Gene therapy for hematopoietic diseases has been hampered by the low frequency of transduction of human hematopoietic stem cells (HSCs) with retroviral vectors pseudotyped with amphotropic envelopes. We hypothesized that transduction could be increased by the use of retroviral vectors pseudotyped with envelopes that recognize more abundant cellular receptors. The levels of mRNA encoding the receptors of the feline retroviruses, RD114 and feline leukemia virus type C (FeLV-C), were significantly higher than the level of gibbon ape leukemia virus (GaLV) receptor mRNA in cells enriched for human HSCs (Lin– CD34+ CD38–). We cotransduced human peripheral blood CD34+ cells with equivalent numbers of FeLV-C and GALV or RD114 and GALV-pseudotyped retroviruses for injection into fetal sheep. Analysis of DNA from peripheral blood and bone marrow from recipient sheep demonstrated that FeLV-C– or RD114-pseudotyped vectors were present at significantly higher levels than GALV-pseudotyped vectors. Analysis of individual myeloid colonies demonstrated that retrovirus vectors with FeLV-C and RD114 pseudotypes were present at 1.5 to 1.6 copies per cell and were preferentially integrated near known genes We conclude that the more efficient transduction of human HSCs with either FeLV-C– or RD114-pseudotyped retroviral particles may improve gene transfer in human clinical trials.
Introduction
Recombinant RNA tumor virus vectors are the only gene transfer vehicles that integrate at high frequency into the genome of a target cell. The transduction of hematopoietic stem cells (HSCs), which continuously give rise to differentiated cells of all hematopoietic lineages, is an alluring prospect for gene therapy of hematopoietic disorders (reviewed by Brenner and Malech,1 Logan et al,2 and Hawley3 ). Recently gene therapy has been used to treat infants with severe combined immunodeficiency (SCID): either X-linked SCID4,5 (which is caused by mutations in the interleukin-2 receptor gamma chain [IL2RG] gene)6 or adenosine deaminase (ADA)–deficient SCID.7 Patient CD34+ progenitor cells were transduced with retrovirus vectors containing IL2RG or ADA and reinfused. In the ADA-deficient patients mild myeloablation was used to improve the engraftment of transduced cells in contrast to earlier studies.8-10 Ten of 11 X-SCID and 3 of 4 ADA-deficient patients produced normal numbers of T lymphocytes and improved immune function.4,6,7 The IL2RG or ADA vectors were present at high levels in T lymphocytes and low levels in cells of the other hematopoietic lineages, suggesting that the overall level of gene transfer to HSCs was low.4,6,7
Although 2 X-SCID patients subsequently developed T-cell leukemia associated with proviral insertion into the LMO2 locus,11-12 adverse events have not been observed in a large number of patients who have received transduced cells in marking studies or trials for chronic granulomatous disease and Gaucher disease.13 In these studies, despite the use of optimized combinations of cytokines and fibronectin to colocalize cells and viral particles,14-18 the frequency of HSC transduction was too low to measure any beneficial effects.19-24
The envelope of the retrovirus particle interacts with specific receptor molecules on the target cell surface and determines which cells can be transduced.25 In mouse models, viruses with an ecotropic envelope recognize a site on a mouse amino acid transport protein that is not conserved among other mammals, restricting ecotropic transduction to mouse cells.26-28 The envelopes of mouse amphotropic29,30 and gibbon ape leukemia viruses (GaLV)29,31 recognize sites on highly related sodium-dependent phosphate transporter proteins. The binding sites are conserved among many mammals, allowing viruses with these envelopes to transduce cells of many species. Amphotropic and GaLV-pseudotyped vectors have been used in all human gene transfer experiments.
We have previously shown that the transduction of mouse HSCs is efficient with ecotropic retrovirus vectors and much less efficient with amphotropic retrovirus vectors.32 The difference in transduction correlates with the level of mRNA encoding the ecotropic and amphotropic retrovirus receptors (ecoR and amphoR) in highly enriched populations of mouse HSCs. Human HSCs (Lin– CD34+ CD38–) also have low levels of amphoR and GaLV receptor (GALVR) mRNA.33 Increasing the levels of amphoR or GALVR improved transduction,34-38 but in mouse models the induction of receptor expression was accompanied by a severe loss in the ability of HSCs to repopulate the hematopoietic system.38
We hypothesized that transduction of human HSCs can be increased by the use of retroviral vectors pseudotyped with envelopes that recognize receptors that are more abundant on primitive hematopoietic cells. The RD114 virus is an endogenous feline retrovirus but is not associated with any disease. The RD114 envelope recognizes a neutral amino acid transporter protein (RDR39 ). Recombinant retrovirus vectors with envelopes derived from the RD114 virus have been used successfully to transduce primitive human, canine, and primate hematopoietic cells, often at high levels.40-45 Feline leukemia virus type C (FeLV-C) causes red-cell aplasia in cats. The FeLV-C envelope recognizes a heme transporter protein (FeLV-C receptor [FLVCR]). FeLV-C–pseudotyped vectors have been shown to transduce canine and human cultured cells.46-48
In this report, we compared the levels of FLVCR, RDR, and GALVR mRNA in populations of human hematopoietic stem and progenitor cells and the efficiency of transduction of human CD34+ cells with FeLV-C–, RD114-, and GALV-pseudotyped retrovirus vectors. In sheep undergoing transplantation in utero with cells exposed to equivalent numbers of FeLV-C and GaLV or RD114- and GALV-pseudotyped vectors, transduction was efficient with FeLV-C– or RD114-pseudotyped vectors and much lower with GaLV vectors. Like other retroviruses, FeLV-C– and RD114-pseudotyped viruses preferentially integrated within 5 kilobases (kb) of a known gene.
Materials and methods
Cells
K562 and HeLa cells were maintained in improved minimal essential medium (IMEM; GIBCO, Gaithersburg, MD) with 10% fetal calf serum (FCS). PG1349 and 293T50 (American Type Culture Collection [ATCC], Manassas, VA) cells were maintained in Dulbecco modified Eagle medium (DMEM; GIBCO) supplemented with 10% newborn calf serum (GIBCO). All cells were cultured at 37°C with 5% CO2.
Analysis of receptor mRNA levels
RNA was extracted from HeLa, K562, and fresh bone marrow cells obtained from Poietics (Gaithersburg, MD) using Trizol (GIBCO) according to the manufacturer's instructions. Fifteen micrograms of each RNA sample was analyzed by sequential Northern blot analysis using probes containing a fragment of the FLVCR cDNA (GenBank accession no. NM14053, base pair [bp] 615 to 1025) fused to a fragment of GALVR cDNA (GenBank accession no. L20859, bp 1654 to 2491), a fragment of the RDR cDNA (GenBank accession no. AF102826, bp 1493 to 2118) fused to the same fragment of GALVR cDNA, and an actin probe as described.51 The relative levels of GALVR and FLVCR or RDR mRNA were determined using a Typhoon 8600 Phoshpoimager (Amersham Pharmacia, Piscataway, NJ).
For analysis of mRNA levels in hematopoietic stem and progenitor cells, 6 independent fresh human bone marrow samples were suspended in ACK lysing buffer (BioWhittaker, Walkersville, MD) to lyse red blood cells and washed with cold phosphate-buffered saline (PBS). Lineage-positive cells were stained with fluorescein isothiocyanate (FITC)–conjugated antibodies for CD3, CD4, CD8, CD11b, CD14, CD15, and CD19 (Caltag Laboratories, Burlingame, CA) and anti-FITC magnetic beads (Miltenyi Biotec, Auburn, CA). The cells were then run through an AutoMACS (Miltenyi Biotec) using the “deplete S” mode. Lineage-negative cells were collected, stained with anti-CD34–phycoerythrin (anti-CD34–PE) antibody (Becton Dickinson, San Jose, CA) and anti-CD38–allophycocyanin (anti-CD38–APC) antibody (Caltag Laboratories) and separated into Lin– CD34+ CD38– and Lin– CD34+ CD38+ cells by fluorescence-activated cell sorting (FACS) using a Vantage SE instrument with a 488 nm air-cooled laser (Becton Dickinson). RNA was extracted from sorted cells with Trizol.
Reverse transcriptase–polymerase chain reaction (RT-PCR) was performed on RNA extracted from HeLa, K562, bone marrow, Lin– CD34+ CD38+, and Lin– CD34+ CD38– cells using an ABI Prism 7700 Sequence Detector (Applied Biosystems, Foster City, CA). Reverse Transcriptase and RNase Inhibitor were added to SYBR Green PCR Master Mix according to manufacturer's instructions for one-step RT-PCR (Applied Biosystems, Warrington, United Kingdom). The primers used are specific for the target sequences and do not amplify sequences from related gene families: GALVR 5′ primer (GCA TAG ATA GCA CCG TGA ATG G), 3′ primer (GCT GAC GGC TTG ACT GAA CTG); RDR 5′ primer (CCT GGA TCA TGT GGT ACG CC), 3′ primer (GCG GGC AAA GAG TAA ACC C); FLVCR 5′ primer (CTCCCGCATCGCCTCAGTGT), 3′ primer (ACT GCA GTT CCA AGC TGA TT); β2-microglobulin 5′ primer (GGA GGG CAT CCA GCG TAC TCC), 3′ primer (CGG ATT GAT GAA ACC CAG ACA C). RT-PCR results were normalized to the amounts of β2-microglobulin mRNA, and the relative levels of GALVR, RDR, and FLVCR in RNA extracted from HeLa, K562, and unfractionated bone marrow cells were established from the results of the Northern blot analysis. The relative levels of FLVCR, RDR, and GALVR mRNA were then normalized to the mean level of GALVR mRNA in RNA extracted from unfractionated bone marrow cells.
Retrovirus production
The generation of the GALV-pseudotyped retrovirus vector MFGs-nlsLacZ containing the LacZ reporter gene with a 21-bp nuclear localization signal (nls) has been described previously.34
RD114-pseudotyped MFGs-LacZ retroviruses were produced by transient cotransfection of 293T cells (which express the Moloney murine leukemia virus [MLV] gag and pol genes) with the pMGFs-LacZ plasmid and pRDF (obtained from Dr Elio Vanin, Baylor University52 ) containing the RD114 envelope gene. FeLV-C pseudotyed MFGs-LacZ retroviruses were produced by transient cotransfection of 293T cells with the pMFGs-LacZ plasmid and p61EC, which contains FeLV-C gag, pol, and env genes.38 Cells were transfected using a CellPhect Transfection Kit (Amersham Pharmacia). Supernatants were collected 40 to 48 hours following transfection, filtered through a 0.45-μm filter, and stored at –80°C. An aliquot of each supernatant was analyzed for approximate virus titer by slot blot analysis and transduction of K562 cells followed by Southern blot analysis. FeLV-C envelope sequences were not detected by PCR analysis of DNA from FeLV-C–transduced cells.
Human CD34+ cell transduction
The transduction of human CD34+ cells was done essentially as described.18 After obtaining informed consent (protocol 94-I-0073 approved by the Institutional Review Board of the National Institute of Allergy and Infectious Diseases) healthy adult volunteers received 5 daily subcutaneous injections with 10 μg/kg granulocyte colony-stimulating factor (G-CSF; Amgen, Thousand Oaks, CA). Peripheral blood stem cells (PBSCs) were collected by apheresis on day 5 (CS3000 Plus; Baxter Healthcare, Fenwal Division, Deerfield, IL), and CD34+ cells were selected from the apheresis product (ISOLEX 300i; Nexell Therapeutics, Irvine, CA) and cryopreserved. Mobilized human peripheral blood CD34+ cells were thawed and cultured overnight in X-VIVO 10 medium plus 1% human serum albumin, 50 ng/mL stem-cell factor (SCF), 50 ng/mL FLT3 ligand, 50 ng/mL thrombopoietin (TPO), and 10 ng/mL interleukin-3 (IL-3) (X-VIVO 10 with cytokines). Six-well plates were treated overnight at 37°C with 20 μg/mL Retronectin (Takara Shuzo, Otsu, Japan). The following day, the wells were blocked with 2% bovine serum albumin in phosphate-buffered saline for 30 minutes at 37°C. The wells were washed with 2.5% HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid) in Hanks balanced salt solution.
After overnight culture, CD34+ cells were collected, washed, and resuspended at a concentration of 2 × 105/mL in mixtures of retroviral supernatants containing equivalent numbers of either RD114 MFGs-LacZ or FeLV-C MFGs-LacZ virus and GALV MFGs-nlsLacZ particles with the addition of cytokines and 4 mg/mL Polybrene (Sigma, St Louis, MO). A total of 1 × 106 cells per well were incubated on the fibronectin-coated plates for 6 hours at 37°C. After exposure to virus particles, the cells were collected, centrifuged for 5 minutes at 500g (1500 rpm), resuspended in fresh X-VIVO 10 with cytokines, returned to the fibronectin-coated plates, and incubated overnight. This procedure was repeated daily for 4 days. As a control, parallel cultures of K562 cells were transduced with the same mixture of virus particles.
Following transduction, CD34+ cells were resuspended in medium containing 10% fetal bovine serum (FBS) and sent by overnight mail for injection into preimmune fetal sheep at 55 to 60 days of gestation by means of the amniotic bubble procedure described previously.53,54 Transduced CD34+ cells and bone marrow cells from repopulated sheep were plated in MethoCult GF H4435 (StemCell Technologies, Vancouver, BC, Canada) and cultured at 37°C and 5% CO2 according to the manufacturer's instructions. Individual culture colony-forming units (CFU-Cs) in 20 μL medium were withdrawn with a Pipetman into 1 mL PBS (GIBCO). After centrifugation, pelleted cells were resuspended in 200 μL of 10 mM Tris (tris(hydroxymethyl)aminomethane) pH 7.4, 0.15 M NaCl, 10 mM EDTA (ethylenediaminetetraacetic acid), 0.01% sodium dodecyl sulfate (SDS), and 1.5 mg/mL proteinase K and digested at 56°C for 2 hours followed by phenol and CHCl3 extraction and ethanol precipitation. DNA was resuspended in 20 μL water.
Analysis of DNA from sheep peripheral blood and bone marrow
Peripheral blood and bone marrow was collected from sheep at 1 to 9 months after birth. DNA was extracted for PCR analysis of the human β-globin gene (engraftment) and MFGs-LacZ and MFGs-nlsLacZ proviruses (transduction). The red blood cells were lysed in 1 × red-cell–lysis buffer (10 × stock: 82.9 g NH3Cl, 10 g KHCO3, and 4 mL of 250 mM EDTA) by incubating the samples on ice for 5 minutes and centrifuging them for 10 minutes at low speed and then repeating. The remaining cells were lysed in cell-lysis buffer (0.32 M sucrose, 10 mM Tris-HCl pH 7.5, and 5 mM MgCl2) by incubating and low-speed contrifugation. Nuclei were resuspended in nuclei dropping buffer (24 mM EDTA and 75 mM NaCl) and the samples vortexed. Proteinase K and SDS were added to concentrations of 1 mg/mL and 1%, respectively. Samples were incubated overnight at 37°C. DNA was extracted using phenol and chloroform, ethanol precipitated, and resuspended in TE (10 mM Tris, pH 7.5; 1 mM EDTA).
PCR for the MFGs-LacZ and MFGs-nlsLacZ proviruses was carried out with the addition of 0.1 μL per reaction of [32P]deoxycytidine triphosphate (32PdCTP, 800 mCi/mmol [370 MBq/mL]; Amersham, Arlington Heights, IL) on 400 ng DNA. Primers used were as follows: β-globin 5′ primer (CAG GGC AGA GCC ATC TAT TG), 3′ primer (ACA TGC CCA GTT TCT ATT GG), fragment size 220 bp; LacZ 5′ primer (GCC GAC ACC AGA CTA AGA AC), 3′ primer (CCT CTT CGC TAT TAC GCC AG), MFGs-LacZ fragment size 289 bp, MFGs-nlsLacZ fragment size 310 bp. The PCR conditions were 94°C for 1 minute, 58°C for 1 minute, 72°C for 2 minutes, and the number of cycles adjusted to allow the detection of 100-fold differences in the amount of product as determined by limiting dilution of control samples. Nondenaturing polyacrylamide gels were used to separate the PCR products. The relative amounts of the β-globin and the 289 bp MFGs-LacZ and 310 bp MFGs-nlsLacZ products were determined using a Typhoon 8600 Phoshpoimager (Amersham Pharmacia).
Linker-mediated PCR (LM-PCR)
The linker-mediated PCR (LM-PCR) technique has been previously described.55 Briefly, up to 100 ng DNA isolated from sheep peripheral blood, bone marrow, or CFU-Cs was digested with MseI or MseI and PstI (New England Biolabs, Beverly, MA). Linkers were attached using FastLink Ligase Kit according to manufacturer's instructions (New England Biolabs). Linker sequences are as follows: Afl3 upper strand: 5′GTA ATA CGA CTC AC TAT AGG GCT CCG CT TAAG GGA C3′; and Afl3 lower strand: 5′TAG TCC CTT AAG CGG AC3′ with a 5′ phosphate and a 3′ amino c7. Both linkers were polyacrylamide gel electrophoresis (PAGE) purified. PCR and nested PCR were performed on 2 μL of the ligation or PCR product, respectively. Primers are as follows: 3′long terminal repeat (3′LTR): 5′GAC TTG TGG TCT CGC TGT TCC TTG G3′; AP1 (primer on linker): 5′GTA ATA CGA CTC ACT ATA GGG C3′; 3′LTR nested: 5′GGT CTC CTC TGA GTG ATT GAC TAC C3′; and Afl3 nested primer on linker: 5′AGG GCT CCG CTT AAG GGA C3′. Nested PCR products were analyzed by electrophoresis on 2% NuSieve agarose gel or 1.5% acrylamide gel.
Nested PCR products were ligated into TA cloning vectors and transformed into competent cells. Cultures were grown overnight and the DNA extracted with Qiagen Mini-prep Spin kits and sequenced by DNA (BioServe, Laurel, MD). Sequences were mapped by BLAT analysis of the human genome sequence on the University of California Santa Cruz (UCSC) Genome Bioinformatics Website.56
Results
Human CD34+ cells show high mRNA levels of FLVCR and RDR
To establish standards for evaluating the levels of FLVCR, RDR, and GALVR mRNA, we performed Northern blot analysis on RNA extracted from HeLa, K562, and HL60 cells and unfractionated bone marrow cells. We developed “double probes” containing regions of the GALVR fused to either FLVCR or RDR cDNA sequences with equivalent GC content to ensure that both probes are labeled to equivalent specific activities. The difference in size between the GALVR mRNA (4.0 kb) and the FLVCR (2.4 kb) and RDR (3.4 kb) mRNAs allows a direct comparison of the relative mRNA levels. FLVCR mRNA was present at 4- to 5-fold lower levels than GALVR mRNA in all cell lines and unfractionated bone marrow cells (Figure 1). RDR mRNA was present at 3-fold lower levels than GALVR mRNA in unfractionated bone marrow cells and at slightly higher levels than GALVR mRNA in HeLa and K562 cells (Figure 1).
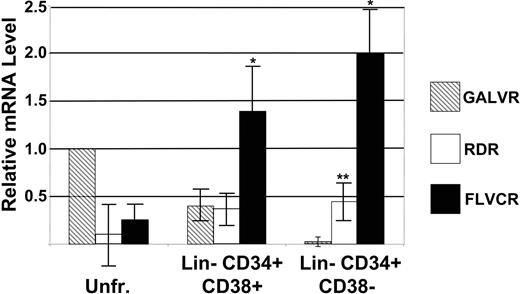
The same HeLa, K562, and bone marrow RNAs were used as standards in real-time PCR analysis to determine the levels of FLVCR, RDR, and GALVR mRNA in RNA extracted from 6 independent FACS-sorted populations of human Lin– CD34+ CD38+ hematopoietic progenitor cells and Lin– CD34+ CD38– cells that are enriched for HSCs. As we have shown previously, the level of GALVR mRNA was highest in unfractionated bone marrow cells, declined in Lin– CD34+ CD38+ cells, and was nearly undetectable in Lin– CD34+ CD38– cells33 (Figure 2 and Table 1). In contrast, FLVCR mRNA levels were lowest in unfractionated bone marrow cells and significantly higher than the levels of GALVR mRNA in Lin– CD34+ CD38+ progenitor cells (P < .001) and Lin– CD34+ CD38– cells (P < .001; Figure 2 and Table 1). In unfractionated bone marrow cells the mean level of RDR mRNA was 6-fold lower than that of GALVR mRNA but not statistically different due to a high degree of variability. RDR mRNA levels were similar to the level of GALVR mRNA in Lin– CD34+ CD38+ cells and significantly higher than the level of GALVR mRNA in Lin– CD34+ CD38– cells (P < .001; Figure 2 and Table 1). These results are similar to our previously published comparison of the relative levels of RDR mRNA in human mobilized peripheral blood and cord blood stem and progenitor cells.18
Relative retrovirus receptor mRNA levels
RNA . | HeLa* . | K562* . | Unfractionated marrow*† . | CD34+ CD38+† . | CD34+ CD38-† . |
|---|---|---|---|---|---|
| GALVR | 1.1 | 2.0 | 1.0 | 0.42 ± 0.1 | 0.04 ± 0.02 |
| FLVCR | 0.19 | 0.24 | 0.27 ± 0.11 | 1.45 ± 0.34‡ | 2.0 ± 0.32‡ |
| RDR | 1.4 | 1.2 | 0.17 ± 0.27 | 0.41 ± 0.12 | 0.45 ± 0.13§ |
RNA . | HeLa* . | K562* . | Unfractionated marrow*† . | CD34+ CD38+† . | CD34+ CD38-† . |
|---|---|---|---|---|---|
| GALVR | 1.1 | 2.0 | 1.0 | 0.42 ± 0.1 | 0.04 ± 0.02 |
| FLVCR | 0.19 | 0.24 | 0.27 ± 0.11 | 1.45 ± 0.34‡ | 2.0 ± 0.32‡ |
| RDR | 1.4 | 1.2 | 0.17 ± 0.27 | 0.41 ± 0.12 | 0.45 ± 0.13§ |
Determined by Northern blot analysis (n = 2).
Determined by RT-PCR (n = 6).
Significantly higher than the level of GALVR mRNA (P < .001).
Significantly higher than the level of GALVR mRNA (P < .001); significantly lower than the level of FLVCR mRNA (P < .001).
Northern blot analysis of FLVCR, RDR, and GALVR mRNA levels in RNA from HeLa, K562, and human bone marrow (HuBM) cells. Total RNA extracted from the indicated cells was separated on agarose gels and transferred to a nylon filter. The filter was sequentially probed with a DNA fragment containing FLVCR and GALVR sequences (A), a DNA fragment containing RDR and GALVR sequences (B), and a DNA fragment containing actin sequences (C). The approximate sizes of the mRNAs are indicated at the right. Blots in panels A and B were exposed for approximately 48 hours; panel C, approximately 10 hours.
Northern blot analysis of FLVCR, RDR, and GALVR mRNA levels in RNA from HeLa, K562, and human bone marrow (HuBM) cells. Total RNA extracted from the indicated cells was separated on agarose gels and transferred to a nylon filter. The filter was sequentially probed with a DNA fragment containing FLVCR and GALVR sequences (A), a DNA fragment containing RDR and GALVR sequences (B), and a DNA fragment containing actin sequences (C). The approximate sizes of the mRNAs are indicated at the right. Blots in panels A and B were exposed for approximately 48 hours; panel C, approximately 10 hours.
RT-PCR analysis of GALVR, RDR, and FLVCR mRNA levels in human bone marrow and hematopoietic progenitor and stem cells. RNA from FACS-sorted bone marrow fractions was analyzed by RT-PCR. The data for unfractionated (Unfr.) bone marrow was normalized to the data from Northern blot analysis of the same RNA samples as in Figure 1. The level of GALVR mRNA in unfractionated bone marrow was set as 1 and compared with the relative amount of RDR and FLVCR mRNA in unfractionated bone marrow, Lin– CD34+ CD38+, and Lin– CD34+ CD38– cells. *Significantly higher than the level of GALVR mRNA (P < .001). **Significantly higher than the level of GALVR mRNA (P < .001); significantly lower than the level of FLVCR mRNA (P < .001). Error bars indicate standard deviation.
RT-PCR analysis of GALVR, RDR, and FLVCR mRNA levels in human bone marrow and hematopoietic progenitor and stem cells. RNA from FACS-sorted bone marrow fractions was analyzed by RT-PCR. The data for unfractionated (Unfr.) bone marrow was normalized to the data from Northern blot analysis of the same RNA samples as in Figure 1. The level of GALVR mRNA in unfractionated bone marrow was set as 1 and compared with the relative amount of RDR and FLVCR mRNA in unfractionated bone marrow, Lin– CD34+ CD38+, and Lin– CD34+ CD38– cells. *Significantly higher than the level of GALVR mRNA (P < .001). **Significantly higher than the level of GALVR mRNA (P < .001); significantly lower than the level of FLVCR mRNA (P < .001). Error bars indicate standard deviation.
Transduction of human CD34+ cells with FeLV-C– and RD114-pseudotyped vectors
FeLV-C– or RD114-pseudotyped MFGs-LacZ retroviral vectors were produced by transient cotransfection of 293T cells. We have previously described a PG13 (GALV-pseudotyped) MFGs-nlsLacZ producer cell line with a titer of approximately 2 × 105 infectious particles per milliliter.34 The relative number of FeLV-C– or RD114-pseudotyped virus particles containing the MFGs-LacZ vector was estimated by slot-blot analysis of equal volumes of viral supernatant (Figure 3). The production of RD114-pseudotyped virus by transient transfection was efficient; more than 50% of transfections generated supernatant with titers above 1 × 105 infectious particles per milliliter. However, the production of high-titer supernantants with the FeLV-C envelope was relatively inefficient, because only 11.5% of the transfections generated titers above 1 × 105 infectious particles per milliliter (Table 2).
Production of retrovirus particles with FeLV-C and RD114 envelopes by transient transfection of 293T cells
Vector . | Envelope plasmid . | No. of transfections . | Titer more than 1 × 105 infectious particles per mL (%) . |
|---|---|---|---|
| MFGs-LacZ | p61EC (FeLV-C)* | 29 | 3 (11.5) |
| MFGs-LacZ | pRDF (RD114)† | 11 | 6 (54.5) |
Vector . | Envelope plasmid . | No. of transfections . | Titer more than 1 × 105 infectious particles per mL (%) . |
|---|---|---|---|
| MFGs-LacZ | p61EC (FeLV-C)* | 29 | 3 (11.5) |
| MFGs-LacZ | pRDF (RD114)† | 11 | 6 (54.5) |
Contains FeLV-C gag pol and env sequences.
Contains only RD114 env sequence.
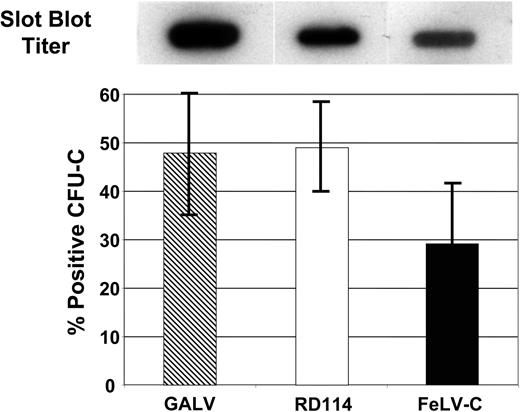
To confirm the estimates of virus particle number, aliquots of supernatant containing either FeLV-C– (estimated titer 0.5 × 105 particles/mL), RD114- (1.0 × 105 particles/mL), or GaLV- (2.0 × 105 particles/mL) pseudotyped virus particles were used to transduce human peripheral blood CD34+ cells, which were then plated in semisolid medium for analysis of CFU-C transduction. FeLV-C, RD114, and GALV-pseudotyped vectors transduced an average of 29.2% ± 13%, 49.0% ± 9.6%, and 47.9% ± 13% of CFU-Cs, respectively, which correlates most closely with the virus titer, but were not significantly different (Table 3).
Transduction of CFU-Cs with GaLV, FeLV-C, and RD114 pseudotyped retrovirus vectors
Pseudotype . | Estimated titer . | % positive CFU-Cs ± SD* . | No. of integrations per colony* . |
|---|---|---|---|
| GaLV | 2.0 × 105 | 47.9 ± 13 | 1.3 ± 0.67 |
| FeLV-C | 0.5 × 104 | 29.2 ± 13 | 1.6 ± 0.71 |
| RD114 | 1.0 × 104 | 49.0 ± 9.6 | 1.5 ± 0.96 |
Pseudotype . | Estimated titer . | % positive CFU-Cs ± SD* . | No. of integrations per colony* . |
|---|---|---|---|
| GaLV | 2.0 × 105 | 47.9 ± 13 | 1.3 ± 0.67 |
| FeLV-C | 0.5 × 104 | 29.2 ± 13 | 1.6 ± 0.71 |
| RD114 | 1.0 × 104 | 49.0 ± 9.6 | 1.5 ± 0.96 |
The differences are not significant (P > .05).
Transduction of human CFU-Cs with GALV, RD114, or FeLV-C pseudotyped retroviral vectors. Slot blot analysis of 1.0 mL virus-containing medium was used to compare the titer of the different virus preparations (top panel). Human CD34+ peripheral blood CD34+ cells were transduced with GALV, RD114, or FeLV-C pseudotyped retrovirus particles. Transduced CFU-Cs were identified by PCR analysis of DNA extracted from each colony. Data shown was pooled from 3 experiments. Error bars indicate standard deviation.
Transduction of human CFU-Cs with GALV, RD114, or FeLV-C pseudotyped retroviral vectors. Slot blot analysis of 1.0 mL virus-containing medium was used to compare the titer of the different virus preparations (top panel). Human CD34+ peripheral blood CD34+ cells were transduced with GALV, RD114, or FeLV-C pseudotyped retrovirus particles. Transduced CFU-Cs were identified by PCR analysis of DNA extracted from each colony. Data shown was pooled from 3 experiments. Error bars indicate standard deviation.
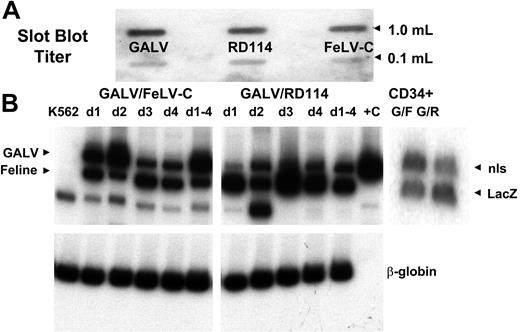
Supernatants were combined such that each aliquot contained equivalent amounts of FeLV-C or RD114 MFGs-LacZ, and GaLV MFGs-nlsLacZ virus particles were used to cotransduce human mobilized peripheral blood CD34+ cells and parallel cultures of control K562 cells. Mobilized peripheral blood cells were chosen because we have previously shown that the relative levels of the receptor mRNAs were similar in bone marrow and mobilized peripheral blood cells18,33 (data not shown) and more accurately simulate current clinical transduction protocols. A 21-bp nls in the MGFs-nlsLacZ vector allows the relative amounts of MFGs-LacZ (FeLV-C or RD114) and MFGs-nlsLacZ (GALV) proviruses to be distinguished using PCR primers spanning the nls region. PCR analysis of DNA extracted from cotransduced K562 cells showed that FeLV-C–, RD114-, and GALV-pseudotyped virus particles integrated into K562 cells over the 4 days of transduction (Figure 4). Similarly, DNA extracted from CD34+ cells at the end of the transduction showed similar levels of transduction with either FeLV-C– or RD114- and GALV-pseudotyped virus particles (Figure 4).
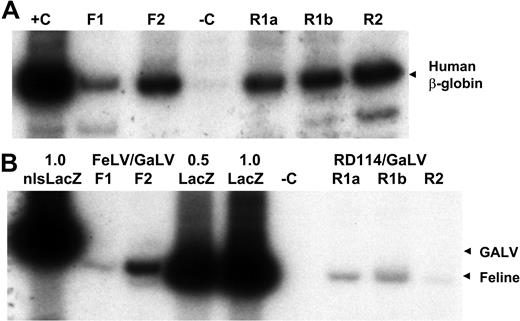
The fetal sheep model was chosen for evaluating the engraftment of the most primitive human hematopoietic cells.53,54 Cotransduced cells representing 9 independent peripheral blood CD34+ –cell collections were transplanted into fetal sheep (5 FeLV-C/GaLV, 4 RD114/GaLV, including 1 set of twins). DNA was extracted from peripheral blood and bone marrow cells collected from live-born animals between 1 to 9 months after birth for analysis of human hematopoietic-cell engraftment by PCR analysis using primers specific for the human β-globin gene. Two of 5 lambs injected with FeLV-C/GaLV-transduced cells demonstrated the presence of human cells in peripheral blood and bone marrow at levels estimated to be between 1% and 5% of the total cells (Figure 5A). Similarly, the set of twin lambs (which shared a placenta and are treated as a single animal) and 1 of 3 additional lambs injected RD114/GaLV-transduced cells demonstrated the presence of human cells in peripheral blood and bone marrow at levels estimated to be between 2% and 10% of the total cells (Figure 5A). The frequency of engrafted animals and the level of human hematopoietic cells in engrafted animals and within the range observed in a large series of sheep transplantations over 10 years indicated that there was no significant effect of the transduction procedure on engraftment.53,54
The relative amounts of the MGFs-LacZ and MFGs-nlsLacZ proviruses in the human cells were measured by PCR using primers that span the 21 bp nls signal. The MFGs-LacZ (FeLV-C) signal was detected within the linear range of amplification in both lambs engrafted with FeLV-C/GaLV-transduced cells, but the MFGs-nlsLacZ band (GaLV) was only detected after additional cycles of PCR (Figure 5B). The MFGs-LacZ (RD114) signal was detected within the linear range of amplification in all 3 lambs engrafted with RD114/GaLV-transduced cells, but again the MFGs-nlsLacZ band (GaLV) was only detected after additional cycles of PCR (Figure 5B). Light scattering properties and magnetic bead selection was performed to isolate peripheral blood cells enriched for human T lymphocytes (CD4 or CD8+), monocytes (CD11+), and neutrophils (CD14+) from 3 animals for DNA analysis. The ratio of MFGs-LacZ to MFGs-nlsLacZ sequences was identical to that seen in the unfractionated blood samples (Figure 6).
LAM-PCR analysis of 15 to 20 proviral integration sites in each of the lymphocyte, monocyte, and neutrophil populations demonstrated 3 to 5 distinct proviral insertion sites common to all 3 cell types in each animal. This is probably an underestimate of the total number of shared integration sites, because the entire repertoire of insertion sites was not analyzed. Taken together, these data indicate that multipotent sheep repopulating cells (SpRCs) had been transduced. Comparison of the β-globin signals and the LacZ signals generated an approximate copy number of 0.1 and 0.05 proviral copies per human cell, respectively (Figure 5).
Analysis of vector insertion sites
DNA extracted from transduced individual CFU-Cs was analyzed by LM-PCR for chromosomal location of proviral insertion sites. Nested PCR products were cloned and sequenced to confirm the presence of the nested primer and LTR sequences and the cell virus junction sequences. The average number of proviral insertions per cell were 1.6 ± 0.71, 1.5 ± 0.96, and 1.3 ± 0.67 copies per cell for FeLV-C–, RD114-, and GaLV-transduced CFU-Cs, respectively. The differences were not significant, indicating that the FeLV-C pol that was present along with the Moloney MLV pol in the FeLV-C–pseudotyped particles did not affect the frequency of integration into the target-cell DNA.
Cotransduction of K562 and human peripheral blood CD34+ cells with FeLV-C and GALV or RD114 and GALV-pseudotyped retrovirus particles. Slot blot analysis of 1.0 mL and 0.1 mL virus-containing medium was used to compare the titer of the different virus preparations (A). These titers were used to adjust the volumes of supernatants to ensure that equivalent numbers of each pseudotype were used. Human CD34+ peripheral blood CD34+ cells were cotransduced with FeLV-C and GALV or RD114 and GALV-pseudotyped virus preparations over a 4-day period. Parallel cultures of K562 cells were cotransduced with the same virus preparations each day (d1, d2, d3, d4). One culture was cotransduced over the 4-day period (d1-4). DNA was extracted from the K562 cultures and a portion of the CD34+ cells for analysis of the integration of the FeLV-C or RD114 and GALV-pseudotyped viruses by PCR using primers that span the nls in the MFG-nlsLacZ vector. (B) The analysis of FeLV-C/GALV- and RD114/GALV-transduced K562 cells is shown in the left and center panels. Sequences from the human β-globin were amplified in the same samples as a control. The analysis of FeLV-C/GALV (G/F)– and RD114/GALV (G/R)–transduced CD34+ cells used for transplantation into fetal sheep is shown in the right panel.
Cotransduction of K562 and human peripheral blood CD34+ cells with FeLV-C and GALV or RD114 and GALV-pseudotyped retrovirus particles. Slot blot analysis of 1.0 mL and 0.1 mL virus-containing medium was used to compare the titer of the different virus preparations (A). These titers were used to adjust the volumes of supernatants to ensure that equivalent numbers of each pseudotype were used. Human CD34+ peripheral blood CD34+ cells were cotransduced with FeLV-C and GALV or RD114 and GALV-pseudotyped virus preparations over a 4-day period. Parallel cultures of K562 cells were cotransduced with the same virus preparations each day (d1, d2, d3, d4). One culture was cotransduced over the 4-day period (d1-4). DNA was extracted from the K562 cultures and a portion of the CD34+ cells for analysis of the integration of the FeLV-C or RD114 and GALV-pseudotyped viruses by PCR using primers that span the nls in the MFG-nlsLacZ vector. (B) The analysis of FeLV-C/GALV- and RD114/GALV-transduced K562 cells is shown in the left and center panels. Sequences from the human β-globin were amplified in the same samples as a control. The analysis of FeLV-C/GALV (G/F)– and RD114/GALV (G/R)–transduced CD34+ cells used for transplantation into fetal sheep is shown in the right panel.
Cotransduction of human S-RCs with FeLV-C and GALV or RD114 and GALV-pseudotyped retrovirus particles. Human CD34+ peripheral blood CD34+ cells were cotransduced with FeLV-C and GALV or RD114 and GALV-pseudotyped virus preparations over a 4-day period and transplanted into fetal sheep. DNA was extracted from sheep peripheral blood cells 6 months after transplantation and analyzed for the presence of human cells and the integration of the FeLV-C or RD114 and GALV-pseudotyped viruses by PCR analysis of 0.4 μg DNA. (A) Sequences from the human β-globin gene were amplified to detect the presence of human cells. F1 and F2 indicate analysis of DNA from 2 lambs infused with human CD34+ cells exposed to FeLV-C (LacZ) and GaLV (nls-LacZ) pseudotyped particles; R1a, R1b, and R2, analysis of DNA from 3 lambs infused with human CD34+ cells exposed to RD114 and GALV-pseudotyped particles; R1a and R1b, DNA from twin lambs infused with the same population of cells; –C, DNA extracted from control sheep; and +C, DNA extracted from K562 cells. (B) The relative amounts of integrated FeLV-C, RD114, and GALV-pseudotyped vectors were measured using primers that span the nls in the MFGs-nlsLacZ vector. 1.0 nls-LacZ indicates 3T3-cell DNA containing a single copy of the MFGs-nlsLacZ vector; 1.0 LacZ, 3T3-cell DNA containing a single copy of the MFGs-LacZ vector; 0.5 LacZ, 3T3-cell DNA containing a 1:1 dilution of this 1.0 LacZ DNA with untransduced 3T3-cell DNA; and –C, DNA extracted from control sheep. For quantitation, only the β-globin band and the 289-bp MFGs-LacZ and 310-bp MFGs-nlsLacZ bands were analyzed.
Cotransduction of human S-RCs with FeLV-C and GALV or RD114 and GALV-pseudotyped retrovirus particles. Human CD34+ peripheral blood CD34+ cells were cotransduced with FeLV-C and GALV or RD114 and GALV-pseudotyped virus preparations over a 4-day period and transplanted into fetal sheep. DNA was extracted from sheep peripheral blood cells 6 months after transplantation and analyzed for the presence of human cells and the integration of the FeLV-C or RD114 and GALV-pseudotyped viruses by PCR analysis of 0.4 μg DNA. (A) Sequences from the human β-globin gene were amplified to detect the presence of human cells. F1 and F2 indicate analysis of DNA from 2 lambs infused with human CD34+ cells exposed to FeLV-C (LacZ) and GaLV (nls-LacZ) pseudotyped particles; R1a, R1b, and R2, analysis of DNA from 3 lambs infused with human CD34+ cells exposed to RD114 and GALV-pseudotyped particles; R1a and R1b, DNA from twin lambs infused with the same population of cells; –C, DNA extracted from control sheep; and +C, DNA extracted from K562 cells. (B) The relative amounts of integrated FeLV-C, RD114, and GALV-pseudotyped vectors were measured using primers that span the nls in the MFGs-nlsLacZ vector. 1.0 nls-LacZ indicates 3T3-cell DNA containing a single copy of the MFGs-nlsLacZ vector; 1.0 LacZ, 3T3-cell DNA containing a single copy of the MFGs-LacZ vector; 0.5 LacZ, 3T3-cell DNA containing a 1:1 dilution of this 1.0 LacZ DNA with untransduced 3T3-cell DNA; and –C, DNA extracted from control sheep. For quantitation, only the β-globin band and the 289-bp MFGs-LacZ and 310-bp MFGs-nlsLacZ bands were analyzed.
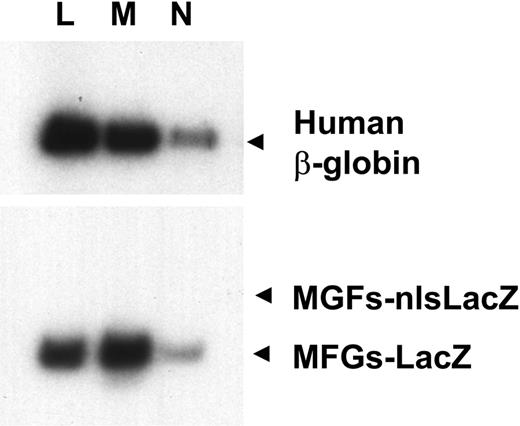
Transduction of lymphoid and myeloid cells. Human lymphocytes (L), monocytes (M), and neutrophils (N) were enriched from the peripheral blood of sheep transplanted with transduced cells by light scattering and cell-surface antigen expression. DNA from these cells was analyzed for the presence of the human β-globin gene and the MFGs-LacZ and MFGs-nlsLacZ proviruses as in Figure 5.
Transduction of lymphoid and myeloid cells. Human lymphocytes (L), monocytes (M), and neutrophils (N) were enriched from the peripheral blood of sheep transplanted with transduced cells by light scattering and cell-surface antigen expression. DNA from these cells was analyzed for the presence of the human β-globin gene and the MFGs-LacZ and MFGs-nlsLacZ proviruses as in Figure 5.
Retrovirus vectors have been shown to preferentially integrate near genes as opposed to randomly throughout the genome.55 We mapped a total of 46 FeLV-C and 44 RD114 proviral insertions in DNA extracted from transplanted sheep bone marrow and CFU-Cs. The CFU-Cs from animals undergoing transplantation all contained either the FeLV-C– or RD114-pseudotyped vector. Similar to previously published results, 41% and 34% of the integrated FeLV-C– and RD114-pseudotyped viruses mapped within 5 kb of a RefSeq gene. These data indicate that the presence of FeLV-C pol did not alter proviral integration (Table 4).
Insertion site analysis
Pseudotype . | No. of insertions cloned . | No. of insertions in unique sequence (%) . | % unique insertions less than 5 kb from known gene . |
|---|---|---|---|
| FeLV-C | 46 | 22 (48) | 41 |
| RD114 | 44 | 18 (41) | 34 |
Pseudotype . | No. of insertions cloned . | No. of insertions in unique sequence (%) . | % unique insertions less than 5 kb from known gene . |
|---|---|---|---|
| FeLV-C | 46 | 22 (48) | 41 |
| RD114 | 44 | 18 (41) | 34 |
Discussion
Unlike X-SCID and ADA-deficient SCID where there is an in vivo selective advantage for corrected cells,57,58 there is no evidence of in vivo selection in disorders like the hemoglobinopathies or chronic granulomatous disease. Gene therapy for these disorders will require a higher level of transduction than is currently available with vectors pseudotyped with GaLV or amphotropic envelopes. This can be accomplished by either improved HSC transduction or the modification of HSCs with drug-selectable genes or growth regulators.59-64 Our observations about superior transduction with RD114-pseudotyped retrovirus particles are consistent with previous data in canine, nonhuman primate, and NOD-SCID mouse xenograft model systems.40-45 Our data are the first demonstration of transduction of primary hematopoietic cells with recombinant FeLV-C retrovirus particles. We conclude that FeLV-C– and RD114-pseudotyped virus particles transduce human stem and progenitor cells more efficiently than GALV-pseudotyped retrovirus particles, which may allow gene therapy to be extended to hematopoietic diseases other than SCID.
Retrovirus transduction is a function of the titer of the virus preparation, the abundance of the receptor, the affinity with which the virus envelope binds to the receptor, and the cell-cycle status of the target cells.35,65-69 Tagged ecotropic and amphotropic murine leukemia virus (MLV) and GaLV envelope proteins have been used to demonstrate that retrovirus binding to target cells depends on the receptor density and envelope binding.65-68 Analysis of a cell line with an inducible GALVR gene showed that a 4- to 5-fold increase in GALVR mRNA led to a 150-fold increase in the frequency of GALV transduction.67 Cytokine treatment of human CD34+ progenitor cells increased GALVR mRNA levels, GALVR density on the cell surface, GaLV envelope binding, and the frequency of transduction with GALV-pseudotyped virus particles. However, the levels of GALVR mRNA in cytokine-treated CD34+ progenitor cells are lower than the levels seen in cultured cells.67 In CD34+ progenitor cells, where the levels of GALVR, RDR, and FLVCR mRNA are most similar, the frequency of transduction was more strongly correlated with the titer of the virus preparation, although the differences were not significant. In human Lin– CD34+ CD38– the level of FLVCR and RDR mRNA is significantly higher than the level of GALVR mRNA, and transduction with equivalent titers of virus correlated with receptor mRNA levels. Because the levels of FLVCR mRNA in Lin– CD34+ CD38– cells is even higher than in HeLa and K562 cells, we hypothesize that FeLV-C might be the ideal envelope for HSC transduction.
Our experimental design in which a single population of target cells is cotransduced with equivalent numbers of virus particles allows us to directly compare the level of retrovirus receptor mRNA with the level of transduction with retrovirus particles with different pseudotypes without the confounding influences of cell-cycle dynamics. The level of retrovirus receptor mRNA correlates with the frequency of transduction of human SpRCs. The relative difference between the frequency of transduction of human SpRCs with FeLV-C– and GALV-pseudotyped virus particles cannot be calculated precisely because the level of GaLV transduction is below the linear range in our PCR assays. However, our data demonstrate a 40-fold difference in the level of FLVCR and GALVR mRNA in human Lin– CD34+ CD38– cells, which is a good estimate of the increase in the frequency of transduction.
The production of FeLV-C–pseudotyped particles for clinical use will require improvements in FeLV-C virus production. Production of FeLV-C–pseudotyped virus by transient transfection of 293 cells was inefficient even when a plasmid containing gag/pol and env plasmid was used to provide the packaging functions. Although we did not detect any FeLV-C env sequences in DNA from sheep bone marrow or blood cells (data not shown), it is clear that the development of a plasmid that expresses only FeLV-C env or a stable producer cell line with separate gag/pol and FeLV-C env sequences will be necessary. These studies are under way (J.L.A., personal communication, November 2004). It is likely that the FeLV-C envelope can be modified to pseudotype lentivirus vectors as has recently been demonstrated for amphotropic MLV and RD114 envelopes.45,70
The integration of FeLV-C–pseudotyped virus was similar to that of typical oncoretrovirus, a preference for gene-rich areas of the genome, and a relatively low copy number per cell. Increased transduction of human HSCs carries with it the increased risk of insertional mutagenesis.71,72 This risk will need to be weighed against the potential benefits and the knowledge that most insertion events in human HSCs appear to be benign at this time.13 These data suggest that outside of the increased frequency of integration events, FeLV-C–pseudotyped viruses should be as safe as the amphotropic or GALV-pseudotyped virus particles approved for use in humans. Safety data in large animal models will be necessary to confirm this.
Prepublished online as Blood First Edition Paper, March 17, 2005; DOI 10.1182/blood-2004-11-4491.
Supported by HL31823 and HL46598 (J.L.A.) and HD43038, HL66058, HL52955, and HL49042 (C.D.P. and E.D.Z.).
An Inside Blood analysis of this article appears at the front of the issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal