Abstract
The chromosomal translocation t(4;11) marks infant acute lymphoblastic leukemia associated with a particularly dismal prognosis. The leukemogenic role of the corresponding fusion gene MLL-AF4 is not well understood. We show that transient inhibition of MLL-AF4 expression with small interfering RNAs impairs the proliferation and clonogenicity of the t(4; 11)–positive human leukemic cell lines SEM and RS4;11. Reduction of mixed-lineage leukemia (MLL)–ALL-1 fused gene from chromosome 4 (AF4) levels induces apoptosis associated with caspase-3 activation and diminished BCL-XL expression. Suppression of MLL-AF4 is paralleled by a decreased expression of the homeotic genes HOXA7, HOXA9, and MEIS1. MLL-AF4 depletion inhibits expression of the stem-cell marker CD133, indicating hematopoietic differentiation. Transfection of leukemic cells with MLL-AF4 siRNAs reduces leukemia-associated morbidity and mortality in SCID mice that received a xenotransplant, suggesting that MLL-AF4 depletion negatively affects leukemia-initiating cells. Our findings demonstrate that MLL-AF4 is important for leukemic clonogenicity and engraftment of this highly aggressive leukemia. Targeted inhibition of MLL-AF4 fusion gene expression may lead to an effective and highly specific treatment of this therapy-resistant leukemia.
Introduction
Chromosomal aberrations giving rise to fusion genes are observed for many different leukemias.1 Such tumor-specific oncogenes would be promising targets for new therapeutic approaches with improved specificity if these oncogenes were important for maintaining the leukemic phenotype. However, in contrast to the development of leukemia, a central role for leukemic persistence has only been established for a minority of fusion genes.
The mixed-lineage leukemia (MLL) gene located on chromosome 11 band q23 is involved in numerous chromosomal aberrations associated with human leukemia.2 The most prevalent among those is the translocation t(4;11)(q21;q23), which fuses MLL with the AF4 gene located on chromosome 4 band q21.3-5 This translocation is the hallmark of a high-risk acute lymphoblastic leukemia (ALL) with a particularly poor prognosis in infants.6
The wild-type MLL gene is a member of the trithorax family and encodes for a 430-kDa protein, which is proteolytically processed into 2 fragments of 300 and 180 kDa heterodimerizing with each other.7-10 The MLL protein has a complex structure that includes an AT hook domain for A-T base-pair–rich DNA binding, a metallothionein domain showing homology to DNA methyltransferase, and methyl-binding domain protein 1 (MBD1), a plant homeodomain (PHD) containing zinc fingers and a Su(var)3-9, enhancer of zeste, trithorax (SET) histone methyl transferase domain.2 MLL and at least some of its leukemic derivatives are involved in mechanisms controlling HOX gene transcription.11-14 Moreover, the HOX genes HOXA7 and HOXA9 in combination with the homeotic gene MEIS1 are necessary for the transformation induced by several different MLL fusion genes.15-17 Such a crucial role has not yet been reported for MLL-AF4. Nevertheless, expression levels of several HOX genes and of MEIS1 are raised in both primary t(4;11) ALL and t(4;11) leukemic cell lines.18-21
The AF4 gene encodes a serine/proline-rich protein containing a nuclear localization signal and a guanosine triphosphate (GTP)–binding domain. It localizes to the nucleus22 and is probably involved in the control of gene transcription. Whereas homozygous inactivation of MLL is embryonally lethal,23 AF4-deficient mice exhibit imperfect T-cell development and modest alterations in B-cell development.24
The t(4;11) translocation generates 2 fusion genes, AF4MLL and MLL-AF4. The significance of either fusion gene for leukemogenesis is currently not completely understood. AF4MLL has recently been shown to interfere with ubiquitin-mediated ALL-1 fused gene from chromosome 4 (AF4) degradation and to transform murine embryonic fibroblasts.25 Ectopic expression of MLL-AF4 in t(4;11)–negative leukemic cell lines, however, inhibits cell-cycle progression and triggers apoptosis.26 Paradoxically, 20% of all t(4;11) Patients with ALL lack AF4MLL on either the transcriptional or genomic level, whereas MLL-AF4 is always detectable despite its proapoptotic activities observed upon ectopic expression.27,28 Interestingly, several studies suggest that MLL-AF4 supports cell survival in the t(4;11) context. Cells with t(4;11) translocation survive extended serum starvation29 and are resistant to CD95-mediated apoptosis.30
To define the role of this fusion oncogene in leukemogenesis more precisely we applied RNA interference (RNAi) to inhibit MLL-AF4 expression in leukemic cells. RNAi is a cellular process leading to the enzymatic cleavage and breakdown of mRNA.31 Cell transfection with double-stranded small interfering RNAs (siRNAs) results in the generation of a cytoplasmically located ribonucleoprotein complex called RNA-induced silencing complex (RISC). Upon activation of this complex by discarding one of the siRNA strands,32,33 the remaining strand guides RISC to complementary RNA sequences, leading to the endonucleolytic cleavage of the target RNA by the RISC component Ago-2.34-36 Exogenously added synthetic siRNAs were shown to act as very potent and sequence-specific agents to silence gene expression,37 demonstrating their great potential not only for the analysis of gene function but also for gene-specific therapeutic approaches.38,39
We used RNAi to specifically inhibit MLL-AF4 expression in t(4;11) cells. We demonstrate that depletion of the MLL-AF4 fusion transcript inhibits clonogenicity and proliferation, induces apoptosis in t(4;11)-positive leukemic cells, and compromises their engraftment in a severe combined immunodeficiency (SCID) mouse xenotransplantation model.
Materials and methods
Cell culture
The human leukemia cell lines SEM,40 RS4;1141 (obtained from the DSMZ, Braunschweig, Germany), and MV4;1142 (obtained from J. Krauter, Medical School Hannover, Germany) carry the chromosomal translocation t(4;11)(q21;q23) but express different MLL-AF4 variants due to different break points. Further leukemic cell lines used in this study were HL60,43 K562,44 Kasumi-1,45 SKNO-1,46 and U937.47 SKNO-1 cells were maintained in RPMI 1640 Glutamax medium (Invitrogen, Karlsruhe, Germany) supplemented with 20% fetal calf serum (FCS; PAN Biotech, Aidenbach, Germany) and 7 ng/mL granulocyte-macrophage colony-stimulating factor (GM-CSF). All other lines were cultivated in RPMI 1640 Glutamax medium supplemented with 10% FCS at 37°C and 5% CO2. Highly purified CD34+-selected cells from healthy donors, which had been sampled for reasons of quality control and stored in liquid nitrogen for several years, were thawed and used according to institutional guidelines after confirmation that these cells were no longer useful for the recipient.
siRNA treatment
Synthetic sense and antisense oligoribonucleotides were synthesized by Alnylam Europe AG (Kulmbach, Germany). MLL-AF4 siRNAs used in this study were siMA6 (sense, 5′-AAGAAAAGCAGACCUACUCCA-3′; antisense, 5′-UGGAGUAGGUCUGCUUUUCUUUU-3′), targeting the MLL exon 9–AF4 exon 4 (e9-e4) MLL-AF4 fusion site present in SEM cells, and siMARS (sense, 5′-ACUUUAAGCAGACCUACUCCA-3′; antisense, 5′-UGGAGUAGGUCUGCUUAAAGUCC-3′), homologous to the exon 10–exon 4 (e10-e4) fusion site variant present in RS4;11 cells. As control siRNAs we used the mismatch control siMM (sense, 5′-AAAAGCUGACCUUCUCCAAUG-3′; antisense, 5′-CAUUGGAGAAGGUCAGCUUUUCU-3′), the acute myeloid leukemia 1/myeloid translocation gene on 8q22 (AML1/MTG8) siRNA siAGF1, and its mismatch control siAGF6.48 siRNA preparations and electroporations were carried out as described previously.48-52 The procedure yields siRNA transfection efficiencies close to 100% for a variety of leukemic cell lines including SEM cells.48-52
Real-time RT-PCR
Total RNA extraction was performed with the RNeasy Kit (Qiagen, Hilden, Germany) as suggested by the manufacturer. Real-time reverse transcriptase–polymerase chain reactions (RT-PCRs) were performed as described.51 The primers for MLL-AF4 (sense, 5′-ACAGAAAAAAGTGGCTCCCCG-3′; antisense, 5′-TATTGCTGTCAAAGGAGGCGG-3′), MLL (sense, 5′-ACAGAAAAAAGTGGCTCCCCG-3′; antisense, 5′-GCAAACCACCCTGGGTGTTA-3′), AF4 (sense, 5′-CAGAAGCCCACGGCTTATGT-3′; antisense, 5′-TATTGCTGTCAAAGGAGGCGG-3′), HOXA7 (sense, 5′-CGCCAGACCTACACGCG-3′; antisense, 5′-CAGGTAGCGGTTGAAGTGGAA-3′), HOXA9 (sense, 5′-CCACCATCCCCGCACA-3′; antisense, 5′-AACAGGGTTTGCCTTGGAAA-3′), MEIS1 (sense, 5′-GCATGCAGCCAGGTCCAT-3′; antisense, 5′-TAAAGCGTCATTGACCGAG-3′), OAS1 (sense, 5′-TCCAAGGTGGTAAAGGGTGG-3′; antisense, 5′-AGGTCAGCGTCAGATCGGC-3′), CD133 (sense, 5′-ATGGCAACAGCGATCAAGG-3′; antisense, 5′-GTACTTTGTTGGTGCAAGCTCT-3′), GAPDH,51 and STAT151 were designed with PRIMER-EXPRESS software (Applied Biosystems, Foster City, CA). If not otherwise indicated, real-time RT-PCR data shown include at least 3 independent experiments with 3 replicates per experiment.
Colony-formation assay
Twenty-four hours after cell electroporation with siRNAs, 10 000 cells were plated in 0.5 mL of RPMI 1640 medium containing 20% FCS and 0.56% methylcellulose in 24-well plates. In the case of RS4;11, cell numbers were increased to 12 000 per well. Colonies consisting of more than 20 cells were counted 14 days after plating. Under these conditions, mock-transfected cells (electroporated without siRNAs) yielded 50 to 100 colonies per well dependent on the cell line examined. Human colony-forming cell assays were performed using MethoCult Methylcellulose-based media (CellSystems, St Katharinen, Germany). After electroporation, 5000 human primary CD34+ cells were plated in duplicate in 35-mm culture dishes with 1 mL of methylcellulose medium. Granulocyte-erythrocyte-megakaryocyte-macrophage colony-forming units (CFU-GEMMs) and granulocyte macrophage colony-forming units (CFU-GMs) were counted 10 days after plating.
MTT test
Cells were electroporated twice within 48 hours and were plated on 96-well plates at a density of 5 × 104 cells in 100 μL/well. Every 24 hours later, 10 μL of MTT (3-(4,5-cimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide) solution (Roche, Mannheim, Germany) was added. After incubation for 4 hours at 37°C, cells were lysed with the solubilization solution according to the manufacturer's instruction. Optic densities were determined at 560 nm and 650 nm as a reference wavelength. Cell numbers were calculated by cell-dilution series.
Cell-cycle analysis, apoptosis assay, and fluorescence-activated cell sorter (FACS) analysis
Cell-cycle analysis was performed as described previously.51 The obtained data were subsequently analyzed and evaluated using ModFit (Verity, Topsham, ME). Apoptosis was examined with a human annexin V/fluorescein isothiocyanate (FITC) kit (Bender MedSystems, Wien, Austria) according to the provider's instructions. Briefly, 2 × 105 to 5 × 105 cells were washed with phosphate-buffered saline (PBS) at the indicated time points after electroporation followed by incubation in the presence of annexin V/FITC solution for 10 minutes at room temperature. The cells were washed again with PBS and stained with propidium iodide. The samples were then immediately analyzed by flow cytometry using a FACSCalibur (Becton Dickinson, Heidelberg, Germany). CD133 surface expression was monitored by staining with a phycoerythrin-conjugated CD133 antibody (Miltenyi Biotec, Bergisch Gladbach, Germany) followed by flow cytometry analysis.
Western blotting
To obtain total cellular protein, proteins present in the flow-through of RNeasy columns were precipitated with 2 volumes of acetone and dissolved in urea buffer (9 M urea, 4% [w/w] 3-[(3-Cholamidopropyl)-dimethylammonio]-propansulfonat [CHAPS], 1% [w/w] dithiothreitol). Total lysates (50 μg for MLL-AF4 detection, 10 μg for all other immunoblots) were analyzed as described.51 The following antibodies were used for immunoblot detection: cleaved caspase-3 (Asp175; 1:1000, no. 9661; Cell Signaling Technology, Beverly, MA); tubulin Ab-4 (1 mg/L, MS-719-P0; NeoMarkers, Fremont, CA); BCL2-related protein, long isoform (BCL-XL) (1:500, no. 556499; BD PharMingen, Heidelberg, Germany); MLLT2 (1:600, no. 10852; Orbigen, San Diego, CA); glyceraldehyde phosphate dehydrogenase (GAPDH; 1:20 000, no. 5G4; HyTest, Turku, Finland).
Xenotransplantation of SCID mice
Female 4- to 5-week-old CB17/Icr-Prkdc scid/Crl mice were obtained from Charles River Germany (Sulzfeld, Germany). SEM cells (2 × 107) were electroporated on day 1 and day 3 either without (Mock) or with 500 nM of the indicated siRNA. On day 4, cells were counted and 2 × 107 cells were intraperitoneally injected into mice. Animals were maintained and treated according to protocols approved by the Regional Board Tübingen.
Histology
Organs were removed and fixed in neutrally buffered 4% formalin at room temperature for 4 to 5 days followed by dehydration, embedding into paraffin, and sectioning. The tissues were stained with hematoxylin (Mayer hemalum solution; Merck, Darmstadt, Germany) and eosin (Eosin Y; Merck) for light microscopy. Light microscopy was performed with a Zeiss Axioplan microscope (Zeiss, Guttingen, Germany) equipped with Plan-Neofluar 20×/1.3 or 40×/1.3 oil-immersion objective lenses. Images were captured using an AxioCam HRc camera (Zeiss), and were analyzed with Axio Vision 4 software provided with the microscope and Adobe Photoshop 6.0 (Adobe Systems, San Jose, CA).
Statistical analyses
Colony-formation assays were analyzed by Student t test assuming unequal variances and 2-tailed distributions. Survival curves were analyzed by log-rank test. Error bars indicate standard deviation.
Results
Efficiency and specificity of MLL-AF4 siRNAs
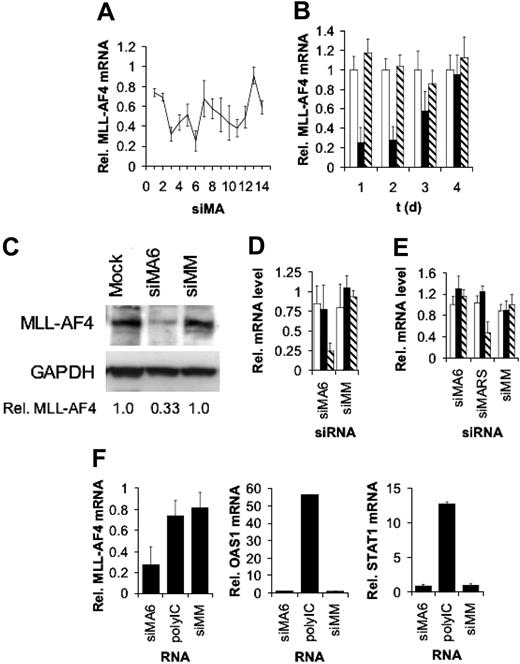
To identify highly efficient MLL-AF4 siRNAs we performed an siRNA scan of the MLL-AF4 fusion site. We synthesized 14 different siRNAs with target sites moved by one single nucleotide each. The efficiencies of the different siRNAs were examined in the t(4;11)-positive leukemic cell line SEM established from a 5-year-old patient with ALL in relapse.40 Of all 14 siRNAs examined, 2 siRNAs, siMA3 and siMA6, diminished MLL-AF4 mRNA levels by more than 60% (Figure 1A). The reduction of MLL-AF4 transcript levels was dose dependent and reached its maximum of 70% with 750 nM siRNA (data not shown). Time course experiments showed that MLL-AF4 mRNA amounts reached their minimum between 24 and 48 hours after siRNA transfection and recovered to normal levels at day 4 (Figure 1B). The decrease in MLL-AF4 mRNA levels was reflected by a concomitant 67% decrease in MLL-AF4 protein, suggesting a half-life shorter than 48 hours (Figure 1C). Moreover, siMA6 affected neither wild-type AF4 nor MLL mRNA levels (Figure 1D), whereas siMA3 substantially reduced AF4 levels (data not shown). The mismatch control siRNA siMM influenced neither MLL-AF4 nor the corresponding wild-type allele transcripts.
The MLL-AF4 fusion site varies between different t(4;11)–positive cell lines. Whereas SEM cells express a transcript containing an e9-e4 fusion, RS4;11 cells express an e10-e4 variant. In spite of a homology of 67%, siMA6 did not diminish levels of the e10-e4 isoform in RS4;11 cells,5 whereas a perfectly homologous siRNA, siMARS, reduced the MLL-AF4 e10-e4 variant in RS4;11 by 60%, without affecting AF4 or MLL expression (Figure 1E).
Activity and specificity of MLL-AF4 siRNAs. (A) SiRNA scan of the MLL-AF4 mRNA fusion site. MLL-AF4 mRNA levels normalized by GAPDH mRNA levels are shown. Target sites of the indicated siRNAs were moved by one single nucleotide from the AF4 part to the MLL part of the fusion site. Total RNA was isolated 24 hours after electroporation with 500 nM of siRNA and analyzed by real time RT-PCR. One of 2 experiments yielding similar results is shown. (B) Time course of MLL-AF4 depletion. Total RNA was isolated at the indicated time points after electroporation with 750 nM siRNA. Real time RT-PCR was performed as in panel A. □ indicates Mock; ▪, siMA6; ▧, siMM. (C) Depletion of MLL-AF4 protein upon siRNA transfection. Total cell lysates were isolated 48 hours after electroporation with 500 nM siRNA. MLL-AF4 was detected with an antibody targeting the C-terminus of AF4. GAPDH served as a loading control and for normalization. Normalized MLL-AF4 protein levels are indicated at the bottom. (D) Effects of the MLL-AF4 siRNA siMA6 and a mismatch control siMM on MLL-AF4, AF4, and MLL mRNA levels in SEM cells. Analysis was performed as in panel A. □ indicates MLL; ▪, AF4; ▧, MLL-AF4. (E) Effects of the MLL-AF4 siRNAs siMA6 and siMARS and a mismatch control siMM on MLL-AF4, AF4, and MLL mRNA levels in RS4;11 cells. Analysis was performed as in panel A. siMA6 is homologous to the MLL-AF4 variant expressed in SEM cells; siMARS targets the variant present in RS4;11 cells. □ indicates MLL; ▪, AF4; ▧, MLL-AF4. (F) MLL-AF4 siRNAs do not induce an interferon response. SEM cells were transfected with the indicated RNAs. PolyIC (7.5 μg/mL) served as a positive control for the induction of the interferon response genes OAS1 and STAT1. Analysis was performed as in panel A.
Activity and specificity of MLL-AF4 siRNAs. (A) SiRNA scan of the MLL-AF4 mRNA fusion site. MLL-AF4 mRNA levels normalized by GAPDH mRNA levels are shown. Target sites of the indicated siRNAs were moved by one single nucleotide from the AF4 part to the MLL part of the fusion site. Total RNA was isolated 24 hours after electroporation with 500 nM of siRNA and analyzed by real time RT-PCR. One of 2 experiments yielding similar results is shown. (B) Time course of MLL-AF4 depletion. Total RNA was isolated at the indicated time points after electroporation with 750 nM siRNA. Real time RT-PCR was performed as in panel A. □ indicates Mock; ▪, siMA6; ▧, siMM. (C) Depletion of MLL-AF4 protein upon siRNA transfection. Total cell lysates were isolated 48 hours after electroporation with 500 nM siRNA. MLL-AF4 was detected with an antibody targeting the C-terminus of AF4. GAPDH served as a loading control and for normalization. Normalized MLL-AF4 protein levels are indicated at the bottom. (D) Effects of the MLL-AF4 siRNA siMA6 and a mismatch control siMM on MLL-AF4, AF4, and MLL mRNA levels in SEM cells. Analysis was performed as in panel A. □ indicates MLL; ▪, AF4; ▧, MLL-AF4. (E) Effects of the MLL-AF4 siRNAs siMA6 and siMARS and a mismatch control siMM on MLL-AF4, AF4, and MLL mRNA levels in RS4;11 cells. Analysis was performed as in panel A. siMA6 is homologous to the MLL-AF4 variant expressed in SEM cells; siMARS targets the variant present in RS4;11 cells. □ indicates MLL; ▪, AF4; ▧, MLL-AF4. (F) MLL-AF4 siRNAs do not induce an interferon response. SEM cells were transfected with the indicated RNAs. PolyIC (7.5 μg/mL) served as a positive control for the induction of the interferon response genes OAS1 and STAT1. Analysis was performed as in panel A.
Neither siMA6 nor siMM induced STAT1 or 2′-5′-oligoadenylate synthase 1 (OAS1) expression (Figure 1F), indicating that these siRNAs did not trigger an interferon response.53 Transfection with polyIC, a strong inductor of interferon response, increased OAS1 transcript levels more than 50-fold and STAT1 mRNA levels more than 10-fold (Figure 1F), demonstrating that interferon response pathways can be induced in these leukemic cells. Because of their high specificity, the MLL-AF4 siRNA siMA6 and the mismatch control siRNA siMM were chosen to prove the significance of MLL-AF4 expression for the leukemic phenotype.
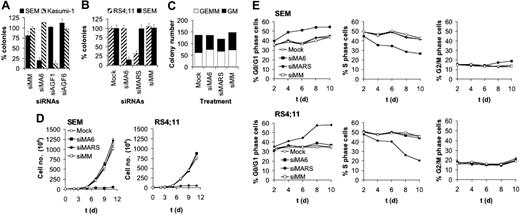
MLL-AF4 depletion inhibits colony formation and proliferation of t(4;11)-positive leukemic cells. (A) Specificity of MLL-AF4 and AML1/MTG8 siRNAs. SEM cells express MLL-AF4, whereas Kasumi-1 cells express AML1/MTG8. Colony numbers of siMA6-treated SEM cells are significantly lower than SEM controls (P < .001). (B) Inhibition of SEM and RS4;11 clonogenicity is dependent on perfect homology to the MLL-AF4 fusion site. Colony numbers of siMA6-treated SEM cells and siMARS-treated RS4;11 cells are significantly lower than the corresponding controls (P < .001). (C) MLL-AF4 siRNAs do not affect colony formation of primary human CD34+ hematopoietic cells. In all figure parts, colony formation is shown after electroporation with 750 nM siRNA. siMA6 indicates MLL-AF4 siRNA targeting the e9-e4 variant expressed in SEM; siMARS, MLL-AF4 siRNA targeting the e10-e4 variant expressed in RS4;11; siAGF1, AML1/MTG8 siRNA; siMM, siAGF6, mismatch control siRNAs. (D) Growth curves of siRNA-treated t(4;11) cell lines. Cells were electroporated every second day with 750 nM siRNA. Cell numbers were determined by MTT assays. (E) Effects of MLL-AF4 siRNAs on the cell-cycle distribution of SEM and RS4;11 cells. The graphs show the percentage of cells in the indicated cycle phase. Cell cycle distribution was determined by flow cytometry at the indicated days using cells from the time course experiments shown in panel D.
MLL-AF4 depletion inhibits colony formation and proliferation of t(4;11)-positive leukemic cells. (A) Specificity of MLL-AF4 and AML1/MTG8 siRNAs. SEM cells express MLL-AF4, whereas Kasumi-1 cells express AML1/MTG8. Colony numbers of siMA6-treated SEM cells are significantly lower than SEM controls (P < .001). (B) Inhibition of SEM and RS4;11 clonogenicity is dependent on perfect homology to the MLL-AF4 fusion site. Colony numbers of siMA6-treated SEM cells and siMARS-treated RS4;11 cells are significantly lower than the corresponding controls (P < .001). (C) MLL-AF4 siRNAs do not affect colony formation of primary human CD34+ hematopoietic cells. In all figure parts, colony formation is shown after electroporation with 750 nM siRNA. siMA6 indicates MLL-AF4 siRNA targeting the e9-e4 variant expressed in SEM; siMARS, MLL-AF4 siRNA targeting the e10-e4 variant expressed in RS4;11; siAGF1, AML1/MTG8 siRNA; siMM, siAGF6, mismatch control siRNAs. (D) Growth curves of siRNA-treated t(4;11) cell lines. Cells were electroporated every second day with 750 nM siRNA. Cell numbers were determined by MTT assays. (E) Effects of MLL-AF4 siRNAs on the cell-cycle distribution of SEM and RS4;11 cells. The graphs show the percentage of cells in the indicated cycle phase. Cell cycle distribution was determined by flow cytometry at the indicated days using cells from the time course experiments shown in panel D.
MLL-AF4 affects leukemic clonogenicity
To study the relevance of MLL-AF4 for leukemic clonogenicity, we transfected t(4;11)-positive SEM cells with siRNAs followed by incubation in semisolid medium. siMA6-mediated depletion of MLL-AF4 reduced the number of colonies 5-fold (Figure 2A). This effect was specific, since colony formation of the t(8;21)–positive leukemic cell line Kasumi-1 was not affected by siMA6. Vice versa, transfection with the AML1/MTG8-specific siRNA siAGF1 compromised Kasumi-1 colony formation without interfering with SEM colony formation.51 None of the mismatch controls (siMM and siAGF6) affected leukemic clonogenicity. Furthermore, neither the t(4;11)–negative leukemic cell lines (HL60, K562, SKNO-1, and U937) nor the t(4;11)-positive cell lines (RS4;11 and MV4;11) expressing MLL-AF4 variants not affected by siMA6 showed impaired colony formation upon siRNA transfection (Figure 2B; data not shown). RS4;11 clonogenicity was more than 2-fold reduced upon siMARS-mediated suppression of the MLL-AF4 e10-e4 variant, thereby demonstrating for another t(4;11) cell line the dependence of clonogenic efficacy on MLL-AF4 (Figure 2B). MLL-AF4 siRNA electroporation of primary human hematopoietic CD34+ cells did not affect the number of GEMM or GM colonies (Figure 2C). This lack of effect cannot be attributed to inefficient siRNA transfections, since both of the cell lines used in this study and the human hematopoietic CD34+ cells can be efficiently transfected with functional siRNAs48,50,52 (Daniela Werth and O.H., unpublished data, 2003).
Suppression of MLL-AF4 inhibits leukemic proliferation and cell-cycle progression
Next, we examined the role of MLL-AF4 in leukemic proliferation in suspension culture. Whereas a single electroporation with siMA6 did not affect the doubling time of t(4;11)-positive SEM cells (data not shown), repeating siRNA electroporation for every second day resulted in a sustained inhibition of proliferation of SEM cells by siMA6 and of RS4;11 cells by siMARS (Figure 2D). Thus, proliferation was only inhibited by the siRNA homologous to the corresponding MLL-AF4 fusion site, demonstrating the specificity of these MLL-AF4 siRNAs. Since MLL-AF4 protein levels decreased within 48 hours of a single siRNA treatment (Figure 1C), the necessity of repeated siRNA electroporations is unlikely to be caused by a long MLL-AF4 half-life. Instead, an extended MLL-AF4 knockdown might be required to down-modulate proliferation-supportive signals provided by, for instance, cell-cell contacts or secreted growth factors. Mock- or control siRNA–electroporated SEM or RS4;11 cells had doubling times of 1.4 days, demonstrating that the repeated electroporation did not seriously affect their proliferation.
The reduced proliferation of t(4;11)-positive cells upon MLL-AF4 depletion was paralleled by changes in the cell-cycle distribution. During a time course of 10 days with repetitive MLL-AF4 siRNA electroporation, the fraction of S-phase cells decreased in both SEM and RS4;11 cells from 50% to 30% and 20%, respectively, with a concomitant increase in the fraction of G0/G1-phase cells (Figure 2E). Notably, siMA6 affected cell-cycle distribution only in SEM cells, whereas siMARS caused those changes only in RS4;11 cells. Thus, depletion of MLL-AF4 negatively interferes with the progression of t(4;11)-positive cells from G1 to S phase. The impaired G1/S transition is not associated with cellular senescence, as senescence-associated β-galactosidase activity did not increase upon MLL-AF4 depletion (data not shown).
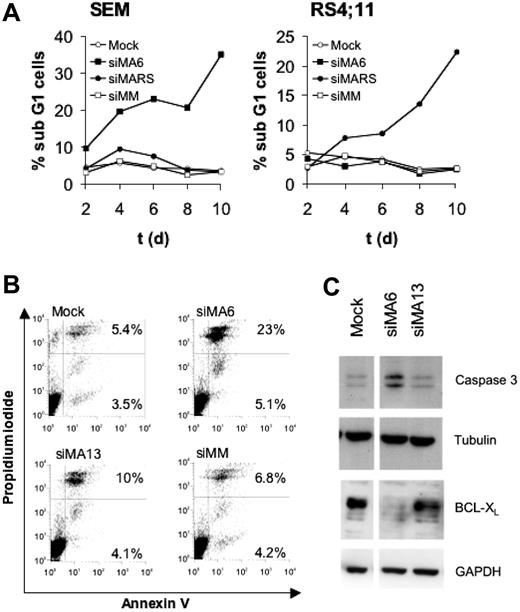
MLL-AF4 depletion induces apoptosis in t(4;11)-positive SEM cells
Cell-cycle analysis of SEM and RS4;11 cells revealed that the continuous depletion of MLL-AF4 for 10 days raised the number of sub-G1 cells 10-fold compared with controls, indicating an increased amount of apoptotic cells (Figure 3A). Staining with annexin V and propidium iodide also demonstrated for SEM cells a 3-fold increase in apoptotic cells upon suppression of MLL-AF4 (Figure 3B). The almost inactive siRNA siMA13 (Figure 1A) only marginally affected the amount of apoptotic cells, suggesting a direct correlation between the extent of MLL-AF4 depletion and the rate of apoptosis. Proteolytic activation of caspase-3 and decreased amounts of the antiapoptotic protein BCL-XL accompanied the siMA6-mediated induction of apoptosis (Figure 3C).
MLL-AF4 depletion induces apoptosis in t(4;11) cells. (A) Effects of MLL-AF4 suppression on the fraction of sub-G1 cells. Cells obtained from the time courses shown in Figure 2D and E were analyzed for DNA content by flow cytometry. (B) Annexin V staining of SEM cells. Annexin V–positive SEM cells were quantified by flow cytometry 4 days after the second electroporation with 750 nM of the indicated siRNA. The percentages of annexin V and annexin V/propidium iodide–positive cells are given in the corresponding quadrants. (C) MLL-AF4 suppression triggers caspase-3 activation and diminishes BCL-XL protein levels. Immunoblots show BCL-XL and proteolytically activated caspase-3. Tubulin and GAPDH served as loading controls.
MLL-AF4 depletion induces apoptosis in t(4;11) cells. (A) Effects of MLL-AF4 suppression on the fraction of sub-G1 cells. Cells obtained from the time courses shown in Figure 2D and E were analyzed for DNA content by flow cytometry. (B) Annexin V staining of SEM cells. Annexin V–positive SEM cells were quantified by flow cytometry 4 days after the second electroporation with 750 nM of the indicated siRNA. The percentages of annexin V and annexin V/propidium iodide–positive cells are given in the corresponding quadrants. (C) MLL-AF4 suppression triggers caspase-3 activation and diminishes BCL-XL protein levels. Immunoblots show BCL-XL and proteolytically activated caspase-3. Tubulin and GAPDH served as loading controls.
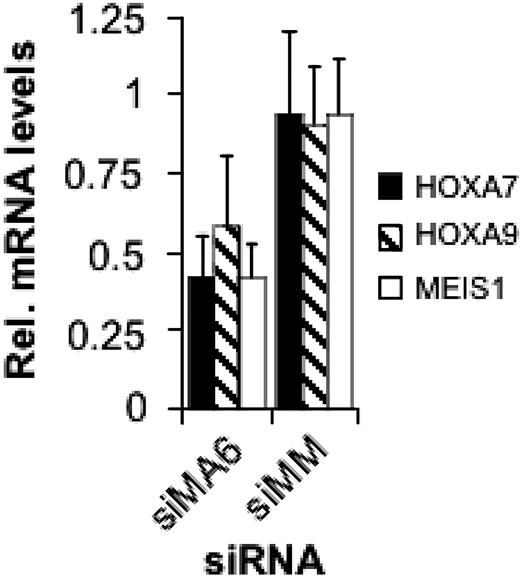
MLL-AF4 suppression decreases expression of HOX genes
Expression of MLL oncoproteins including MLL-AF4 is associated with increased expression of several HOX genes including HOXA9 and of the homeotic gene MEIS1.15,18,19,21 In ALL lines containing rearranged MLL genes, HOXA7, HOXA9, and MEIS1 levels are higher compared with ALL lines with wild-type MLL.20 Therefore, we examined the expression of these 3 homeotic genes in dependence on the MLL-AF4 level. After 2 consecutive transfections of SEM cells with the MLL-AF4 siRNA siMA6, HOXA7 and MEIS1 mRNA levels decreased by 60% and HOXA9 levels by 40% (Figure 4). Thus, MLL-AF4 causes an increased expression of these homeotic genes.
MLL-AF4 depletion affects CD133 expression and myeloid differentiation
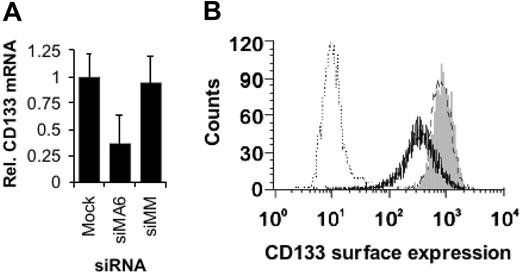
MLL fusion genes such as MLLENL have been shown to interfere with hematopoietic differentiation in a Hoxa9- and Meis1-dependent fashion.17 Furthermore, absence of HOXA7 and HOXA9 expression results in B-cell development even in the presence of MLL-AF4.54 Therefore, the decreased HOXA7, HOXA9, and MEIS1 expression upon MLL-AF4 suppression might result in an, at least partially, reactivated hematopoietic differentiation. Expression of CD133 (prominin), a marker for hematopoietic stem and early progenitor cells, correlates with mixed-lineage leukemia.18 Moreover, CD133 expression is controlled by the methylation status of a cytosine-phosphate-guanosine (CpG) island,55 raising the possibility that CD133 is a direct target gene of MLL-AF4. We analyzed the consequences of MLL-AF4 depletion for the expression of CD133 in SEM cells. Inhibition of MLL-AF4 expression resulted in 2-fold reduced CD133 mRNA levels and in a more than 2-fold reduced surface expression of CD133 (Figure 5A-B), which may indicate the onset of hematopoietic differentiation.
MLL-AF4 suppression inhibits HOXA7, HOXA9, and MEIS1 gene expression. Total RNA was isolated 48 hours after the second electroporation with 500 nM of the indicated siRNA and analyzed by real-time RT-PCR.
MLL-AF4 suppression inhibits HOXA7, HOXA9, and MEIS1 gene expression. Total RNA was isolated 48 hours after the second electroporation with 500 nM of the indicated siRNA and analyzed by real-time RT-PCR.
MLL-AF4 is important for the leukemic engraftment of t(4;11)-positive cells
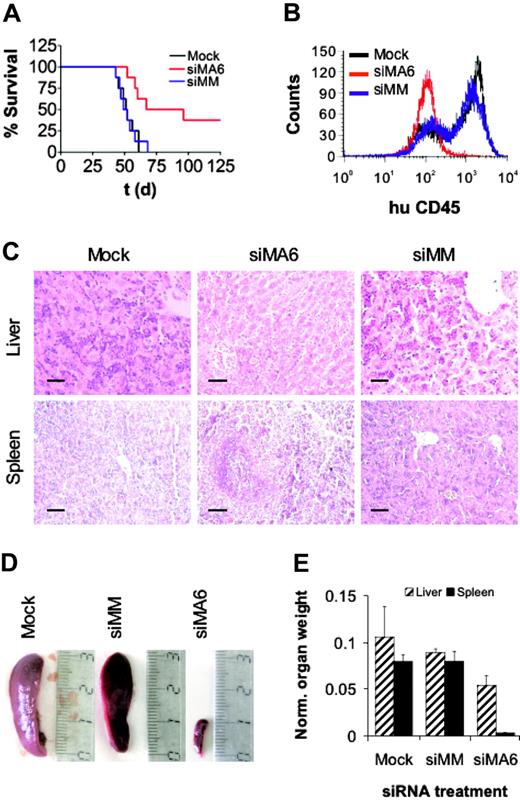
Leukemic cell growth in SCID mice has been shown to be associated with high-risk B-ALL.56 Therefore, we used a t(4;11)–SCID mouse model to ask whether siRNA-mediated depletion of MLL-AF4 affects leukemic engraftment and the development of leukemia in vivo.57 Intraperitoneal transplantation of either mock- or control siRNA–treated SEM cells into SCID mice resulted in a 100% leukemia-associated mortality within 70 days after transplantation with a median survival of 52 days (Figure 6A). Xenotransplantation of MLL-AF4-depleted SEM cells yielded a median survival of 82 days and an overall survival of 38% at day 125 (P < .01). Animals succumbing to the disease showed ovarian tumors; massive leukemic blast infiltration in bone marrow (Figure 6B), spleen, and liver (Figure 6C); and concomitant hepatosplenomegaly (Figure 6D-E), whereas the organs of surviving animals of the siMA6 group showed no signs of leukemic infiltration up to 228 days after transplantation. In conclusion, siRNA-mediated suppression of MLL-AF4 reduced the leukemic engraftment of t(4;11)–positive cells in SCID mice that received a xenotransplant. Since leukemic engraftment crucially depends on leukemia-initiating cells,58 these findings may also indicate a possible function of MLL-AF4 for the persistence of this cell type.
MLL-AF4 depletion facilitates hematopoietic differentiation. (A) Reduction of CD133 mRNA levels upon MLL-AF4 suppression. Total RNAs were isolated 24 hours after the third electroporation of SEM cells with 500 nM of the indicated siRNA and were analyzed by real-time RT-PCR. The columns represent the averages of 2 independent experiments with 3 replicates each. (B) MLL-AF4 siRNAs diminish CD133 surface expression. Three days after the third electroporation with 500 nM of the indicated siRNA, SEM cells were analyzed by flow cytometry. One of 2 experiments yielding similar results is shown. Gray peak indicates mock-transfected cells; broken line, siMM-transfected cells; solid line, siMA6-transfected cells; dotted line, isotype control.
MLL-AF4 depletion facilitates hematopoietic differentiation. (A) Reduction of CD133 mRNA levels upon MLL-AF4 suppression. Total RNAs were isolated 24 hours after the third electroporation of SEM cells with 500 nM of the indicated siRNA and were analyzed by real-time RT-PCR. The columns represent the averages of 2 independent experiments with 3 replicates each. (B) MLL-AF4 siRNAs diminish CD133 surface expression. Three days after the third electroporation with 500 nM of the indicated siRNA, SEM cells were analyzed by flow cytometry. One of 2 experiments yielding similar results is shown. Gray peak indicates mock-transfected cells; broken line, siMM-transfected cells; solid line, siMA6-transfected cells; dotted line, isotype control.
MLL-AF4 suppression diminishes leukemic engraftment. (A) Survival curves of SCID mice that received a transplant of SEM cells. Prior to transplantation, SEM cells were electroporated twice with the indicated siRNAs. Pretreatment with the MLL-AF4 siRNA siMA6 extended median survival and increased overall survival significantly compared with mock or control siRNA siMM pretreatment (P < .01 according to log-rank test). In each treatment arm, 8 mice received a transplant. (B) FACS analysis of bone marrow. Bone marrow cells of animals were stained with α-human CD45 antibody and analyzed by flow cytometry. siRNAs electroporated prior to transplantation are indicated. (C) Liver and spleen histologies. Original magnification × 200; scale bar, 50 μm. Mice that received a transplant of mock or siMM-pretreated cells were moribund at the time of analysis. The animal that received a transplant of siMA6-pretreated cells was killed 228 days after transplantation without any sign of leukemia-associated morbidity. (D) Comparison of spleen size. The siRNAs used for the electroporation are indicated on top. (E) Graphic representation of organ weights. Organ weights were normalized to whole body weight. Normalized liver and spleen weights of surviving animals of the siMA6 group were significantly smaller than those from the mock or siMM group (P < .05 and P < .001, respectively).
MLL-AF4 suppression diminishes leukemic engraftment. (A) Survival curves of SCID mice that received a transplant of SEM cells. Prior to transplantation, SEM cells were electroporated twice with the indicated siRNAs. Pretreatment with the MLL-AF4 siRNA siMA6 extended median survival and increased overall survival significantly compared with mock or control siRNA siMM pretreatment (P < .01 according to log-rank test). In each treatment arm, 8 mice received a transplant. (B) FACS analysis of bone marrow. Bone marrow cells of animals were stained with α-human CD45 antibody and analyzed by flow cytometry. siRNAs electroporated prior to transplantation are indicated. (C) Liver and spleen histologies. Original magnification × 200; scale bar, 50 μm. Mice that received a transplant of mock or siMM-pretreated cells were moribund at the time of analysis. The animal that received a transplant of siMA6-pretreated cells was killed 228 days after transplantation without any sign of leukemia-associated morbidity. (D) Comparison of spleen size. The siRNAs used for the electroporation are indicated on top. (E) Graphic representation of organ weights. Organ weights were normalized to whole body weight. Normalized liver and spleen weights of surviving animals of the siMA6 group were significantly smaller than those from the mock or siMM group (P < .05 and P < .001, respectively).
Discussion
The role of MLL-AF4 in leukemogenesis is not well understood. Dependent on the experimental system used, contradictory results about its leukemic properties were obtained. For instance, in Drosophila MLL-AF4 leads to a retarded cell cycle and larval lethality.59 Ectopic expression of MLL-AF4 in the myelomonocytic leukemia cell line U937 inhibits proliferation and cell-cycle progression and is associated with an increased rate of apoptosis.26 In contrast, t(4;11)-positive ALLs are very resistant against induction of apoptosis.29,30 Additionally, gene expression profiling revealed for t(4;11) ALL blasts increased expression levels of HOX and other homeotic genes including HOXA9 and MEIS1.15,18-21 The t(4;11)-positive leukemic cell lines such as RS4;11, MV4;11, or SEM engraft very efficiently and give rise to a rapid development of aggressive leukemias in murine xenotransplantation models.57,60 Our data suggest that MLL-AF4 is crucially involved in all these processes. Inhibition of MLL-AF4 expression diminished both leukemic proliferation in suspension culture and colony formation of t(4;11) cell lines. This reduced clonogenicity and proliferation was accompanied by an increase in apoptosis. Furthermore, depletion of MLL-AF4 caused a decrease in HOXA7, HOXA9, and MEIS1 expression, which in turn may lead to apoptosis and hematopoietic differentiation.17,61 Finally, siRNA-mediated MLL-AF4 suppression seriously compromised the leukemic engraftment in SCID mice that received a xenotransplant. Since efficient engraftment in SCID mice predicts an increased probability of relapse in patients with ALL,56 these data suggest that interfering with MLL-AF4 functions may improve patient outcome.
There is only one leukemia-originated cell line known to express the CD133 antigen. This cell line was designated MUTZ-2 and has been derived from a patient with AML who had leukemic blasts exhibiting a French-American-British (FAB)–AML-M2 morphology.62 In contrast to this CD34+ cytokine responsive, AML cell line SEM is lacking CD34. Thus, the antigenic profile of SEM is CD133+/CD34- and is even more immature compared with the CD133+/CD34+ myeloid cell line MUTZ-2. Therefore, the diminished expression of the stem-cell marker CD133 upon siRNA treatment suggests that interfering with MLL-AF4 functions may, at least to a limited extent, facilitate hematopoietic differentiation.
In agreement with our data, recent studies showed that peptide-mediated disruption of the interaction between MLL-AF4 and AF9 triggers apoptosis in t(4;11) leukemia cell lines, further underlining the significance of MLL-AF4 for sustaining a leukemic phenotype.63 The discrepancy between ectopic expression and depletion in leukemic cells with endogenous fusion gene expression has also been noted for other fusion proteins such as MLL-AF9 or AML1/MTG8, where ectopic expression has antiproliferative consequences, whereas depletion of endogenously expressed fusion protein suggests proliferation-supporting functions.51,64-66
Because of its exclusive expression in t(4;11) leukemic cells, and because of its central role in the maintenance of leukemia including a supportive function for SCID-leukemia initiating cells, MLL-AF4 would be a very promising target for a molecularly defined treatment of this highly aggressive leukemia. Currently, no small molecule inhibitors are available for this fusion protein. We show that siRNAs homologous to the fusion site efficiently suppress MLL-AF4. Moreover, we demonstrate that 2 different variants of this fusion gene can be targeted with high efficacy and exclusive specificity. This specificity also proves that the observed antileukemic properties of these siRNAs are directly due to MLL-AF4 suppression and not to off-target effects such as unintended inhibition of other genes or induction of interferon response. Furthermore, the successful targeting of 2 different MLL-AF4 variants has implications for the treatment of possible escape mutants. RNAi resistance-conferring point mutations in the fusion site may simply be counteracted with an siRNA containing an adapted sequence. Finally, the observed reduction of leukemic engraftment upon MLL-AF4 depletion suggests that MLL-AF4 siRNAs may also impair leukemia-initiating cell functions. Although the problem of systemic siRNA delivery to hematopoietically relevant organs is not yet solved,67 our results suggest that MLL-AF4 siRNAs may provide a specific, but still flexible, and thus promising therapeutic tool for the treatment of t(4;11) ALL.
Prepublished online as Blood First Edition Paper, July 26, 2005; DOI 10.1182/blood-2005-03-1283.
Supported by a grant from the Deutsche José Carreras Leukämie-Stiftung (DJCLS-R03/10; J.G. and O.H.).
Two of the authors (H.-P.V. and P.H.) are employed by a company whose potential product was studied in the present work.
M.T. designed and performed the experiments, A.G. contributed to the experiments, H.-P.V. and P.H. contributed to the siRNA scans and siRNA design, and J.G. and O.H. conceived the experiments and coordinated the study.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We gratefully acknowledge the superb technical assistance of Heidemarie Riehle. We would like to thank Rolf Marschalek, Robert Slany, Arndt Borkhardt, and Uta Fuchs for their support in immunoblotting MLL-AF4 and Bernd Knöll for carefully reading the manuscript.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal