Abstract
Macrophages and myeloid dendritic cells (DCs) represent alternative differentiation options of bone marrow progenitors and blood monocytes. This choice profoundly influences the immune response under normal and pathological conditions, but the underlying transcriptional events remain unresolved. Here, we show that experimental activation of the transcription factors PU.1 and MafB in transformed chicken myeloid progenitors triggered alternative DC or macrophage fate, respectively. PU.1 activation also was instructive for DC fate in the absence of cytokines in human HL-60 cell-derived myeloid progenitor and monocyte clones. Differentiation of normal human monocytes to DCs led to a rapid increase of PU.1 to high levels that preceded phenotypic changes, but no MafB expression, whereas monocyte-derived macrophages expressed MafB and only moderate levels of PU.1. DCs inducing levels of PU.1 inhibited MafB expression in monocytes, which appeared to be required for DC specification, since constitutive MafB expression inhibited DC differentiation. Consistent with this, PU.1 directly bound to MafB, inhibited its transcriptional activity in macrophages, and repressed its ability to induce macrophage differentiation in chicken myeloid progenitors. We propose that high PU.1 activity favors DCs at the expense of macrophage fate by inhibiting expression and activity of the macrophage factor MafB.
Introduction
Dendritic cells (DCs) serve a crucial function in the immune system as the major antigen presenting cells with the unique ability to activate naive T cells,1 a capacity that has made them the major target of therapeutic manipulation in vaccination and cancer immunotherapy protocols.1 They can develop from myeloid progenitors in the bone marrow or circulating blood monocytes.2 When exiting the blood stream, monocytes can give rise to inflammatory macrophages or to antigen presenting DCs.3 A disturbance of this balance in favor of DC differentiation has been observed in autoimmune disease,4 whereas tumors can skew it toward the macrophage option.5
Cytokine conditions favoring outgrowth of DCs in vitro have been well defined in mouse and human cells, most commonly culture of bone marrow progenitors with granulocyte-macrophage colony-stimulating factor (GM-CSF), CD34+ progenitors with GM-CSF and tumor necrosis factor α (TNFα), or stimulation of blood monocytes with GM-CSF and interleukin-4 (IL-4).2 By contrast M-CSF,5 IL-6,5,6 IL-10,7 and interferon-γ (IFN-γ)8 favor macrophage differentiation at the expense of DC fate.
In contrast to the cytokine requirements, the transcriptional events controlling the choice of macrophage versus DC fate have remained unresolved. Some transcription factor knockout mice show defects in DC differentiation9,10 but also in other lineages, making it difficult to determine at what stage the differentiation block occurred. Certain transcription factor oncogenes have been shown to be capable of transforming DCs or their progenitors, but how this relates to the normal differentiation program remains unclear.11,12 Constitutive overexpression of myeloid transcription factors in heterogeneous populations of hematopoietic progenitors also has defined factors incompatible with alternative Langerhans cell or granulocyte fates under GM-CSF differentiation conditions.13 However, the transcription factors that instruct the commitment of myeloid-restricted progenitors or monocytes to DC versus macrophage fate are unknown.
To address this question we have used a hormone-inducible transcription factor and 2 different cell-culture systems that make it possible to analyze uniform clonal-cell populations expressing a transduced gene. In one of these, primary transformed avian myeloblasts can differentiate into granulocytes or macrophages under appropriate conditions.14 We have previously shown that the expression of the monocyte/macrophage-specific bZip transcription factor MafB in this system favored macrophage differentiation, whereas expression of the myeloid- and lymphoid-specific Ets family factor PU.1 did not.14
Gene inactivation studies in mice have revealed that PU.1 is required for the correct development of both the lymphoid and the myeloid lineages, including neutrophils, macrophages, osteoclasts, DCs, and mast cells.15-21 Pu.1–/– mice do not generate myeloid DCs.15,17 However, because of the early general defect of myeloid differentiation in Pu.1–/– mice, this could be due to the lack of a common myeloid progenitor rather than a function in DC differentiation, especially since retroviral reconstitution experiments in PU.1-null mice give rise to monocyte/macrophage but not DC differentiation.21-23 It therefore remained a crucial question whether PU.1 has a direct role in specifying DC fate and how it might influence the immunologically important choice between macrophage and DC differentiation.
Here, we now show in 2 different cell systems and in the absence of any DC-promoting cytokines that the activation of PU.1 in clonal populations of myeloblasts triggers the rapid induction of DC fate in the absence of macrophage differentiation, whereas MafB overexpression results in macrophage, but not DC, differentiation. We observed that primary human monocytes moderately increase PU.1 expression when differentiated into macrophages but dramatically up-regulate PU.1 when subjected to a DC differentiation protocol. Consistent with this, PU.1 activation in a clonal population of monocytes also triggered DC fate. We observed that macrophages but not DCs express MafB, and that PU.1-induced DC differentiation in monocytes inhibits MafB expression. Furthermore, preventing MafB down-regulation by constitutive expression interfered with DC differentiation. We propose that monocytes and macrophages tolerate moderate levels of PU.1, whereas DC differentiation at the expense of macrophage fate is instructed by high levels of PU.1 and requires down-regulation of MafB.
Materials and methods
Virus constructs and production
Recombinant E26ts21, E26ts21-MafB, or E26ts21–PU.1-estrogen receptor hormone binding domain fusion protein (PUER) retroviruses were produced as described and used to infect hematopoietic progenitor cells from 2-day chick embryos.14,24
MFG-iGFP was obtained by inserting an internal ribosomal entry site–enhanced green fluorescent protein (IRES-eGFP) cassette from pIRES-eGFP (Clontech) into MFG, a MoMuLV-based vector.25 MFG-PUER-iGFP and MFG-MafB-iGFP constructs were generated by inserting the PUER or the MafB cassettes respectively into MFG-iGFP. Recombinant retroviruses were produced in Phoenix (ΦNX) packaging cells and used to infect HL-60 cells (see “Virus infection and cell culture”).
Virus infection and cell culture
Clones of E26 virus–transformed hematopoietic cells were obtained and cultured as described.14 ts21-PUER clones were stimulated with 1 μM β-estradiol (βE) (Sigma-Aldrich, Saint Quentin Fellevier, France), and ts21-MafB clones were shifted to 42°C to induce differentiation.
HL-60 cells were cultured and infected as described. Sixty hours after infection, cells were analyzed for GFP expression. MFG-PUER-iGFP or MFG-iGFP cells were fluorescence-activated cell-sorter scanner (FACS) sorted for GFP-positive cells to 90% purity and plated in semisolid methylcellulose medium to isolate individual GFP-positive clones after 2 weeks. MFG-MafB-iGFP–infected cells were used unsorted and stimulated with 200 ng/mL of calcium ionophore A23187 (Sigma) to induce their differentiation to DCs.
Differentiation of human primary cells
Human blood from healthy donors was collected according to institutional guidelines (Site Transfusionnel, Marseille, France). Informed consent was provided according to the Declaration of Helsinki. Peripheral blood mononuclear cells (PBMCs) were isolated by Ficoll-Paque (Amersham Bioscience, Orsay, France) gradient centrifugation. DCs and macrophages were generated from monocytes purified by adherence in the presence of 10 ng/mL of GM-CSF and IL-4 or M-CSF respectively (all from Peprotech, London, United Kingdom) as described.26
Immunostaining and FACS analysis
Human cells were stained with mouse antihuman antibodies against CD1a phycoerythrin (PE), Langerin, CD86 (Immunotech, Marseille, France), CD11b and CD14 antigens (BD PharMingen, San Diego, CA) and phycoerythrin (PE)–conjugated secondary antimouse antibodies (Immunotech). Chicken cells were stained with mouse antichicken major histocompatibility complex II (MHCII) antibody (CIa-1) (Southern Biotechnology, Birmingham, AL) and 47/83 antibody.27
To detect cytoplasmic proteins, cells were fixed in 4% paraformaldehyde for 15 minutes and permeabilized with 0.3% saponin (Sigma-Aldrich), then stained with rabbit antihuman or antichicken S100 (Interchim) or mouse antichicken MHCII antibody (CIa-1), followed by fluorescein isothiocyanate (FITC) or Alexa-conjugated secondary antimouse or rabbit antibodies (BD PharMingen, Pont de Claix, France). Cells were counterstained with the nuclear dye DAPI (4′6-diamidino-2-phenylindole 2HCl) (5 μg/mL for 45 minutes; Sigma-Aldrich).
For cytochemical analysis, cells were cytocentrifuged onto glass slides, methanol-fixed for 4 minutes, and then stained with May-Grünwald-Giemsa reagent as indicated by the manufacturer (Sigma-Aldrich).
Functional assays
Allogenic splenocytes for the mixed lymphocyte reaction were isolated by Ficoll-Paque (Amersham Biosciences) density gradient centrifugation of homogenized spleens from 3- to 4-week-old chickens (INRA, Lyon, France). Monocytes were removed by adherent cell depletion. Duplicates of 105 lymphocyte-enriched splenocytes were cultured in 96-well plates and 200 μL medium for 5 days with an increasing number of stimulating cells that had been pretreated for 45 minutes with 25 μg/mL mitomycin C (Sigma-Aldrich). Cells were labeled for 16 hours with [3H]-thymidine (1 μCi (0.037 MBq)/well; Perkin-Elmer, Courteboeuf, France) and lysed in H2O. Insoluble material was collected onto fiberglass filter paper (Packard Instrument), and radioactivity was quantified as cpm with a beta counter (Wallac Oy 1450 Microbeta, Perkin-Elmer). Phagocytocis assays were performed as previously described.14
Reverse transcriptase–polymerase chain reaction
Total RNA was extracted from cells using RNeasy Mini Kit (Qiagen, Hilden, Germany) and DNase treated (RNase free DNase Roche, Meylon, France). One microgram of total RNA was denatured 5 minutes at 65°C and reverse transcribed with SuperScript II (Life Technologies, Bethesda, MD) for 50 minutes at 42°C in a final volume of 20 μL as specified by the manufacturers. PCR amplifications were performed using 2 μL of the reverse transcription products in 25 μL of reaction with puReTaq Ready-to-go PCR Beads (Amersham) for 3 minutes at 94°C followed by 30 (MafB) or 18 (β-actin) cycles of 45 seconds at 94°C, 45 seconds at 55°C (β-actin) or 65°C (MafB), and 2 minutes at 72°C and terminated by 10 minutes at 72°C. PCR products were resolved on agarose gels containing ethidium bromide. Primers used: MafB, 5′CCCGGCTGGCCCGCGAGAGAG3′/5′CTAGGAGGCGGCGCAGGCGT3′; β-actin, 5′TACCACTGGCATCGTGATGGACT3′/5′TCCTTCTGCATCCTGTCGGCAAT3′.
Protein interaction and Western blot analysis
GST pull-down assays were performed as described.14 Proteins were extracted from purified monocytes and in vitro–differentiated DCs or macrophages from the same donors. Western analysis was performed as described.14 Primary antibodies were mouse anti–human PU.1 (1:400) and mouse antitubulin (1:20 000; Sigma).
Transient transfection assays
Qt6 cells or HD11 macrophages were seeded at 2.5 × 105 cells/35 mm plate and transfected by CaPo4 precipitation and lipofectamin 2000 (Life Technologies), respectively, as described.14 Luciferase activity was assayed 48 hours after transfection and normalized to β-galactosidase activity from 0.5 μg cotransfected RSV-β-gal plasmid.14 PU.1 was expressed from the cytomegalovirus (CMV) promoter in RC/CMV (Life Technologies). The Maf-responsive luciferase reporter construct has been described.14
Results
Activation of PU.1 in E26 myeloblast clones induces DC morphology and marker expression
To assess the transcriptional basis of differentiation options in myeloid progenitor cells, we have used a differentiation system taking advantage of inducible transcription factor activation in a homogeneous clonal population of primary cells. Avian myeloblasts transformed by the E26 retrovirus have an extended self-renewal capacity and can be maintained in culture for several weeks but are not immortalized nor cytogenetically abnormal.14 Since gene inactivation studies in mice suggested a role for PU.1 in late myeloid differentiation and gain of function experiments indicated that PU.1 function is strongly context dependent,13,21-24,28-32 we wanted to assess the consequence of PU.1 activation in myeloblasts. For this purpose we infected yolk sac hematopoietic progenitors with a bicistronic vector expressing a β-estradiol (βE)–inducible fusion protein of PU.1 (E26ts21-PUER) or control virus (E26ts21) and plated the cells in methylcellulose medium. Individual expanded myeloblast colonies had an indistinguishable myeloblast phenotype in control and PUER clones, as judged by surface marker expression and dependence on the avian myeloid cytokine cMGF.14 All studied PUER clones expressed the transgene (data not shown).
To analyze the effect of PU.1 activation in avian myeloblasts, we cultured equal numbers of cells from control and PUER clones either in absence or presence of 1 μM βE. As early as 24 hours after PU.1 activation, we observed dramatic morphological changes in the βE-treated PUER-expressing cells. By 48 hours of βE treatment, they had assumed an irregular-shaped morphology, reminiscent of immature veiled DC as judged by phase contrast (Figure 1D) and MGG staining (Figure 1H). No changes were observed in untreated PUER-expressing cells (Figure 1C,G) or in control cells irrespective of βE treatment (Figure 1A-B,E-F). To test whether these morphological changes were indeed due to DC differentiation, we analyzed the cells for expression of DC markers. Most myeloid cells express low levels of MHCII on the plasma membrane, but immature DCs typically show high levels of intracellular MHCII in lysosomes.33 As expected for myeloblasts, both control (Figure 1I,J) and untreated PUER clones (Figure 1K) expressed low levels of MHCII on the cell surface but not intracellularly. In contrast, immunofluorescence on βE-treated PUER clones revealed typical intracellular MHCII-positive vesicles in most cells after PU.1 activation (Figure 1L). To further document the DC phenotype, we also tested the expression of the DC-specific calcium binding protein S100.34 Nearly all cells from βE-treated PUER clones (Figure 1P) but none from untreated (Figure 1O) or control clones (Figure 1M-N) were positive for S100. The analysis of a total of 6 individual PUER clones in several separate experiments consistently revealed the same changes in phenotype and marker expression after βE induction (Figure 1Q). Together, these results indicated that activation of PU.1 in myeloblasts stimulated DC differentiation.
Activation of PU.1 in primary myeloblast clones induces DC phenotype. Clonal E26ts21-PUER virus (C-D, G-H, K-L, O-P)– or E26ts21 control virus (A-B, E-F, I-J, M-N)–transformed chicken myeloblasts were cultured for 48 hours either in the absence (-βE; A, C, E, G, I, K, M, O) or presence of 1 μM βE(+βE; B, D, F, H, J, L, N, P). (A-D) Phase contrast micrographs of cell cultures visualized under a Leica DMiL optical microscope equipped with a 40×/0.5 objective lens (Leica, Wetzlar, Germany). (E-H) May-Grünwald-Giemsa staining (MGG) of cytocentrifuged cells (shown at original magnification, ×40). (I-L) Immunofluorescence detection of MHCII with an FITC-conjugated secondary antibody. Images were viewed under a Leica DMRBE fluorescence microscope equipped with a 40×/1.0 objective lens (Leica) and a Nikon DVM1200 digital camera (Nikon, Champigny sur Marne, France). Lucia software version 4.61 was used for image processing (Nikon). (M-P) Immunofluorescence detection of S100 with an FITC-conjugated secondary antibody. Nuclei were counterstained with DAPI. (Q) Cells containing MHCII lysosomal vesicles (▪)or staining positive for S100 (▦) were counted for a total of at least 100 cells in 6 independent PUER clones cultured with or without βE.
Activation of PU.1 in primary myeloblast clones induces DC phenotype. Clonal E26ts21-PUER virus (C-D, G-H, K-L, O-P)– or E26ts21 control virus (A-B, E-F, I-J, M-N)–transformed chicken myeloblasts were cultured for 48 hours either in the absence (-βE; A, C, E, G, I, K, M, O) or presence of 1 μM βE(+βE; B, D, F, H, J, L, N, P). (A-D) Phase contrast micrographs of cell cultures visualized under a Leica DMiL optical microscope equipped with a 40×/0.5 objective lens (Leica, Wetzlar, Germany). (E-H) May-Grünwald-Giemsa staining (MGG) of cytocentrifuged cells (shown at original magnification, ×40). (I-L) Immunofluorescence detection of MHCII with an FITC-conjugated secondary antibody. Images were viewed under a Leica DMRBE fluorescence microscope equipped with a 40×/1.0 objective lens (Leica) and a Nikon DVM1200 digital camera (Nikon, Champigny sur Marne, France). Lucia software version 4.61 was used for image processing (Nikon). (M-P) Immunofluorescence detection of S100 with an FITC-conjugated secondary antibody. Nuclei were counterstained with DAPI. (Q) Cells containing MHCII lysosomal vesicles (▪)or staining positive for S100 (▦) were counted for a total of at least 100 cells in 6 independent PUER clones cultured with or without βE.
Activation of MafB or PU.1 in E26 myeloblasts induces alternative macrophage or DC fates
Since murine hematopoietic progenitors infected with a PU.1 retrovirus had been shown previously to give rise to macrophages under appropriate conditions,21,23 we expected that activation of PU.1 in myeloblasts might also induce macrophage differentiation. To investigate this possibility, we assessed macrophage and DC phenotype in avian myeloblast clones after PU.1 activation (E26ts21-PUER) or overexpressing the bZIP factor MafB (E26ts21-MafB), which we had previously shown to induce macrophage differentiation in this system.14 MafB-induced macrophages were adherent, had a typical vacuolated phenotype (Figure 2D), and expressed the avian monocyte/macrophage marker 47.8327 (Figure 2E). In contrast, cells differentiated by PU.1 activation had a nonvacuolated phenotype with dendritic protrusions (Figure 2A) and were negative for 47.83 (Figure 2B). Quantification of cells with macrophage or DC morphology in 3 independent clones each revealed nearly complete differentiation and no subpopulation of DCs in MafB-induced macrophages (Figure 2C) and no macrophages in the PU.1-induced DC clones (Figure 2F). In contrast to PU.1-induced DCs (Figure 1), MafB-differentiated macrophages also were negative for intracellular MHCII lysosomal vesicles and showed no significant S100 expression (data not shown). Together these results indicate that MafB or PU.1 activation in E26 myeloblasts induce alternative macrophage or DC differentiation without mixed-lineage phenotypes.
MafB and PU.1 induce alternative macrophage or DC phenotype and function in myeloblasts. E26ts21 MafB–expressing myeloblasts (MafB) were induced to differentiate for 48 hours by temperature shift to 42°C (which releases a vMyb-imposed differentiation block), E26ts21-PUER (PU.1), and E26ts21-control (ctrl) were induced by 1 μM βE treatment. (A,D) May-Grünwald-Giemsa (MGG) staining of cytocentrifuged cells (objective lens ×40). (B,E) FACS analysis for the chicken monocyte/macrophage marker 47.83. (C,F) Cells with DC (▪) or macrophage (MΦ) morphology (▦) were counted for a total of at least 100 cells in 3 independent PUER (C) and MafB (F) clones from 2 separate experiments. (G) Mixed lymphocyte reaction (MLR) of myeloid cells differentiated by PU.1 or MafB activation. Ctrl, PU.1, and MafB clones were induced (ind.) or not induced (–) to differentiate, treated with 25 μg/mL mitomycin C, and incubated in duplicates in the absence (–) or presence (spleno.) of 105 monocyte-depleted allogenic splenocytes at different stimulating cell–splenocyte ratios for 5 days. [3H]-thymidine incorporation was measured after 16 hours of labeling. (H) Stimulating activity of 6 independent clones in 3 different experiments was analyzed and is expressed as proliferation index at a stimulating cell–splenocyte ratio of 1. (I) Phagocytic capacity of myeloid cells differentiated by PU.1 or MafB activation. Induced ctrl, PU.1, and MafB clones were incubated for 2 hours with phycoerythrin (PE)–conjugated latex beads and analyzed by FACS. Phagocytic activity before and after induction of differentiation was determined for 6 independent clones in 3 different experiments and expressed as the product of percent positive cells (as a measure of phagocytosing cells) and their mean fluorescence intensity (as a measure of the number of phagocytosed beads). In H-I, ▦ indicates uninduced cells; ▪, induced. Error bars indicate the standard error of the mean. Images in panels A and D were acquired using a Leica DMiL optical microscope equipped with a 40×/1.0 objective lens and a Leica MPS30 camera. Contrast and exposure were enhanced with Adobe Photoshop 5.0 software (Adobe, San Jose, CA) using equal treatment on all panels.
MafB and PU.1 induce alternative macrophage or DC phenotype and function in myeloblasts. E26ts21 MafB–expressing myeloblasts (MafB) were induced to differentiate for 48 hours by temperature shift to 42°C (which releases a vMyb-imposed differentiation block), E26ts21-PUER (PU.1), and E26ts21-control (ctrl) were induced by 1 μM βE treatment. (A,D) May-Grünwald-Giemsa (MGG) staining of cytocentrifuged cells (objective lens ×40). (B,E) FACS analysis for the chicken monocyte/macrophage marker 47.83. (C,F) Cells with DC (▪) or macrophage (MΦ) morphology (▦) were counted for a total of at least 100 cells in 3 independent PUER (C) and MafB (F) clones from 2 separate experiments. (G) Mixed lymphocyte reaction (MLR) of myeloid cells differentiated by PU.1 or MafB activation. Ctrl, PU.1, and MafB clones were induced (ind.) or not induced (–) to differentiate, treated with 25 μg/mL mitomycin C, and incubated in duplicates in the absence (–) or presence (spleno.) of 105 monocyte-depleted allogenic splenocytes at different stimulating cell–splenocyte ratios for 5 days. [3H]-thymidine incorporation was measured after 16 hours of labeling. (H) Stimulating activity of 6 independent clones in 3 different experiments was analyzed and is expressed as proliferation index at a stimulating cell–splenocyte ratio of 1. (I) Phagocytic capacity of myeloid cells differentiated by PU.1 or MafB activation. Induced ctrl, PU.1, and MafB clones were incubated for 2 hours with phycoerythrin (PE)–conjugated latex beads and analyzed by FACS. Phagocytic activity before and after induction of differentiation was determined for 6 independent clones in 3 different experiments and expressed as the product of percent positive cells (as a measure of phagocytosing cells) and their mean fluorescence intensity (as a measure of the number of phagocytosed beads). In H-I, ▦ indicates uninduced cells; ▪, induced. Error bars indicate the standard error of the mean. Images in panels A and D were acquired using a Leica DMiL optical microscope equipped with a 40×/1.0 objective lens and a Leica MPS30 camera. Contrast and exposure were enhanced with Adobe Photoshop 5.0 software (Adobe, San Jose, CA) using equal treatment on all panels.
PU.1- and MafB-differentiated cells express alternative DC or macrophage functions
We wanted to assess whether PU.1-induced DCs and MafB-induced macrophage phenotypes were also reflected in typical DC or macrophage functions.
A characteristic functional hallmark of DCs is their ability to stimulate a strong proliferative response of allogenic T cells in a mixed lymphocyte reaction (MLR). To test whether PU.1-induced DCs have the capacity to stimulate T cells, we incubated increasing numbers of cells from a PUER clone that had been cultured in the absence or presence of βE for 2 days with 105 splenocytes from an allogenic chicken. At a stimulating cell–splenocyte ratio of 1:10 to 1:100, splenocyte proliferation was induced at a level similar to the response elicited by MafB-induced macrophages at a stimulating cell–splenocyte ratio of 1:1 (Figure 2G, PU.1, MafB). At this ratio PU.1-induced DCs elicited a 12-fold stronger response than MafB macrophages. βE-treated control cells or untreated PUER cells induced no significant T-cell proliferation (Figure 2G, ctrl, PU.1). Comparable results were obtained with 6 different clones (Figure 2H).
As a measure of macrophage function, we assessed phagocytic activity in PU.1-induced DCs and MafB-induced macrophages by incubating the cells with fluorescent beads and quantifying phagocytosed beads in FACS analysis. Whereas in comparison to control myeloblasts PU.1-induced DCs showed only a small increase in their phagocytic ability, as expected for immature DCs,26 MafB-induced clones had a dramatically higher phagocytic activity that is typical of macrophages (Figure 2I).
Together, these results indicate that PU.1 activation induced differentiation of functional DCs with a high capacity to simulate T cells in an allogenic MLR, but low phagocytic activity, whereas MafB overexpression resulted in the differentiation of typical functional macrophages with low antigen presenting but high phagocytic activity.
PU.1-induced DCs have typical phenotypic and functional maturation capacity
Upon activation, which can be mimicked in culture by lipopolysaccharide (LPS), immature DCs undergo dramatic phenotypic and functional changes.35 They rapidly shuttle MHCII molecules from lysosomes to the cell surface,33 develop numerous dendritic protrusions, and change their cell-cell adhesion properties.35 By FACS analysis we observed that LPS activation of βE-treated PUER cells resulted in an increased surface expression of MHCII (Figure 3B-C). This observation was confirmed by immunofluorescence, which revealed a relocalization of MHCII from intracellular vesicles to the cell surface and development of large dendrites after LPS treatment of PU.1-induced DCs (Figure 3E-F). Furthermore, LPS treatment of βE-induced PUER clones resulted in the DC typical clustering in culture (Figure 3H-I). None of these LPS effects were observed in βE-treated control clones, untreated PUER clones, or in MafB-induced macrophage clones (data not shown). Mature DCs also have an enhanced capacity to stimulate T cells compared to nonactivated DCs. We observed that LPS activation of βE-treated PUER cells indeed resulted in an increased capacity to stimulate allogenic T cells in an MLR as compared to nonactivated PU.1-induced DCs or to LPS-activated MafB-induced macrophages (Figure 3J). On the other hand, LPS stimulation of MafB macrophages induced macrophage typical release of NO, whereas LPS treatment of PU.1-induced DCs did not (data not shown and Kelly et al14 ). These results indicated that PU.1-induced DCs are capable of characteristic phenotypic and functional DC maturation and show no macrophage-typical response to LPS challenge.
PU.1-induced DCs are capable of maturation in response to LPS. E26ts21-PUER clones were cultured for 72 hours in the absence (A,D,G) or presence (B,E,H) of 1 μM βE, or treated for the last 48 hours with 10 ng/mL LPS in addition (C,F,I). (A-C) FACS analysis of MHCII expression on the cell surface. (D-F) Immunofluorescence detection of MHCII by confocal microscopy. Note weak surface expression and round morphology of myeloblasts (D), lysosomal MHCII vesicles in veiled immature DCs (E), and strong surface expression on prolonged membrane protrusion (×40) typical of mature DCs (F). Images in panels D-F were obtained using a Zeiss Axiovert-200 confocal microscope equipped with a 63×/1.4 objective lens and an integrated camera (Zeiss, Jena, Germany). LSM 510 version 3.2 software was used for image acquisition. Contrast and exposure were enhanced through Adobe Photoshop 5.0 using equal treatment on all panels. (G-H) Phase-contrast images of the cells in culture were obtained with a confocal Zeiss Axiovert 200 microscope equipped with a motorized 63×/1.4 objective lens. Images were acquired with LSM10 software version 3.2 (Zeiss, Jena, Germany). Note the formation of clusters typical of DC maturation after LPS stimulation (I). (J) Mixed lymphocyte reaction with PUER (▪) or MafB clones (▦) induced (ind.) to differentiate by βE addition or temperature shift, respectively, and in the absence or presence of LPS stimulation as indicated, at a stimulating cell–splenocyte ratio of 0.3 and expressed as proliferation index. Data are representative of 3 independent experiments.
PU.1-induced DCs are capable of maturation in response to LPS. E26ts21-PUER clones were cultured for 72 hours in the absence (A,D,G) or presence (B,E,H) of 1 μM βE, or treated for the last 48 hours with 10 ng/mL LPS in addition (C,F,I). (A-C) FACS analysis of MHCII expression on the cell surface. (D-F) Immunofluorescence detection of MHCII by confocal microscopy. Note weak surface expression and round morphology of myeloblasts (D), lysosomal MHCII vesicles in veiled immature DCs (E), and strong surface expression on prolonged membrane protrusion (×40) typical of mature DCs (F). Images in panels D-F were obtained using a Zeiss Axiovert-200 confocal microscope equipped with a 63×/1.4 objective lens and an integrated camera (Zeiss, Jena, Germany). LSM 510 version 3.2 software was used for image acquisition. Contrast and exposure were enhanced through Adobe Photoshop 5.0 using equal treatment on all panels. (G-H) Phase-contrast images of the cells in culture were obtained with a confocal Zeiss Axiovert 200 microscope equipped with a motorized 63×/1.4 objective lens. Images were acquired with LSM10 software version 3.2 (Zeiss, Jena, Germany). Note the formation of clusters typical of DC maturation after LPS stimulation (I). (J) Mixed lymphocyte reaction with PUER (▪) or MafB clones (▦) induced (ind.) to differentiate by βE addition or temperature shift, respectively, and in the absence or presence of LPS stimulation as indicated, at a stimulating cell–splenocyte ratio of 0.3 and expressed as proliferation index. Data are representative of 3 independent experiments.
Activation of PU.1 is instructive for human DC differentiation in the absence of DC-promoting cytokines
To analyze whether our results were transferable to human cells, we used the human promyelocytic cell line HL-60, which is similar to avian E26 myeloblasts,14 in that it can be induced to differentiate to macrophages or granulocytes,36 and has no cytokine requirements for its growth or differentiation. To obtain clonal PUER-expressing populations of these cells, we constructed a mammalian retrovirus expressing PUER followed by an IRES-driven GFP cassette (MFG-PUERiGFP) and a GFP-only control virus (MFG-iGFP; Figure 4A). After infection, GFP-expressing cells were enriched by FACS sorting and used to isolate individual PUER-iGFP- and iGFP-infected clones that were verified to still express an inducible PUER transgene (data not shown). After 6 days of induction with 1 μM βE almost all the cells from PUER-iGFP, but not from control clones, acquired a typical DC morphology (Figure 4B). Further characterization showed that these cells expressed the human DC marker CD1a and had up-regulated CD11b, an antigen expressed on several mature myeloid cells, including monocytes/macrophages and DCs, as well as the DC costimulatory molecule CD86, but did not express langerin, a marker of Langerhans cell (LC)–type DCs (Figure 4C). Furthermore, the obtained DCs showed a typical maturation phenotype, as demonstrated by the up-regulation of the activation antigens CD40 and CD83 after LPS stimulation for 48 hours (Figure 4D).
Activation of PU.1 in human promyelocytic cells is instructive for DC fate in the absence of cytokines. Human HL-60 cells were infected with a PUER-expressing (MFG-PUERiGFP) or control retrovirus (MFG-iGFP) and individual clones were isolated (“Materials and methods”). (A) Schematic representation of used retroviral vectors. The constructs are based on an MFG vector containing an IRES-eGFP cassette. LTR indicates long terminal repeat; hER, human estrogen receptor hormone binding domain. (B) Phase contrast images of HL-60 clones expressing PUERiGFP or iGFP control retroviruses and cultured for 6 days in the absence (-βE) or presence (+βE) of 1 μM βE. (C) FACS analysis of surface antigens CD1a, CD11b, CD86, and Langerin on an HL-60 clone expressing PUERiGFP and cultured for 6 days in the absence (-βE) or presence (+βE) of 1 μM βE. (D) HL60PUER clones were induced by 1 μM βE treatment to differentiate into DCs for 48 hours then cultured in the presence or absence of 20 ng/mL LPS for an additional 48 hours under the continuous presence of βE. Expression of the activation antigens CD40 and CD83 was analyzed by FACS. The average percentage of positive cells of 6 independent clones was plotted. Error bars indicate standard error of the mean. (E) Kinetics of DC differentiation after βE treatment. PUERiGFP-expressing clones were cultured in the absence (○) or presence (▪)of1 μM βE and stained for the expression of CD1a at the indicated time points after treatment. The average percentage of CD1a-positive cells of 4 independent clones was plotted. Error bars indicate standard error of the mean. (F) FACS profiles of CD1a and GFP expression in an iGFP and PUERiGFP HL-60 clone stimulated for the indicated number of days with 1 μM βE.
Activation of PU.1 in human promyelocytic cells is instructive for DC fate in the absence of cytokines. Human HL-60 cells were infected with a PUER-expressing (MFG-PUERiGFP) or control retrovirus (MFG-iGFP) and individual clones were isolated (“Materials and methods”). (A) Schematic representation of used retroviral vectors. The constructs are based on an MFG vector containing an IRES-eGFP cassette. LTR indicates long terminal repeat; hER, human estrogen receptor hormone binding domain. (B) Phase contrast images of HL-60 clones expressing PUERiGFP or iGFP control retroviruses and cultured for 6 days in the absence (-βE) or presence (+βE) of 1 μM βE. (C) FACS analysis of surface antigens CD1a, CD11b, CD86, and Langerin on an HL-60 clone expressing PUERiGFP and cultured for 6 days in the absence (-βE) or presence (+βE) of 1 μM βE. (D) HL60PUER clones were induced by 1 μM βE treatment to differentiate into DCs for 48 hours then cultured in the presence or absence of 20 ng/mL LPS for an additional 48 hours under the continuous presence of βE. Expression of the activation antigens CD40 and CD83 was analyzed by FACS. The average percentage of positive cells of 6 independent clones was plotted. Error bars indicate standard error of the mean. (E) Kinetics of DC differentiation after βE treatment. PUERiGFP-expressing clones were cultured in the absence (○) or presence (▪)of1 μM βE and stained for the expression of CD1a at the indicated time points after treatment. The average percentage of CD1a-positive cells of 4 independent clones was plotted. Error bars indicate standard error of the mean. (F) FACS profiles of CD1a and GFP expression in an iGFP and PUERiGFP HL-60 clone stimulated for the indicated number of days with 1 μM βE.
To further investigate whether PU.1 was indeed instructive for DC differentiation, we studied the kinetics of the differentiation process. We observed that βE induction of PUER-iGFP but not of iGFP-virus–infected HL-60 cells resulted in continuous up-regulation of CD1a up to 74% positive cells after 6 days of induction in 6 different individual clones (Figure 4E). Close analysis of FACS profiles at different time points of induction revealed that CD1a-positive cells arose from the continuous up-regulation of the antigen in a previously negative population rather than from the increase of a small pre-existing CD1a-positive population (Figure 4F). This was strong evidence that activation of PU.1 is instructive for DC differentiation rather than selecting for the outgrowth of a pre-existing small population of precommitted DCs.
Together, these results indicated that in multipotent human myeloid cells PU.1 activation is instructive for DC differentiation in the absence of cytokines.
Activation of PU.1 in monocytes induces DC differentiation
Monocytes exiting the bloodstream can differentiate to a significant extent toward DCs,3 a differentiation pathway that plays a key role in the coordination of the innate and adaptive immune response, and can be disturbed under several pathological conditions (Introduction). A number of cell culture conditions have been established that mimic this option and induce differentiation of blood monocytes to DCs in vitro.1 We therefore wanted to determine whether activation of PU.1 can also redirect monocytes into a DC differentiation pathway.
HL-60 cells have been reported to differentiate into monocytes in the presence of vitamin D3 (D3).37 We used this protocol to derive PUER-iGFP–infected HL-60 monocytes. After 20 hours in 100 nM D3 nearly all cells from PUER-iGFP clones had a monocyte phenotype as verified by morphology, the induction of CD11b, nonspecific esterase (NSE) activity, and the absence of the DC-specific marker CD1a (data not shown). To test whether these monocytes could be diverted into a DC differentiation pathway by PU.1 activation, we incubated them under D3 conditions in the absence or presence of βE and monitored DC marker expression after 5 days of culture. Whereas expression of CD11b, a marker present on both monocytes/macrophages and DCs, did not change after PU.1 induction (Figure 5A), PUER monocytes rapidly assumed DC morphology after βE treatment (data not shown) and expressed the DC markers CD1a (Figure 5B) and S100 (Figure 5C-D).
PU.1 activation redirects monocytic differentiation toward DC fate. Monocyte/macrophage differentiation of HL-60 PUER clones was induced by treatment with 100 nM 1α,25-dihydroxyvitaminD3 (D3) for 20 hours. Results are representative of 2 separate experiments with 4 independent clones. (A-B) HL-60 PUER monocyte clones were either stimulated (+; ▪) or not stimulated (–; ▦) for 5 days with 1 μM βE in the continuous presence of D3. Cells were analyzed for the expression of CD11b (A) and CD1a (B) by FACS. The average for 4 independent clones is shown. Error bars indicate standard error of the mean. (C-D) Unstimulated (C) or 5-day βE-stimulated (D) HL-60 PUER monocytes maintained in the continuous presence of 100 nM D3 were analyzed for the expression of S100 protein by immunofluorescence with Alexa Red secondary antibody. Nuclei were counterstained with DAPI. (E) Primary human monocytes (Mo) were differentiated to macrophages (MΦ) or DCs with M-CSF or GM-CSF and IL-4, respectively. Western blot analysis for PU.1 expression was performed on total cell extracts from 2 different donors after 7 days of culture. (F) Monocytes were subjected to DC differentiation conditions as in panel E, collected at different time points of culture, and analyzed for expression of PU.1 by Western blot. Tubulin expression was used as internal control for protein concentration of Western extracts in panels E and F. (G) Monocytes in panel F were analyzed for expression of the DC marker CD1a and the monocyte/macrophage marker CD14 antigens by FACS staining. (H) Kinetics of CD1a, CD14, and PU.1 levels during monocyte-to-DC differentiation. CD1a (▴) and CD14 (▪) are represented as percent positive cells and PU.1 levels (○) as percent of maximal expression (16 hours) normalized to tubulin.
PU.1 activation redirects monocytic differentiation toward DC fate. Monocyte/macrophage differentiation of HL-60 PUER clones was induced by treatment with 100 nM 1α,25-dihydroxyvitaminD3 (D3) for 20 hours. Results are representative of 2 separate experiments with 4 independent clones. (A-B) HL-60 PUER monocyte clones were either stimulated (+; ▪) or not stimulated (–; ▦) for 5 days with 1 μM βE in the continuous presence of D3. Cells were analyzed for the expression of CD11b (A) and CD1a (B) by FACS. The average for 4 independent clones is shown. Error bars indicate standard error of the mean. (C-D) Unstimulated (C) or 5-day βE-stimulated (D) HL-60 PUER monocytes maintained in the continuous presence of 100 nM D3 were analyzed for the expression of S100 protein by immunofluorescence with Alexa Red secondary antibody. Nuclei were counterstained with DAPI. (E) Primary human monocytes (Mo) were differentiated to macrophages (MΦ) or DCs with M-CSF or GM-CSF and IL-4, respectively. Western blot analysis for PU.1 expression was performed on total cell extracts from 2 different donors after 7 days of culture. (F) Monocytes were subjected to DC differentiation conditions as in panel E, collected at different time points of culture, and analyzed for expression of PU.1 by Western blot. Tubulin expression was used as internal control for protein concentration of Western extracts in panels E and F. (G) Monocytes in panel F were analyzed for expression of the DC marker CD1a and the monocyte/macrophage marker CD14 antigens by FACS staining. (H) Kinetics of CD1a, CD14, and PU.1 levels during monocyte-to-DC differentiation. CD1a (▴) and CD14 (▪) are represented as percent positive cells and PU.1 levels (○) as percent of maximal expression (16 hours) normalized to tubulin.
To investigate whether PU.1-induced DC differentiation of monocytes reflects the normal transcriptional events during cytokine-mediated differentiation of primary monocytes, we isolated monocytes from human PBMCs and subjected them alternatively to a macrophage or DC differentiation protocol with M-CSF or GM-CSF and IL-4, respectively. The cultures obtained after 7 days of differentiation were lysed and subjected to Western blot analysis. Comparing cells derived from the same donor, PU.1 expression was very low in monocytes, moderately increased in macrophages, and dramatically up-regulated in differentiated DCs (Figure 5E). The same differences in expression levels were already seen after 24 and 48 hours of culture and consistently observed in cells differentiated from 7 different blood donors (data not shown). To test whether this up-regulation of PU.1 in DCs was a consequence of the differentiated phenotype or preceded it, as would be expected in the case of an instructive role, we assessed PU.1 and surface-marker expression at various times of culture under DC differentiation conditions. We observed that PU.1 levels were already increased as early as 6 hours and reached high levels at 16 hours of incubation under DC conditions (Figure 5F-H), whereas the DC surface marker CD1a was not expressed at 6 hours, weakly positive at 16 hours, and only fully expressed at 48 hours (Figure 5G-H), and the monocyte/macrophage marker CD14 only started to be down-regulated at 48 hours (Figure 5G-H). This indicated that a dramatic increase in PU.1 levels is an early event of normal monocyte-to-DC differentiation that precedes the expression of the DC phenotype.
Together, these results suggested that induction of high PU.1 activity in monocytes instructs DC differentiation.
MafB repression is required for DC differentiation
MafB is expressed at high levels in avian and mammalian monocytes and macrophages,38,39 and we have shown here that MafB-directed monocyte/macrophage fate or PU.1-directed DC fate appear to be alternative and exclusive differentiation options (Figure 2). Consistent with these results, we observed high levels of MafB mRNA in normal human monocyte-derived macrophages (Figure 6A lane 1) but not in DCs (Figure 6A lane 2). MafB mRNA also was undetectable in uninduced HL-60 PUER clones or in DCs generated by PU.1 induction (Figure 6B lanes 1-2). We therefore hypothesized that PU.1-induced DC differentiation involves inhibition of MafB expression and/or activity. To test this hypothesis, we induced PU.1 in HL-60 PUER monocyte clones. Interestingly, the high levels of MafB mRNA expressed by the HL-60–derived monocytes (Figure 6B lane 3) were strongly repressed by activation of DC-inducing levels of PU.1 (Figure 6B lane 4).
MafB repression is required for DC differentiation. (A) MafB expression was monitored by RT-PCR in blood PBMC-derived macrophages (MΦ) and DCs. Data are representative of at least 2 separate experiments; actin RT-PCR was used as a normalization control. (B) MafB expression was monitored by RT-PCR in untreated promyelocytic HL-60 PUER clones (Myel) (lane 1) or differentiated to DC by βE treatment (lane 2), monocytic HL-60 PUER clones (Mo) cultured in the presence of D3 in the absence of βE (lane 3), or redirected to DC fate by βE-induced PU.1 activation (lane 4). Culture conditions were as described for Figure 5. (C-D) Constitutive MafB expression inhibits DC differentiation. FACS analysis of CD1a, CD80, and CD86 DC marker expression in GFP+ and GFP– cells of MFG-MafB-iGFP virus (MafB) or MFG-iGFP control viruses (ctrl) infected HL-60 cells 96 hours after induction of DC differentiation with 200 ng/mL A23187 calcium ionophore (C). Percentages of positive cells per quadrant are indicated. Differential DC marker expression in infected (GFP+, ▪) and uninfected (GFP–, ▦) cells was plotted as ratio of positive-to-negative cells for MafB virus and control virus-infected cultures (D).
MafB repression is required for DC differentiation. (A) MafB expression was monitored by RT-PCR in blood PBMC-derived macrophages (MΦ) and DCs. Data are representative of at least 2 separate experiments; actin RT-PCR was used as a normalization control. (B) MafB expression was monitored by RT-PCR in untreated promyelocytic HL-60 PUER clones (Myel) (lane 1) or differentiated to DC by βE treatment (lane 2), monocytic HL-60 PUER clones (Mo) cultured in the presence of D3 in the absence of βE (lane 3), or redirected to DC fate by βE-induced PU.1 activation (lane 4). Culture conditions were as described for Figure 5. (C-D) Constitutive MafB expression inhibits DC differentiation. FACS analysis of CD1a, CD80, and CD86 DC marker expression in GFP+ and GFP– cells of MFG-MafB-iGFP virus (MafB) or MFG-iGFP control viruses (ctrl) infected HL-60 cells 96 hours after induction of DC differentiation with 200 ng/mL A23187 calcium ionophore (C). Percentages of positive cells per quadrant are indicated. Differential DC marker expression in infected (GFP+, ▪) and uninfected (GFP–, ▦) cells was plotted as ratio of positive-to-negative cells for MafB virus and control virus-infected cultures (D).
To test whether this MafB down-regulation was required for DC differentiation rather than a mere consequence of it, we tested the effect of constitutively expressed MafB on the induction of DC differentiation. Toward this end, we infected HL-60 cells with an MFG retrovirus expressing a MafB-iresGFP cassette (MafB-iGFP) or an iresGFP control virus (iGFP) and treated the cells with the calcium ionophore A23187, which has been previously shown to induce DC differentiation in HL-60 cells.40 We monitored the effect of viral transgene expression on DC differentiation by comparing the expression of the DC markers CD1a, CD80, and CD86 in infected GFP+ cells and noninfected GFP– cells. Whereas in iGFP control virus-infected cultures these markers were expressed at equal or higher levels in GFP+ cells than in GFP– cells, in the MafB-iGFP–infected culture, the expression of DC markers was strongly inhibited in GFP+ cells compared to GFP– cells (Figure 6C-D). This provided strong evidence that expression of the monocyte/macrophage transcription factor MafB interferes with DC differentiation and suggested that its down-regulation is essential for the redirection of monocyte/macrophage fate toward a DC differentiation pathway.
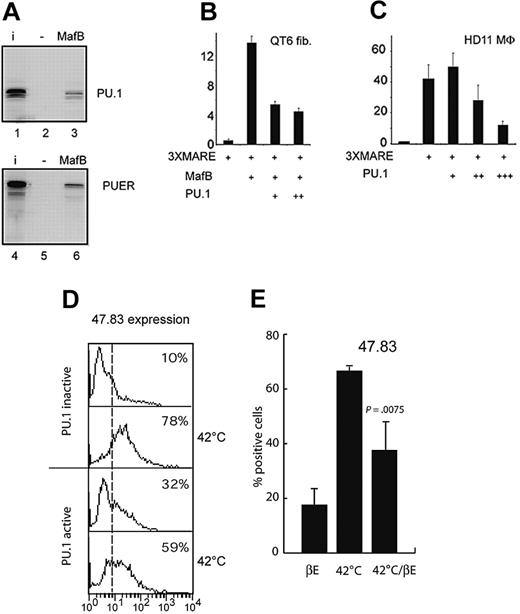
PU.1 directly binds to MafB and inhibits its function
To further elucidate the mechanism of MafB repression during DC differentiation, we tested whether PU.1 could directly interfere with MafB activity. Toward this end, we initially analyzed whether PU.1 physically interacted with MafB. As shown in Figure 7A, in vitro–translated full-length PU.1 bound to GST-MafB fusion protein but not to a GST-only control, an interaction potential that also was conserved for the PUER fusion protein used in this study. Since we had previously shown that MafB did not interact with the PU.1 DNA binding domain,14 the interaction surface must be in the N-terminal region of PU.1. To test the functional consequence of this interaction, we analyzed the effect of PU.1 on MafB transactivation activity. Toward this end, we cotransfected expression constructs for PU.1 and MafB together with a Maf-responsive reporter plasmid into QT6 fibroblasts. PU.1 inhibited MafB transactivation on a Maf response element (MARE) reporter by several-fold (Figure 7B), whereas MafB had no effect on PU.1 transactivation (data not shown). Furthermore, high PU.1 levels also could inhibit endogenous Maf activity in HD11 macrophages (Figure 7C), which express high levels of MafB.39 To test the biologic consequences of MafB inhibition by PU.1, we took advantage of the chicken E26ts21-PUER clones, where independent inducible control of Maf and PU.1 activities is possible. We have shown previously that at the nonpermissive temperature (42°C) endogenous MafB expression is strongly and rapidly up-regulated in ts21E26 myeloblasts39 and directs macrophage differentiation in the system (Figure 2).14 Consistent with the observed inhibition of MafB activity by PU.1, we observed that temperature shift–induced macrophage differentiation was significantly reduced in E26ts21-PUER clones, when cells were pretreated with βE and thus contained active PU.1 (Figure 7D). To exclude that this effect was due to the inability of MafB to overcome commitment to the DC pathway rather than to direct PU.1 inhibition, we activated MafB and PU.1 at the same time by simultaneously shifting the temperature to 42°C and adding βE. Also in this case we observed significantly reduced macrophage differentiation when PU.1 was activated (Figure 7E). Together, these results indicate that PU.1 can directly inhibit MafB transactivation capacity and macrophage-inducing function and suggest that the balance of the 2 factors is important for the ability of PU.1 to direct DC versus macrophage fate.
PU.1 binds to MafB and inhibits its activity. (A) Analysis of PU.1 interaction with MafB in GST pull-down assays. In vitro–translated 35S-methionine-labeled PU.1 (lanes 1-3) or PUER (lanes 4-6) were incubated with an affinity matrix-bound GST-fusion protein of full-length MafB (lanes 3 and 6) or GST only control (lanes 2 and 5), washed, resuspended in sodium dodecyl sulfate (SDS) sample buffer, and analyzed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and autoradiography. (B) Effect of PU.1 on MafB transactivation activity. QT6 fibroblasts were cotransfected with 0.5 μg of a synthetic luciferase reporter construct containing 3 multimerized Maf response elements (3xMARE), 0.25 μg MafB expression construct, and 0.25 μg(+)or0.5 μg(++) PU.1 expression construct as indicated. (C) Effect of PU.1 on endogenous Maf transactivation activity in macrophages. HD11 macrophages (MΦ), expressing high levels of endogenous MafB39, were cotransfected with 0.5 μg of a synthetic luciferase reporter construct containing 3 multimerized Maf response elements (3xMARE), and 0.25 μg (+), 0.5 μg (++), or 1 μg (+++) PU.1 expression construct as indicated. Luciferase activities are expressed as fold activation over a reporter containing no response elements. Assays were performed in duplicate, normalized to β-galactosidase activity, and filled up with empty expression vector to a constant amount of expression plasmid. Error bars indicate standard error of the mean. Data are representative of at least 2 separate experiments. (D) PU.1 represses MafB-mediated macrophage differentiation. E26ts21-PUER clones were either treated (PU.1 active) or not (PU.1 inactive) with 1 μM βE for 48 hours, then shifted to 42°C or continued to be cultured at 37°C for an additional 48 hours. Expression of the macrophage-specific marker 47.83 was analyzed by FACS. (E) E26ts21-PUER clones were simultaneously treated with 1 μM βE and shifted to 42°C for 48 hours (42°C/βE) and compared to controls grown with βE or at 42°C only by FACS analysis for expression of 47.83. Results represent the average of at least 3 different clones. Error bars indicate standard error of the mean, and the statistical significance of the effect of 1 μM βE at 42°C is indicated.
PU.1 binds to MafB and inhibits its activity. (A) Analysis of PU.1 interaction with MafB in GST pull-down assays. In vitro–translated 35S-methionine-labeled PU.1 (lanes 1-3) or PUER (lanes 4-6) were incubated with an affinity matrix-bound GST-fusion protein of full-length MafB (lanes 3 and 6) or GST only control (lanes 2 and 5), washed, resuspended in sodium dodecyl sulfate (SDS) sample buffer, and analyzed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and autoradiography. (B) Effect of PU.1 on MafB transactivation activity. QT6 fibroblasts were cotransfected with 0.5 μg of a synthetic luciferase reporter construct containing 3 multimerized Maf response elements (3xMARE), 0.25 μg MafB expression construct, and 0.25 μg(+)or0.5 μg(++) PU.1 expression construct as indicated. (C) Effect of PU.1 on endogenous Maf transactivation activity in macrophages. HD11 macrophages (MΦ), expressing high levels of endogenous MafB39, were cotransfected with 0.5 μg of a synthetic luciferase reporter construct containing 3 multimerized Maf response elements (3xMARE), and 0.25 μg (+), 0.5 μg (++), or 1 μg (+++) PU.1 expression construct as indicated. Luciferase activities are expressed as fold activation over a reporter containing no response elements. Assays were performed in duplicate, normalized to β-galactosidase activity, and filled up with empty expression vector to a constant amount of expression plasmid. Error bars indicate standard error of the mean. Data are representative of at least 2 separate experiments. (D) PU.1 represses MafB-mediated macrophage differentiation. E26ts21-PUER clones were either treated (PU.1 active) or not (PU.1 inactive) with 1 μM βE for 48 hours, then shifted to 42°C or continued to be cultured at 37°C for an additional 48 hours. Expression of the macrophage-specific marker 47.83 was analyzed by FACS. (E) E26ts21-PUER clones were simultaneously treated with 1 μM βE and shifted to 42°C for 48 hours (42°C/βE) and compared to controls grown with βE or at 42°C only by FACS analysis for expression of 47.83. Results represent the average of at least 3 different clones. Error bars indicate standard error of the mean, and the statistical significance of the effect of 1 μM βE at 42°C is indicated.
Discussion
The differentiation choice of myeloid progenitor cells and monocytes toward the macrophage or DC differentiation pathway plays an important role under several normal and pathological conditions of the immune response. Here, we have shown that the transcription factors MafB and PU.1 induce alternative macrophage or DC fates in myeloblasts. PU.1 activation is instructive for the differentiation of functionally competent DCs in the absence of any DC-promoting cytokines. Furthermore, PU.1 expression is strongly induced upon DC differentiation from normal monocytes, and PU.1 activation in monocytes can redirect them toward DC fate. This redirection involves the direct repression of the macrophage factor MafB, which is not expressed in normal DCs and inhibits DC differentiation upon overexpression.
Our results suggest that the biologic effect of cytokines promoting DC differentiation, especially GM-CSF, may be mediated by PU.1 activation. Consistent with this, PU.1 is increased in response to GM-CSF in alveolar macrophages and can rescue the biologic consequences of GM-CSF deficiency.41 This suggests that the positive effect of GM-CSF on DC differentiation may be due to the induction of increased levels and/or activity of PU.1. It also has been shown that PU.1 can replace the requirement for TNFα in a Langerhans cell (LC) differentiation protocol from CD34+ cord blood progenitors, suggesting that TNFα signaling may also increase PU.1 levels and/or activity during LC differentiation.13 Together with our results that PU.1 expression completely bypasses all cytokine requirements, these observations suggest that PU.1 has a central role in integrating DC-promoting cytokine signaling.
Retroviral reconstitution experiments of PU.1 in knockout hematopoietic progenitors favors macrophage differentiation.21-23,28 We have observed here that PU.1 can be coexpressed at moderate levels with MafB in macrophages, but that high levels of PU.1 activity suppress MafB expression and activity and redirect myeloid progenitors or monocytes to DC fate. Finally, we have shown that MafB favors macrophage differentiation and inhibits DC fate. We therefore hypothesize that the co-expression of MafB and moderate levels of PU.1 specifies macrophage fate, whereas high PU.1 levels induce DC differentiation and disfavor macrophage fate by inhibiting MafB expression and activity (Figure 8). This would predict that the macrophage-promoting conditions where PU.1 induces macrophage differentiation21-23,28 are conducive to high levels of MafB and moderate levels of PU.1 that are not sufficient to repress MafB.
Model of DC versus macrophage fate specification by PU.1 and MafB. Both MafB and PU.1 levels increase during differentiation to monocytes and macrophages, which tolerate the co-expression of MafB with low or moderate levels of PU.1. At high levels, however, PU.1 represses MafB and triggers DC fate.
Model of DC versus macrophage fate specification by PU.1 and MafB. Both MafB and PU.1 levels increase during differentiation to monocytes and macrophages, which tolerate the co-expression of MafB with low or moderate levels of PU.1. At high levels, however, PU.1 represses MafB and triggers DC fate.
PU.1-deficient mice show broad hematopoietic defects both in the lymphoid lineage and in myeloid differentiation with mast cell, granulocyte, osteoclast, macrophage, and DC lineages affected to various extent.15-21 This broad effect indicates an early function in myeloid cells, consistent with the ability of PU.1 to induce myeloid lineage commitment in early multipotent hematopoietic progenitors.24 In addition, reconstitution and gain-of-function experiments have revealed that PU.1 also participates in cell fate decisions at several stages later during myeloid differentiation.13,21-24,28-32 This often involves cooperative or competitive interactions with other transcription factors that frequently depend on the relative levels of PU.1 and these partners. In early progenitors, PU.1 inhibits erythroid differentiation via an antagonistic interaction with GATA-1,29 which conversely also represses PU.1 activity.30 Similarly, the granulocyte inducer C/EBPα inhibits PU.1 transactivation and its ability to replace TNFα in LC differentiation.31 On the other hand PU.1 is required for the redirection of B cells into the macrophage lineage by C/EBPα or C/EBPβ.32 Reconstitution experiments of lineage-negative Pu.1–/– progenitors have shown that low levels of PU.1 result in preferential B-cell differentiation, whereas high levels favor macrophage development.23 Similar reconstitution experiments in IL-3–dependent Pu.1–/– progenitors indicate that PU.1 cooperates with GATA-2 at moderate expression levels to induce mast-cell fate, whereas higher levels of PU.1 suppress GATA-2 expression and favor macrophage differentiation.21 Finally, the relative levels of C/EBPα and PU.1 appear to be important for the differentiation choice between granulocytes and macrophages.28 Here, we report that high levels of PU.1 induce DC differentiation at the expense of macrophage fate, which involves repression of the macrophage-inducer MafB. These observations suggest that the biologic consequence of PU.1 activity is strongly dependent both on the expression levels and the context of coexpressed transcriptional regulators. We propose that increasingly higher expression levels of PU.1 during myeloid differentiation may successively exclude specific differentiation options by suppressing other lineage-specific transcription factors with different tolerance levels for PU.1 activity. Together, these observations are consistent with our previously proposed cocktail party model of transcription factor action in lineage specification, where factors are assumed to engage in changing physical and functional relationships at the successive stages of a differentiation process.42
We have shown that multipotent myeloid progenitor cells with the option to differentiate toward granulocytes and macrophages can be directed toward DC fate by PU.1 activation, supporting the existence of a common GM/DC progenitor cell.43 This mechanism of DC differentiation appears to be conserved in monocytic cells. Blood monocytes can differentiate to macrophages or DCs in culture5,26,44 or in vivo,3 and the balance of macrophage versus DC differentiation is influenced by the microenvironment encountered upon emigration from the bloodstream. Macrophage and DC homeostasis can be disturbed in pathological inflammatory and autoimmune conditions, which in some cases has been shown to be a major causative factor of disease development.4 A reciprocal disturbance of this balance is observed during tumorigenesis, where DC differentiation is frequently inhibited,5 possibly due to tumor-derived growth factors, such as M-CSF, IL-10, and IL-6, that can favor macrophage differentiation of infiltrating monocytes.5-7
Besides their importance in these pathological conditions, DCs hold great promise as a therapeutic agent in novel immunotherapy and vaccination strategies. Most of these protocols are based on blood monocyte–derived DCs. Therefore, a detailed understanding of the transcriptional program underlying DC differentiation from monocytes will open up new avenues to the manipulation of DC generation in culture and, most importantly, potentially also of monocyte/DC homeostasis in vivo. Manipulation of transcription factor activity in monocytes could provide the means to circumvent imbalanced cytokine levels under pathological conditions and reestablish homeostatic DC/macrophage ratios or influence them according to therapeutic needs. Strategies to control PU.1 and MafB activity in monocytes promise to present a first step in this direction.
Prepublished online as Blood First Edition Paper, December 14, 2004; DOI 10.1182/blood-2004-04-1448.
Supported by the Centre National de la Recherche Scientifique (CNRS) and the Fondation pour la Recherche Médicale (FRM) (Y.B.), the Association pour la Recherche sur le Cancer (ARC) and the Societé Francaise d'Hématologie (SFH) (S.S.), the Agence Nationale de Recherche sur le SIDA (ANRS) and the SFH (U.P.M.), and the Boehringer Ingelheim Fonds (BIF) and the SFH (S.T.). Funded by grants to M.H.S. from the ATIPE program of the CNRS, the La Ligue Nationale contre le Cancer (LNCC), ARC, ANRS, and a convention of the CNRS and the CNRST (Centre National pour la Recherche Scientifique et Technique, Morocco).
Y.B. and S.S. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank M. Dvorak and J. Ghysdael for 47.83 and PU.1 antibodies, respectively; F. Rossi for the MFG vector; G. Nolan for the ΦNX packaging cell line; and I. Lafon for technical help.


![Figure 2. MafB and PU.1 induce alternative macrophage or DC phenotype and function in myeloblasts. E26ts21 MafB–expressing myeloblasts (MafB) were induced to differentiate for 48 hours by temperature shift to 42°C (which releases a vMyb-imposed differentiation block), E26ts21-PUER (PU.1), and E26ts21-control (ctrl) were induced by 1 μM βE treatment. (A,D) May-Grünwald-Giemsa (MGG) staining of cytocentrifuged cells (objective lens ×40). (B,E) FACS analysis for the chicken monocyte/macrophage marker 47.83. (C,F) Cells with DC (▪) or macrophage (MΦ) morphology (▦) were counted for a total of at least 100 cells in 3 independent PUER (C) and MafB (F) clones from 2 separate experiments. (G) Mixed lymphocyte reaction (MLR) of myeloid cells differentiated by PU.1 or MafB activation. Ctrl, PU.1, and MafB clones were induced (ind.) or not induced (–) to differentiate, treated with 25 μg/mL mitomycin C, and incubated in duplicates in the absence (–) or presence (spleno.) of 105 monocyte-depleted allogenic splenocytes at different stimulating cell–splenocyte ratios for 5 days. [3H]-thymidine incorporation was measured after 16 hours of labeling. (H) Stimulating activity of 6 independent clones in 3 different experiments was analyzed and is expressed as proliferation index at a stimulating cell–splenocyte ratio of 1. (I) Phagocytic capacity of myeloid cells differentiated by PU.1 or MafB activation. Induced ctrl, PU.1, and MafB clones were incubated for 2 hours with phycoerythrin (PE)–conjugated latex beads and analyzed by FACS. Phagocytic activity before and after induction of differentiation was determined for 6 independent clones in 3 different experiments and expressed as the product of percent positive cells (as a measure of phagocytosing cells) and their mean fluorescence intensity (as a measure of the number of phagocytosed beads). In H-I, ▦ indicates uninduced cells; ▪, induced. Error bars indicate the standard error of the mean. Images in panels A and D were acquired using a Leica DMiL optical microscope equipped with a 40×/1.0 objective lens and a Leica MPS30 camera. Contrast and exposure were enhanced with Adobe Photoshop 5.0 software (Adobe, San Jose, CA) using equal treatment on all panels.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/105/7/10.1182_blood-2004-04-1448/6/m_zh80070576480002.jpeg?Expires=1763591646&Signature=vo7mD9VZffRJc70BR~LDbUyLNAexKPTf7lH6YWl51ZkMO2spR6DFLwD6PjdIocbvpuCx3do5BBbqmwLLxw~mn1zqK3u6qFwWZFpNCniYBDaV5GyopJbCjpF~2u9jTnypNoysddZJsFLAMog2YohuNJyE2PxvGagwUeuk5fyU8CDBHxsiN5gnqesEQRyj6JQdEN5jS~POgBXqayxM2JwGXfJC3DYI~WmTD3IADay-vhaSbv7D4xnCGM-6yBVK9OMog38ramT8AbPBgUQaQQkO7EKgsYqvfp0aSiV49G05tzKRzzlFxTNjhNh4UOiqNMti5g5mLqh4FkL7RAjsqfQhdw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal