-kit+CD41+ population and that, although CD41+ cells are found in Runx1-/- embryoid bodies (EBs), c-kit+CD41+ cells are not. Consistent with this, we see a significant reduction in the percentage of c-kit+CD41+ cells in the yolk sac, and no significant reduction in CD41+ cells (16.3 ± 2.6 versus 13.0 ± 8.2). The level of c-kit+ cells in the AGM region does not change upon Tek-cre deletion, presumably because most c-kit+ cells in this tissue are not hematopoietic cells; in fact, Runx1 is only expressed in approximately 20% of c-kit+ cells.14 Mikkola et al25 also reported no decrease in c-kit+ cells in Runx1-/- EBs. In summary, the deletion of Runx1 in Tek+ cells essentially recapitulates Runx1 deficiency in all 3 hematopoietic sites (yolk sac, fetal liver, AGM).
To assess the excision efficiency we crossed Tg(Tek-cre) mice with Rosa 26 reporter mice (R26R)26 and compared the expression of lacZ to that in ROSA26 mice which ubiquitously express lacZ.27 Approximately 30% of total fetal liver cells and 85% of Ter119- fetal liver cells from ROSA26 mice are β-gal+ (Figure 1G). Seventy percent of Ter119- fetal liver cells in R26R Tg(Tek-cre) animals are β-gal+, indicating that approximately 80% of these cells had excised Neo and activated lacZ. Similar (70%-80%) excision percentages were calculated from the c-kit+, CD31+, or Mac1+ populations (Figure 1G). The data are consistent with excision occurring in the majority of definitive hematopoietic progenitors, or in cells that give rise to those progenitors. Because CFU-C activity, c-kit+CD41+ cells, and intra-aortic clusters were essentially absent in the yolk sac, AGM region, vitelline, and umbilical arteries, the hematopoietic defect is likely to have originated in these sites of hematopoietic cell emergence.
Runx1 function is required in the endothelium
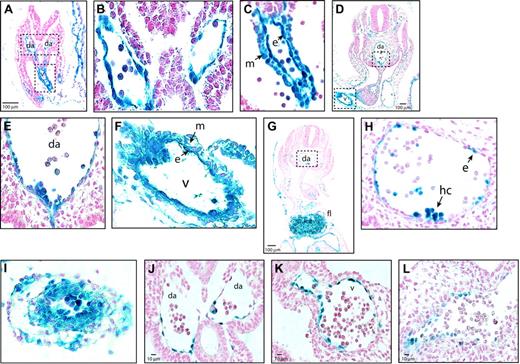
To identify the cells in the AGM region, vitelline, and umbilical arteries in which Cre-mediated excision could have occurred, we analyzed Tek expression in these sites. Between E8.5 and E10.5, all endothelial cells in Tg(Tek-lacZ) mice were Tek+ (β-gal+) (Figure 2A-F). Tek-lacZ expression in the dorsal aorta (Figure 2B,E) and umbilical artery (not shown) was confined to the endothelium and intra-aortic clusters. In E10.5 R26R Tg(Tek-cre) fetuses, β-gal+ cells were also restricted to endothelial cells and cells within the intra-aortic clusters in the dorsal aorta, although not all endothelial cells were β-gal+ (Figure 2G-H). In the vitelline artery of Tg(Tek-lacZ) animals, both the endothelium and underlying mesenchymal cells were β-gal+ at E8.5 and E10.5 (Figure 2C,F), and R26R marking experiments confirmed that excision occurs in both cell types (Figure 2I). The Tek-lacZ expression pattern and R26R marking experiments indicate that Runx1 excision could occur in the endothelium of the dorsal aorta and in either the endothelium or in mesenchymal cells surrounding the vitelline artery. However, because Runx1 expression in the vitelline artery is restricted to the endothelium between E8.5 (Figure 2K) and E10.5 (Figure 2L) and is not found in the mesenchyme, the consequence of Runx1 deficiency in the vitelline artery would only manifest itself in the endothelium. Although excision could also occur in the Tek+ intra-aortic clusters, this is unlikely because cluster formation was severely depressed or absent in Runx1f/lz Tg(Tek-cre) fetuses (Figure 1D).
Sites of Runx1 excision. (A) Tek expression in the para-aortic splanchnopleure of E8.5 Tg(Tek-lacZ) embryos. (B) Dorsal aorta from boxed region in panel A, illustrating that β-gal+ cells are confined to the endothelium. (C) Vitelline artery, from boxed region in panel A, showing Tek-lacZ expression in endothelial and mesenchymal cells. (D) Section through the AGM region of an E10.5 Tg(Tek-lacZ) embryo. (E) Ventral aspect of the dorsal aorta from the region boxed in panel D. (F) Vitelline artery from boxed region in panel D (40 ×). (G) Transverse section through the AGM region of an E10.5 R26R Tg(Tek-cre) fetus. (H) Detailed view of the dorsal aorta from the region boxed in panel G. Examination of approximately 1000 endothelial cells from 30 sections determined that 52% were β-gal+ and had therefore undergone R26R excision by Tek-cre. (I) Vitelline artery of an E10.5 R26R Tg(Tek-cre) fetus, showing that excision occurred in both mesenchymal and endothelial cells. (J) Dorsal aortae of an E8.5 Runxl lz/+ embryo, showing β-gal+ cells are confined to the endothelium. By E10.5 many ventral para-aortic mesenchymal cells are also Runx1+.9 (K) Vitelline artery, E8.5 Runxl lz/+ embryo showing β-gal+ cells confined to the endothelium. (L) Vitelline artery, E10.5 Runxl lz/+ embryo (40 ×). Da indicates dorsal aorta; v, vitelline artery; m, mesenchymal cell; e, endothelial cell; hc, hematopoietic cluster.
Sites of Runx1 excision. (A) Tek expression in the para-aortic splanchnopleure of E8.5 Tg(Tek-lacZ) embryos. (B) Dorsal aorta from boxed region in panel A, illustrating that β-gal+ cells are confined to the endothelium. (C) Vitelline artery, from boxed region in panel A, showing Tek-lacZ expression in endothelial and mesenchymal cells. (D) Section through the AGM region of an E10.5 Tg(Tek-lacZ) embryo. (E) Ventral aspect of the dorsal aorta from the region boxed in panel D. (F) Vitelline artery from boxed region in panel D (40 ×). (G) Transverse section through the AGM region of an E10.5 R26R Tg(Tek-cre) fetus. (H) Detailed view of the dorsal aorta from the region boxed in panel G. Examination of approximately 1000 endothelial cells from 30 sections determined that 52% were β-gal+ and had therefore undergone R26R excision by Tek-cre. (I) Vitelline artery of an E10.5 R26R Tg(Tek-cre) fetus, showing that excision occurred in both mesenchymal and endothelial cells. (J) Dorsal aortae of an E8.5 Runxl lz/+ embryo, showing β-gal+ cells are confined to the endothelium. By E10.5 many ventral para-aortic mesenchymal cells are also Runx1+.9 (K) Vitelline artery, E8.5 Runxl lz/+ embryo showing β-gal+ cells confined to the endothelium. (L) Vitelline artery, E10.5 Runxl lz/+ embryo (40 ×). Da indicates dorsal aorta; v, vitelline artery; m, mesenchymal cell; e, endothelial cell; hc, hematopoietic cluster.
Our data support and extend conclusions from previous studies that demonstrated that endothelial cells purified from the yolk sac or embryo proper can give rise to hematopoietic cells in vitro,7,28,29 and that endothelial cells from Runx1-deficient embryos lack this capacity,9,10 by showing that Runx1 function in the endothelium per se is required for definitive hematopoiesis. We cannot rule out a requirement for Runx1 in the ventral para-aortic mesenchyme of the AGM region where it is also expressed. The hematopoietic block was incomplete, as evidenced by low but detectable fetal liver CFU-Cs and CD45+ cells, as well as the 2 animals that survived, and could therefore be due to hematopoietic development directly from a Runx1+ mesenchymal cell pool14 or a subaortic patch.30 However, incomplete Tek-cre excision in the endothelium might also have caused or contributed to the observed leakiness of the hematopoietic block.
Like Runx1, the stem cell leukemia gene (Scl) is required for the formation of all blood lineages in the embryo but not for maintaining HSCs in the adult.31-33 Conditional disruption of Scl during embryogenesis using a Tek-cre transgene did not result in the very early embryonic lethality associated with Scl deletion, and fetal liver hematopoiesis was initiated, revealing that Scl is required prior to the stage when Tek-cre ablation takes effect.34 In contrast, our data show that Runx1 is required later than Scl and specifically in Tek+ cells.
Prepublished online as Blood First Edition Paper, September 20, 2005; DOI 10.1182/blood-2005-05-1955.
Supported by the Public Health Service (grant CA58343) (N.A.S.). The Dartmouth transgenic, flow, and histology services are supported in part by the Core Grant of the Norris Cotton Cancer Center (CA23108).
Z.L. and M.J.C. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Marella de Bruijn for her help with AGM dissections, Tomomasa Yokomizo for protocols and advice, and Sergei Tevosian for R26R mice.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal