Abstract
Melphalan, a DNA cross-linker, is one of the most widely used and effective drugs in the treatment of multiple myeloma (MM). In this report, we demonstrate that enhanced interstrand cross-link (ICL) repair via the Fanconi anemia (FA)/BRCA pathway contributes to acquired drug resistance in melphalan-resistant myeloma cell lines, and disruption of this pathway reverses drug resistance. Using the alkaline comet assay (single-cell gel electrophoresis), we observed that melphalan-resistant cells have reduced ICL formation and enhanced ICL repair compared with melphalan-sensitive cells. Cell-cycle studies demonstrated that enhanced ICL repair released cells from melphalan-induced cell-cycle delay. Using siRNA to knock down FANCF in 8226/LR5 and U266/LR6 drug-resistant cells demonstrated a direct relationship between ICL repair capacity and drug sensitivity. Overexpression of FANCF in 8226/S and U266/S drug-sensitive cells partially reproduced the drug-resistant phenotype. These data show that enhanced DNA repair via the Fanconi anemia/BRCA pathway is involved in acquired melphalan resistance. Our findings provide for a new target to enhance response to DNA cross-linking agents in cancer treatment. (Blood. 2005;106:698-705)
Introduction
DNA cross-linking agents, including melphalan, induce interstrand cross-links (ICLs) and are considered as important drugs in cancer treatment.1 Unfortunately, although most patients respond to standard- and high-dose melphalan therapy, essentially all patients relapse due to acquired drug resistance. We previously reported that acquired melphalan resistance is associated with reduced melphalan-induced DNA cross-links, and elevated levels of glutathione.2 Cross-resistance to other DNA cross-linkers (such as cisplatin, nitrogen mustard, and radiation) was observed in melphalan-resistant myeloma cells.
More recently, we reported that the acquired melphalan-resistant 8226/LR5 myeloma cell line consistently expressed different genes compared with the parental drug-sensitive 8226/S cell line.3 Results showed significant increases in the expression of FANCF and RAD51C, which are involved in the Fanconi anemia (FA)/BRCA pathway and ICL repair,4,5 in melphalan-resistant cells compared with drug-sensitive cells. Fanconi anemia has at least 11 complementation groups, and 9 Fanconi anemia genes (FANCA,6 FANCB,7 FANCC,8 FANCD1,9 FANCD2,10 FANCE,11 FANCF,12 FANCG,13 and FANCAL14 ) have been cloned. Studies have shown that Fanconi proteins participate in cell-cycle control,15 regulation of detoxification,16 and survival signal transduction.17-20 Of 9 known Fanconi proteins, 7 (A, B, C, E, F, G, L) form a heteromeric complex and monoubiquitinate FANCD2, which, in turn, interacts with well-known DNA-damage-response proteins, including ataxia telangiectasia (ATR), BRCA1, Nijmegen breakage syndrome 1 (NBS1), and RAD51, known to be involved in DNA ICL repair.4,21-24 Disruption of the FA/BRCA pathway results in chromosome instability and hypersensitivity to DNA cross-linking agents.4
Several recent reports suggest that enhanced ICL repair contributes to melphalan resistance. It has been reported that the rate of removal of ICL is associated with melphalan resistance in leukemic cells.25 In another study, similar results were reported for primary multiple myeloma specimens.26 These investigators compared the formation and repair of ICL in plasma cells from melphalan-naive and treated myeloma patients, and found that in vitro sensitivity to melphalan in plasma cells correlated with ICL repair capacity.26
In this study, we show that melphalan-resistant myeloma cell lines have elevated gene expression involving the FA/BRCA pathway, reduced formation of ICLs, and enhanced removal of ICLs. Enhanced ICL repair capacity reduced melphalan-induced growth inhibition. Furthermore, silencing FANCF in drug-resistant cells with siRNA reversed drug resistance, and, conversely, overexpression of FANCF in drug-sensitive cells enhanced cell survival following melphalan treatment. These data show that the FA/BRCA pathway mediates ICL repair and contributes to acquired melphalan resistance in myeloma cell lines. We propose that disruption of the FA/BRCA pathway may prevent acquired resistance to melphalan, and possibly other DNA cross-linking agents, and improve treatment outcome for myeloma and other cancers.
Materials and methods
Cell lines and drugs
The RPMI 8226 and U266 human multiple myeloma cell lines were obtained from the American Type Culture Collection (Rockville, MD). Cell lines were grown in RPMI 1640 medium (CellGro; MediaTech, Herndon, VA) supplemented with 10% heat-inactivated fetal bovine serum (FBS; Omega Scientific, Tarzana, CA), 1% penicillin/strepromycin, and 100 mM l-glutamine (Gemini Bio-Products, Calabasas, CA). The melphalan-resistant cells 8226/LR5 and U266/LR6 were passaged weekly in media containing 5 μMor6 μM melphalan, respectively.2
Methyl-thiazol tetrazolium (MTT) cytotoxicity assay
Cells were seeded at 8000 to 25 000 cells/well in 96-well plates (Becton Dickinson, Lincoln Park, NJ). To establish a dose response to melphalan (Sigma, St Louis, MO), cells were incubated with melphalan for 96 hours in 2-fold serial dilutions ranging from 1 × 10-4 M to 3.9 × 10-7 M. The melphalan-induced growth inhibition assays were performed as previously described.27
Melphalan-induced apoptosis
Cells were continuously treated with 50 μM or 100 μM melphalan or vehicle control (acid-ethanol) for 20 hours. Annexin V-fluorescein isothiocyanate (FITC) (Biovision, Palo Alto, CA) staining was used to measure apoptotic cells as described previously.28
Real-time quantitative reverse-transcriptase-polymerase chain reaction (RT-PCR)
Total RNA was extracted using the RNeasy Mini Kit (Qiagene, Valencia, CA) and used for cDNA synthesis (Invitrogen first-strand cDNA synthesis kit; Invitrogen, Frederick, MD). Expression of 11 genes involved in the FA/BRCA pathway, BRCA1, BRCA2, FANCA, FANCC, FANCD2, FANCE, FANCF, FANCG, FANCL, RAD51, and RAD51C, was analyzed using a custom-designed microfluid card (ABI, Foster City, CA). The gene-expression level was normalized using the endogenous control gene GAPDH and the relative gene-expression level was determined using 2-(-delta delta C (T)) (ΔΔCT) methods.29 Real-time PCR reactions were performed using ABI 7900 Sequence Detection System (ABI).
Western blot analysis
Cells were washed twice with ice-cold PBS, and resuspended in ice-cold lysis buffer (50 mM Tris [tris(hydroxymethyl)aminomethane]-HCl, pH 7.40, 0.1% nonidet P40 [NP40], 1 M NaCl) supplemented with protease and phosphatase inhibitors (10 μg/mL aprotinin, 25 μg/mL leupeptin, 10 μg/mL pepstatin A, 2 mM phenylmethylsulfonyl fluoride [PMSF], 0.1 M NaP2O4, 25 mM NaF, and 2 mM sodium orthovanadate). After sonication, the lysates were quantified using Biorad reagent (Biorad, Hercules, CA). Lysates (50 μg) were separated by 4% to 12% NuPage gel (Invitrogen) and transferred to polyvinylidenefluoride (PVDF) membrane. The antibodies used for the Western blotting were raised against FANCF (Santa Cruz Biotechnology, Santa Cruz, CA), FANCD2 (Novus Biologicals, Littleton, CO), and β-actin (Sigma).
Alkaline comet assay
The alkaline comet assay was used to detect melphalan-induced DNA cross-links in drug-sensitive (8226/S and U266/S) and drug-resistant (8226/LR5 and U266LR6) myeloma cells as described previously.3 To examine dose-response effect, 2 × 105 cells were treated with 25 μM, 50 μM, and 100 μM melphalan or vehicle control for 2 hours, washed in PBS, and cultured in drug-free medium for another 3 hours. Samples were then irradiated at 9 Gy (MARK I model 68A irradiator; J. L. Sheperd and Associates, San Fernando, CA), and alkaline comet assay was performed according to the manufacturer's instructions (Travegene, Gaithersburg, MD). Per slide, 50 images were randomly captured by fluorescence microscopy and images were quantified using Loats Associates comet analysis software (Loats Associates, Westminster, MD). The percent cross-linking was calculated as previously described3 : Relative cross-linking = (1 - [(comet tail moment drug treated - comet tail moment control)/(comet tail moment 9 Gy - comet tail moment control)]) × 100.
To examine the ICL removal, 8226/S, 8226/LR5, U266/S, and U266/LR6 were incubated in 25 μM, 50 μM, 25 μM, or 60 μM melphalan, respectively, or vehicle control for 2 hours, after which they were cultured in drug-free medium for various times. Cells were collected at 8 hours and 23 hours after a 2-hour drug treatment to perform the comet assay. The data shown are the means of 3 independent experiments (n = 50 images for each dose of each independent experiment).
Cell-cycle analysis
Propidium iodide (PI; Sigma) staining was used to measure dose-response effects of melphalan on cell-cycle progression. Both 8226/S and 8226/LR5 were treated with 10 μM and 25 μM melphalan for 2 hours and cultured in drug-free medium, and cells were collected at 24 and 48 hours. Cells were then fixed with 70% ethanol at 4°C overnight; resuspended with 500 μL PBS containing 25 μg/mL propidium iodide (PI) and 1.25 mg/mL Rnase A (Invitrogen); and incubated at 37°C for 30 minutes in the dark. Bromodeoxyuridine (BrDU)/PI staining was performed as previously described to evaluate cell-cycle progression over time following exposure to melphalan.30 Briefly, at specified time points following a 2-hour drug treatment, cells were incubated with 10 μM bromodeoxyuridine (BrDU; Sigma) at 37°C for 30 minutes, fixed in 70% ethanol, denatured (2 M HCl), and neutralized (0.1 M sodium borate). Cells were then stained with anti-BrDU FITC antibody (BD Pharmingen, San Diego, CA); resuspended with 500 μL PBS containing 25 μg/mL propidium iodide (PI) and 1.25 mg/mL Rnase A; and incubated at 37°C for 30 minutes in the dark. Nuclear staining was analyzed using a FACScan flow cytometer (Becton Dickinson, San Jose, CA). Watson (Pragmatic, Ashland, OR) model from FlowJo 4.4.4 software was used for DNA content analysis.
Transient transfection of FANCF or siRNA to inhibit FANCF
FANCF siRNA (catalog no. M-014206-00) and cyanin 3 (Cy3)-labeled Luciferase GL2 siRNA (catalog no. D-001110-01-05) were purchased from Dharmacon (Chicago, IL). FANCF was digested from the cDNA clone (kindly provided by Dr Grover C. Bagby, Oregon Health and Science University [OHSU] Cancer Institute, Portland, OR) by NotI and BamHI (NewEngland Biolab, Beverly, MA), gel purified using Qiagene kit, cloned into pQCXIP vector (BD Biosciences Clontech, San Jose, CA), and confirmed by sequencing. For transient expression, cell lines were transfected by electroporation using Nucleofector (Amaxa, Gaithersburg, MD). Briefly, 8226 and 8226/LR cells and U266 and U266/LR6 cells were resuspended in Nucleofector solution V and R (Amaxa), respectively, to the final concentration of 5 × 107 cell/mL. For each transfection, 8 μg plasmid or siRNA duplex (final concentration 1 μM) was mixed together with 5 × 106 cells. The A20 and T16 programs were used for 8226 and 8226/LR5 transfection and U266 and U266/LR6 transfection, respectively. Cells were then cultured in 5 mL of 37°C prewarmed medium.
Statistical analysis
Analysis of variance (ANOVA) was used to compare ICL formation in drug-sensitive and -resistant cells. All other statistical comparisons were made using the Student t test.
Results
Resistant cells have less melphalan-induced growth inhibition and apoptosis
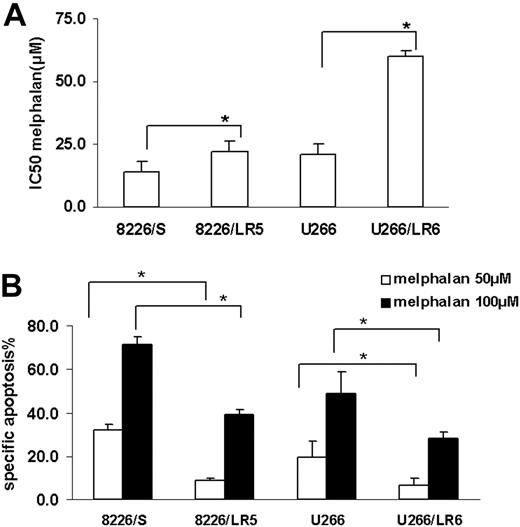
MTT and apoptosis assay were used to compare melphalan sensitivity in the drug-sensitive (8226/S, U266/S) and drug-resistant (8226/LR5, U266/LR6) myeloma cell lines. Similar to our previously published reports,2,3 melphalan-resistant cells 8226/LR5 and U266/LR6 have increased survival (Figure 1A) and decreased drug-induced apoptosis (Figure 1B) compared with their drug-sensitive parental cells 8226/S and U266/S.
Melphalan-resistant myeloma cells have less drug-induced growth inhibition and apoptosis compared with melphalan-sensitive cells. (A) MTT assay. Median inhibitory concentration (IC50) values are the mean of 3 independent experiments and SD. Student t test was used for statistical analysis. *P < .05. (B) Apoptosis assay. Melphalan treatment causes less apoptosis in drug-resistant 8226/LR5 and U266/LR6 cells compared with drug-sensitive 8226/S and U266/S cells. The mean values and standard deviations from a representative experiment performed in triplicate are shown. Student t test was used for statistical analysis. *P < .05.
Melphalan-resistant myeloma cells have less drug-induced growth inhibition and apoptosis compared with melphalan-sensitive cells. (A) MTT assay. Median inhibitory concentration (IC50) values are the mean of 3 independent experiments and SD. Student t test was used for statistical analysis. *P < .05. (B) Apoptosis assay. Melphalan treatment causes less apoptosis in drug-resistant 8226/LR5 and U266/LR6 cells compared with drug-sensitive 8226/S and U266/S cells. The mean values and standard deviations from a representative experiment performed in triplicate are shown. Student t test was used for statistical analysis. *P < .05.
Enhanced expression of the FA/BRCA pathway in melphalan-resistant 8226/LR5 and U266/LR6 cells. (A) Real-time RT-PCR was performed using ABI low-density array card. Fold values were obtained by externally standardizing against identical amplifications in drug-sensitive 8226/S cells and by internally standardizing against GAPDH in each cell line. Data shown are mean value and SD. (B) FANCD2-L relative expression was elevated after melphalan treatment. Experiments were repeated 3 times. (Top) Representative blots are shown. β-Actin blot served as loading control in all blots. (Bottom) The FANCD2-L/S ratio was quantified using densitometry. The mean values and standard deviations from 3 independent experiments are shown. Student t test was used for statistical analysis. *P < .05.
Enhanced expression of the FA/BRCA pathway in melphalan-resistant 8226/LR5 and U266/LR6 cells. (A) Real-time RT-PCR was performed using ABI low-density array card. Fold values were obtained by externally standardizing against identical amplifications in drug-sensitive 8226/S cells and by internally standardizing against GAPDH in each cell line. Data shown are mean value and SD. (B) FANCD2-L relative expression was elevated after melphalan treatment. Experiments were repeated 3 times. (Top) Representative blots are shown. β-Actin blot served as loading control in all blots. (Bottom) The FANCD2-L/S ratio was quantified using densitometry. The mean values and standard deviations from 3 independent experiments are shown. Student t test was used for statistical analysis. *P < .05.
Melphalan-resistant myeloma cells have enhanced expression of the FA/BRCA pathway
Using the Affymetrix oligonucleotide microarray (Affymetrix, Santa Clara, CA) to examine gene expression profile (GEP), we recently reported that the melphalan-resistant myeloma cell line 8226/LR5 showed significant increases in the expression of the FANCF and RAD51C genes and reduced DNA cross-links compared with the drug-sensitive 8226/S cells.3 Because the Affymetrix HG-133A chip does not include the FANCD2 and FANCL genes, we used a custom-designed real-time PCR microfluid card to examine 11 genes of the FA/BRCA pathway in 8226/S and U266/S myeloma cells and their drug-resistant variants, 8226/LR5 and U266/LR6, respectively. The results revealed that expression of multiple components of the FA/BRCA pathway were up-regulated in drug-resistant 8226/LR5 and U266/LR6 cells compared with drug-sensitive 8226/S and U266/S cells, respectively (Figure 2A). The expression levels of BRCA1, BRCA2, FANCA, FANCC, FANCF, FANCL, and RAD51C were at least 2-fold increased in 8226/LR5 compared with 8226/S. The expression of FANCL in U266/LR6 is at least 3-fold higher than U266/S drug-sensitive cells (Figure 2A).
The full integrity of the upstream FA complex promotes monoubiquitination of FANCD2 following DNA cross-linker treatment, therefore formation of the monoubiquitinated FANCD2 (FANCD2-L) isoform has been used to clinically diagnose FA patients.31 To determine whether the upstream portion of the FA/BRCA pathway is intact, we examined FANCD2-L at various time points after drug treatment in the 4 cell lines. The results revealed that the expression of FANCD2-L was significantly increased within 2 hours of melphalan treatment (Figure 2B), and a maximum increase of FANCD2-L occurred within 24 hours after melphalan treatment (Figure 2B lower panel). These data suggest that proteins involved in the upstream FA complex (FANCA, B, C, E, F, G, L) are functionally complete in all 4 cell lines. Data also suggest that increased formation of FANCD2-L may be due to the enhanced expression of the FA complex proteins, and elevated levels of the FA/BRCA pathway may contribute to enhanced ICL repair capacity in drug-resistant cells. To examine this possibility, we compared DNA ICL formation and removal in drug-sensitive and -resistant myeloma cells.
Melphalan-resistant cells have reduced DNA ICL formation and enhanced ICL removal compared with drug-sensitive cells
We recently reported that, within 2 hours following drug exposure, melphalan-induced cross-links were significantly reduced in the drug-selected 8226/LR5 cell line.3 In this study, using alkaline comet assay, we compared the kinetics of DNA ICL formation and removal between melphalan-sensitive (8226/S, U266/S) and melphalan-resistant (8226/LR5, U266/LR6) cells. When cells were treated with 25 μM, 50 μM, and 100 μM melphalan, cross-links were significantly reduced in 8226/LR5 and U266/LR6 compared with drug-sensitive 8226/S and U266/S cells, respectively (Figure 3A-B, ANOVA P < .05). To examine the ICL removal, melphalan-treated cells were examined at 10-hour and 25-hour time points. Similar amounts of ICL were induced in drug-sensitive and drug-resistant cells at the 10-hour time point, when 8226/S, 8226/LR5, U266/S, and U266/LR6 were treated with 25 μM, 50 μM, 25 μM, and 60 μM melphalan, respectively (Figure 3C). At 25 hours after melphalan treatment, 37% and 22% of ICLs were removed in 8226/LR5 and U266/LR6 cells, respectively, while no significant ICL removal was observed in drug-sensitive 8226/S and U266/S cells (Figure 3C, t test P < .05).
Melphalan-resistant cells have reduced ICL formation and enhanced ICL removal. (A) The alkaline comet assay was used to detect melphalan-induced cross-links. When cells were treated with vehicle control without irradiation, no comet tail moment was observed. When cells were irradiated, the comet tail moment was increased to 50. (B) Within 3 hours following a 2-hour exposure to 25 μM, 50 μM, and 100 μM melphalan, fewer ICL formations were observed in 8226/LR5 and U266/LR6 compared with 8226/S and U266/S, respectively. *P < .05 (ANOVA). (C) Similar amounts of cross-links are formed in drug-sensitive and drug-resistant cells at 10 hours by increasing the dose of melphalan treatment for 8226/LR5 (50 μM) and U266/LR6 (60 μM) compared with 8226/S (25 μM) and U266/S (25 μM), respectively. Within 24 hours after treatment, the remaining ICLs in drug-resistant 8226/LR5 (30.4%) are significantly less than the percentage of drug-sensitive 8226/S cells (55.2% ICLs) within 10 hours. The remaining ICLs in U266/LR6 cells (54.9%) is significantly less compared with U266/S cells (75.6%). Student t test was used for statistical analysis. *P < .05. Error bars denote standard deviation.
Melphalan-resistant cells have reduced ICL formation and enhanced ICL removal. (A) The alkaline comet assay was used to detect melphalan-induced cross-links. When cells were treated with vehicle control without irradiation, no comet tail moment was observed. When cells were irradiated, the comet tail moment was increased to 50. (B) Within 3 hours following a 2-hour exposure to 25 μM, 50 μM, and 100 μM melphalan, fewer ICL formations were observed in 8226/LR5 and U266/LR6 compared with 8226/S and U266/S, respectively. *P < .05 (ANOVA). (C) Similar amounts of cross-links are formed in drug-sensitive and drug-resistant cells at 10 hours by increasing the dose of melphalan treatment for 8226/LR5 (50 μM) and U266/LR6 (60 μM) compared with 8226/S (25 μM) and U266/S (25 μM), respectively. Within 24 hours after treatment, the remaining ICLs in drug-resistant 8226/LR5 (30.4%) are significantly less than the percentage of drug-sensitive 8226/S cells (55.2% ICLs) within 10 hours. The remaining ICLs in U266/LR6 cells (54.9%) is significantly less compared with U266/S cells (75.6%). Student t test was used for statistical analysis. *P < .05. Error bars denote standard deviation.
Melphalan has less effect on cell-cycle progression in 8226/LR5 cells compared with drug-sensitive 8226/S cells
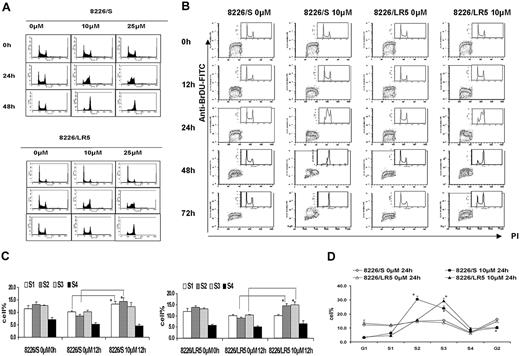
To measure dose-response effects of melphalan on cell-cycle progression, both 8226/S and 8226/LR5 cells were treated with 10 M or 25 μM melphalan for 2 hours, cultured in drug-free medium, and collected at 24 hours and 48 hours. Serial DNA histograms revealed that within 24 hours, high-dose melphalan treatment (25 μM) resulted in a greater accumulation of both 8226/S and 8226/LR5 cells in early S phase compared with low-dose (10 μM) treatment (Figure 4A). High-dose melphalan (25 μM)-treated cells required more time to eliminate the melphalan-induced G2 block, and fewer cells were released from the drug-induced growth inhibition, compared with low-dose melphalan-treated 8226/S and 8226/LR5 cells (Figure 4A). However, the 8226/LR5 cells showed less growth inhibition compared with 8226/S cells, when cells were treated with either 10 μM or 25 μM melphalan (Figure 4A). BrDU/PI staining was performed to evaluate cell-cycle progression over time following drug exposure (Figure 4B). Within 12 hours after 10-μM melphalan treatment, 8226/S cells predominantly accumulated in early S phase (S1, S2) compared with 8226/LR5 cells, which accumulated in late S phase (S3, S4) (Figure 4B-C). Figure 4D shows further detail regarding S phase cell-cycle accumulation at 24 hours using FlowJo 4.4.4 analysis. At the 48-hour post-10-μM melphalan treatment time point, the DNA content of 8226/LR5 cells accumulated at G2/M and G1 phase, while the DNA content of the drug-sensitive 8226/S cells accumulated at late S and G2/M phases and less G1 accumulation was observed. By the 72-hour time point, 8226/LR5 cells had recovered from the drug-induced cell-cycle delay; whereas, the drug-sensitive 8226/S cells remained arrested in the G2/M phase (Figure 4B).
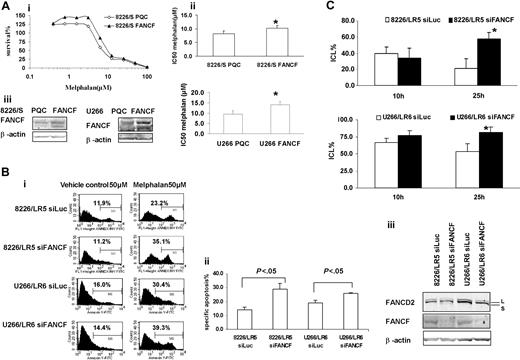
Overexpression or silencing of FANCF shows a direct relationship among FANCF expression, DNA repair capacity, and drug sensitivity to melphalan
To further examine the contribution of the FA/BRCA pathway to ICL repair and melphalan resistance, we transiently overexpressed FANCF in 8226/S and U266/S cells, and knocked down FANCF in 8226/LR5 and U266/LR6 cells. Overexpression of FANCF demonstrated by Western blot analysis in both drug-sensitive myeloma cell lines enhanced cell survival following melphalan treatment (Figure 5A). Conversely, knockdown of FANCF in melphalan-resistant 8226/LR5 and U266/LR6 cells reversed drug resistance (Figure 5B). We also performed the alkaline comet assay to determine whether knocking down FANCF was sufficient to reduce DNA ICL repair in the drug-resistant cell line. As shown in Figure 5C, a reduction in FANCF reduced the repair capacity of the 8226/LR5 and U266/LR6 cell lines. Taken together, these data show that despite increased expression in multiple genes in the FA/BRCA pathway, knocking down FANCF is sufficient to reverse, at least partially, the acquired drug-resistant phenotype in 8226/LR5 and U266/LR6 myeloma cells.
Discussion
DNA cross-linking agents, including melphalan, are important in the treatment of multiple myeloma and other cancers. The precise mechanisms contributing to melphalan resistance in multiple myeloma cells are poorly understood. This study demonstrates that overexpression of FA/BRCA pathway genes contributes to acquired resistance to melphalan for 2 multiple myeloma cell lines. In addition, this study provides evidence that the FA/BRCA pathway contributes to drug resistance via enhanced ICL repair, and release of cells from melphalan-induced growth inhibition.
We previously reported that acquired melphalan-resistant myeloma cells were cross-resistant to irradiation and also cross-resistant to other DNA cross-linkers, such as cisplatin and nitrogen mustard.2 This information, together with our recently published microarray data,3 led us to hypothesize that the FA/BRCA pathway may be involved in repair of melphalan-induced ICL and contributes to the drug-resistant phenotype. Using a custom-designed real-time PCR microfluid card, we examined 11 genes involved in the FA/BRCA pathway. Expression of these genes was greater in drug-resistant cells compared with their drug-sensitive counterparts; however, greater differences were observed between the 8226 sensitive and resistant cells compared with the U266 sensitive and resistant cells. This may, in part, be due to the higher basal expression of FANC/BRCA genes in U266/S cells compared with 8226/S cells. The expression data correlated well with enhanced cell survival and reduced apoptosis following drug treatment. We observed that multiple components of the FANC/BRCA pathway were up-regulated in drug-resistant cells compared with drug-sensitive cells. These data suggest that genes involved in this pathway may be coregulated, and further studies are warranted to delineate the transcriptional regulation of this pathway.
8226/LR5 cells release earlier from melphalan-induced cell-cycle inhibition compared with drug-sensitive 8226/S cells. (A) Flow cytometric analysis of cell-cycle phases. DNA histograms of untreated and melphalan-treated 8226/S (top) and 8226/LR5 cells (bottom) show the dose-response effects of melphalan on cell-cycle progression. Representative data are shown. (B) Time course analysis of cell-cycle progression in 8226/S and 8226/LR5 cells with and without melphalan treatment. The dot plots depict BrDU incorporation (S phase cells) detected with an FITC anti-BrDU antibody on the y-axis, and propidium iodide (PI) to detect DNA content on the x-axis. The distribution of DNA content is represented by the inset histograms. Shown is 1 of 3 representative experiments. 8226/LR5 cells are able to progress through the cell cycle following melphalan exposure; whereas 8226/S cells arrest at G2/M (72 hours). (C) Quantitative analysis of the delay of progression through the cell cycle. To study cell-cycle progression delay in early and late S phases, DNA content was divided into 6 sections and quantified as G1, S1, S2, S3, S4, and G2 phase using FlowJo 4.4.4 software Watson (Pragmatic) model. Within 12 hours following melphalan treatment, 8226/S and 8226/LR5 cells significantly accumulated at early S phase compared with non-drug-treated cells. (D) At 24 hours after melphalan treatment, 8226/S cells significantly accumulated at S2 compared with 8226/LR5, while 8226/LR5 cells significantly accumulated at late S phase (S3) compared with 8226/S cells. The mean values and standard deviations from 3 independent experiments are shown. Student t test was used for statistical analysis. *P < .05.
8226/LR5 cells release earlier from melphalan-induced cell-cycle inhibition compared with drug-sensitive 8226/S cells. (A) Flow cytometric analysis of cell-cycle phases. DNA histograms of untreated and melphalan-treated 8226/S (top) and 8226/LR5 cells (bottom) show the dose-response effects of melphalan on cell-cycle progression. Representative data are shown. (B) Time course analysis of cell-cycle progression in 8226/S and 8226/LR5 cells with and without melphalan treatment. The dot plots depict BrDU incorporation (S phase cells) detected with an FITC anti-BrDU antibody on the y-axis, and propidium iodide (PI) to detect DNA content on the x-axis. The distribution of DNA content is represented by the inset histograms. Shown is 1 of 3 representative experiments. 8226/LR5 cells are able to progress through the cell cycle following melphalan exposure; whereas 8226/S cells arrest at G2/M (72 hours). (C) Quantitative analysis of the delay of progression through the cell cycle. To study cell-cycle progression delay in early and late S phases, DNA content was divided into 6 sections and quantified as G1, S1, S2, S3, S4, and G2 phase using FlowJo 4.4.4 software Watson (Pragmatic) model. Within 12 hours following melphalan treatment, 8226/S and 8226/LR5 cells significantly accumulated at early S phase compared with non-drug-treated cells. (D) At 24 hours after melphalan treatment, 8226/S cells significantly accumulated at S2 compared with 8226/LR5, while 8226/LR5 cells significantly accumulated at late S phase (S3) compared with 8226/S cells. The mean values and standard deviations from 3 independent experiments are shown. Student t test was used for statistical analysis. *P < .05.
Real-time PCR data showed at least 2-fold increase of FANCA, FANCC, FANCF, and FANCL in 8226/LR5 compared with 8226/S cells. Increased expression of FANCD2-L isoform in 8226/LR5 compared with 8226/S cells indicated that the upstream FA complex might facilitate FANCD2-L formation. In comparison, while some of the FA genes were less changed in U266/LR6 compared with the 8226/S-8226/LR5 pair, the increased posttranslational modification of FANCD2 may be related to the 3-fold increase of FANCL, which has E3 ubiquitin ligase activity. Within 24 hours after melphalan treatment, the FANCD2-L/S ratio isoform in 8226/LR5 cells was significantly increased compared with drug-treated 8226/S cells.
We also observed a significant delay in cell-cycle progression within 24 hours after melphalan treatment with most of the cells accumulating in S phase. This observation is consistent with the observation reported by Rothfuss and Grompe.21 They noticed that in human fibroblasts, FANCD2-L is specifically expressed during the S phase after drug treatment.21 It has also been reported that the FANCD2-L/S ratio increased within 4 hours after treatment with photoactivated psoralen-inducing DNA ICLs, and a 3-fold increase of FANCD2-L occurred within 24 hours after melphalan treatment.21 Since the FANCD2 monoubiquitination event is important for normal cellular recovery from exposure to DNA cross-linkers,32 our data suggest that the increased formation of the FANCD2-L isoform, which is activated by melphalan-induced ICL during the S phase, is also essential for melphalan resistance in myeloma cell lines.
It has been suggested that the ICL-induced stalled replication fork is the major signal that activates the FA pathway and induces G2/M arrest in FA cells.21-23,33,34 In human myeloma cell line 8226, melphalan treatment induces a cell-cycle progression delay and G2/M arrest.35 Although equivalent doses of melphalan induce more ICL in drug-sensitive 8226/S cells compared with drug-resistant 8226/LR5 cells, a 2-fold increase in FANCD2-L was observed in 8226/LR5 compared with 8226/S (Figure 2B, 25-hour time point). Because only the monoubiquitinated FANCD2 (FANCD2-L) can be translocated to DNA damage foci to perform repair function, our data suggest that the efficiency of FANCD2-L formation is important for drug resistance. This efficiency in FANCD2-L formation in drug-sensitive and -resistant cell lines may be due to enhanced expression of upstream FA complex.
Overexpression of FANCF in 8226/S and U266/S reduced melphalan response, whereas silencing FANCF in 8226/LR5 and U266/LR6 enhanced melphalan response, respectively. (Ai) MTT assay. Overexpression of FANCF in drug-sensitive 8226/S and U266/S cells enhanced cell survival. The data are presented as percent survival above control cells. The experiment was repeated at least 3 times. Representative results are shown. (ii) IC50 is the mean of 3 independent experiments and SD. Student t test was used for statistical analysis. *P < .05. The experiments were repeated 3 times. (iii) Empty vector-transfected 8226-PQC and U266-PQC cells served as controls for FANCF-transfected 8226-FANCF and U266-PQC cells. β-Actin blot served as loading control. (Bi) Apoptosis assay. Transfection of siFANCF partially reversed melphalan resistance in 8226/LR5 and U266/LR6 cells. Annexin-V-FITC staining was used. The percentage of annexin-V-FITC+ cells is labeled, and specific apoptosis has been calculated. The mean values and standard deviations from a representative experiment performed in triplicate are shown. Student t test was used for statistical analysis. *P < .05. (Biii) Western blot analysis of FANCD2 and FANCF showed reduced protein expression in 8226/LR5 and U266/LR6 cells transfected with siFANCF compared with cells transfected with siLuc as a control. β-Actin blot served as loading control. (C) Capacity to repair ICLs was reduced in FANCF knockdown LR5 and LR6 cells compared with control siLuc-transfected cells, LR5 siLuc (top) and LR6 siLuc (bottom), respectively. The mean values and standard deviations from 3 independent experiments are shown. Student t test was used for statistical analysis. *P < .05.
Overexpression of FANCF in 8226/S and U266/S reduced melphalan response, whereas silencing FANCF in 8226/LR5 and U266/LR6 enhanced melphalan response, respectively. (Ai) MTT assay. Overexpression of FANCF in drug-sensitive 8226/S and U266/S cells enhanced cell survival. The data are presented as percent survival above control cells. The experiment was repeated at least 3 times. Representative results are shown. (ii) IC50 is the mean of 3 independent experiments and SD. Student t test was used for statistical analysis. *P < .05. The experiments were repeated 3 times. (iii) Empty vector-transfected 8226-PQC and U266-PQC cells served as controls for FANCF-transfected 8226-FANCF and U266-PQC cells. β-Actin blot served as loading control. (Bi) Apoptosis assay. Transfection of siFANCF partially reversed melphalan resistance in 8226/LR5 and U266/LR6 cells. Annexin-V-FITC staining was used. The percentage of annexin-V-FITC+ cells is labeled, and specific apoptosis has been calculated. The mean values and standard deviations from a representative experiment performed in triplicate are shown. Student t test was used for statistical analysis. *P < .05. (Biii) Western blot analysis of FANCD2 and FANCF showed reduced protein expression in 8226/LR5 and U266/LR6 cells transfected with siFANCF compared with cells transfected with siLuc as a control. β-Actin blot served as loading control. (C) Capacity to repair ICLs was reduced in FANCF knockdown LR5 and LR6 cells compared with control siLuc-transfected cells, LR5 siLuc (top) and LR6 siLuc (bottom), respectively. The mean values and standard deviations from 3 independent experiments are shown. Student t test was used for statistical analysis. *P < .05.
Approximately one third of the DNA-melphalan adducts are DNA interstrand cross-links.36 By studying gene-specific formation of DNA monoadducts and interstrand cross-links, Souliotis et al37 reported that maximum monoadducts were formed within 2 hours as a result of rapid binding of melphalan to nucleophilic site (N7-guanine) in one DNA strand, whereas DNA ICLs accumulate slowly and reach maximal levels within 8 hours through binding of a second chloroethyl group to another N7-guanine site in the cDNA strand. In our study, we were able to induce equivalent maximum ICLs in drug-sensitive and -resistant cells by increasing the melphalan dose exposure for drug-resistant 8226/LR5 and U266/LR6 cells compared with 8226/S and U266/S drug-sensitive cells. Interestingly, within 24 hours after melphalan treatment, approximately 37% and 22% of the ICLs were removed in the 8226/LR5 and U266/LR6 cells, respectively, while the drug-sensitive 8226/S and U266/S cells showed no significant removal of ICLs in this time frame. Our results strongly suggest that the FA/BRCA pathway contributes to the removal of ICLs during this time period. Furthermore, we demonstrated that the capacity to remove ICLs was significantly reduced in FANCF knockdown 8226/LR5 and U266/LR6 cells compared with control 8226/LR5 siLuc and U266/LR6 siLuc cells. Taken together, these data show that FANC/BRCA is involved in melphalan-induced ICL repair and contributes to drug resistance.
Hypermethylation of FANCF has been reported in ovarian, oral, lung, and cervical cancers,38-41 and demethylation of FANCF restores the FA/BRCA pathway in ovarian tumors leading to cisplatin resistance.38 In this study, we examined the promoter methylation status of FANCF in both 8226/S and 8226/LR5 cells. Although there is a 2-fold increase of FANCF expression in 8226/LR5 compared with 8226/S, no changes in cytosine-phosphate-guanosine (CpG) methylation were found (data not shown). Mechanisms associated with increased expression of FANCF in 8226/LR5 cells are undergoing further study.
We have shown that silencing FANCF in 8226/LR5 and U266/LR6 cells partially reversed drug resistance. We also showed that overexpression of FANCF in drug-sensitive cells could enhance drug resistance. It has been reported that FANCF plays an important role in stabilizing subunits of the FA complex and contributes to the proper function of this pathway.42 It has also been shown that the cellular level of FANCA, FANCC, and FANCG depends on the expression of FANCF.43 Overexpression of FANCF in parental 8226 and U266 cell lines induced less resistance compared with acquired melphalan-resistant cell lines, suggesting that other members of the FANC/BRCA complex contribute to melphalan resistance. Taniguchi et al reported that forced expression of FANCF in cisplatin-sensitive ovarian parental cells conferred a relative cisplatin resistance, however the resistance is less than the acquired cisplatin-resistant cells.38 Their data also show that overexpression of FANCF in acquired cisplatin-resistant cells increased resistance to cisplatin.38 They suggested that acquired cisplatin-resistant cells might have additional mechanisms involved in cisplatin resistance beyond the re-expression of FANCF. For example, overexpression of FANCC has been reported to inhibit FAS-induced apoptosis.44 This suggests that FA proteins might have other functions beyond assembly of the complex and participate in the drug-resistant phenotype. Taken together, we suggest that repeated exposure to DNA cross-linking agents increases expression of multiple components of the FA pathway, which contribute to the acquired drug resistance. Further investigations will analyze the role of individual FA proteins to the overall melphalan-resistance phenotype in myeloma cells.
Studying cell-cycle kinetics to diagnose Fanconi anemia patients has been reported.45 Exposing peripheral blood mononuclear cells ex vivo to DNA cross-linker treatment induces a significant G2 block in patients with FA.45 In our study, we observed that 10-μM melphalan treatment induces a cell-cycle progression delay in S phase within 24 hours and a G2 block within 48 hours. It has been shown that fibroblasts from FA patients require 3 times longer to recover from the ICL-induced cell-cycle arrest compared with fibroblasts from healthy subjects.33 In 8226/S cells, removal of melphalan-induced ICL is required for cells to progress from G2 block.35 We observed a higher degree of G2/M blockade in drug-sensitive 8226/S cells compared with 8226/LR5 cells following melphalan treatment. We also observed that 8226/LR5 cells require less time for cell-cycle progression from G2/M to the G1 phase compared with 8226/S cells; this is related to increased expression of the FA/BRCA pathway genes and successful removal of ICLs in 8226/LR5 cells compared with 8226/S cells. Overall, our data demonstrate that the FA/BRCA pathway contributes to melphalan resistance via enhanced ICL repair and reduced ICL-induced growth arrest via release of cells from G2/M block.
In summary, to our knowledge, our study is the first to show that enhanced expression of the FA/BRCA pathway is involved in melphalan-induced DNA ICL repair and is an important mechanism for acquired melphalan resistance in myeloma cells. This pathway also likely contributes to cross-resistance to other DNA cross-linking agents and irradiation. We propose that the FA/BRCA pathway represents a new target for preventing acquired drug resistance and improving cancer treatment.
Prepublished online as Blood First Edition Paper, March 31, 2005; DOI 10.1182/blood-2004-11-4286.
Supported by Multiple Myeloma Research Foundation (Q.C.), the Peninsula Myeloma Research Foundation (W.S.D.), and National Institutes of Health grant CA76292 (W.S.D.). Supported in part by the Microscopy Core, Molecular Biology Core, Flow Cytometry Core, Microarray Core, and Molecular Imaging Core Facilities at the H. Lee Moffitt Cancer Center & Research Institute.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Dr Grover C. Bagby (OSHU Cancer Institute, Portland, OR) and Dr Zhi-Wei Li (H. Lee Moffitt Cancer Center, Tampa, FL) for helpful comments.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal