Abstract
The HOX family of homeobox genes plays an important role in normal and malignant hematopoiesis. Dysregulated HOX gene expression profoundly effects the proliferation and differentiation of hematopoietic stem cells (HSCs) and committed progenitors, and aberrant activation of HOX genes is a common event in human myeloid leukemia. HOXB6 is frequently overexpressed in human acute myeloid leukemia (AML). To gain further insight into the role of HOXB6 in hematopoiesis, we overexpressed HOXB6 in murine bone marrow using retrovirus-mediated gene transfer. We also explored structure-function relationships using mutant HOXB6 proteins unable to bind to DNA or a key HOX-binding partner, pre–B-cell leukemia transcription factor-1 (PBX1). Additionally, we investigated the potential cooperative interaction with myeloid ecotropic viral integration site 1 homolog (MEIS1). In vivo, HOXB6 expanded HSCs and myeloid precursors while inhibiting erythropoiesis and lymphopoiesis. Overexpression of HOXB6 resulted in AML with a median latency of 223 days. Coexpression of MEIS1 dramatically shortened the onset of AML. Cytogenetic analysis of a subset of HOXB6-induced AMLs revealed recurrent deletions of chromosome bands 2D-E4, a region frequently deleted in HOXA9-induced AMLs. In vitro, HOXB6 immortalized a factor-dependent myelomonocytic precursor capable of granulocytic and monocytic differentiation. These biologic effects of HOXB6 were largely dependent on DNA binding but independent of direct interaction with PBX1.
Introduction
The HOX family of homeobox genes encodes DNA-binding proteins that play a crucial role in early body morphogenesis.1 HOX genes also play a role in hematopoietic development, with important effects on stem cell renewal, lineage commitment, and differentiation.2-6 HOX gene expression is tightly regulated during blood differentiation, and perturbations in the normal pattern of expression can have dramatic phenotypic effects.3,4,7,8
HOX gene dysregulation is a common event in human acute myeloid leukemia (AML) and may portend a worse prognosis.9-12 One emerging theory proposes that HOX genes act as downstream mediators of a variety of oncogenic insults,13 supporting a central role for HOX genes in leukemogenesis.11 Consistent with these observations, overexpression of several HOX cDNAs including Hoxb8, HOXB3, Hoxa9, and HOXA10 in murine marrow cells produces AML.4,8,14,15 Invariably, these leukemias occur with long latencies, suggesting additional oncogenic events are required for a full leukemic phenotype.
The DNA-binding specificity and avidity of the HOX proteins is greatly enhanced by physical interactions with TALE (three amino acid loop extension) homeodomain proteins including pre–B-cell leukemia transcription factor 1 (PBX1), myeloid ecotropic viral integration site 1 homolog (MEIS1), and PBX-regulating protein-1 (PREP1).16,17 Products of 3′ HOX genes preferentially associate with PBX1, whereas products of 5′ HOX genes associate with MEIS1.16,17 HOX proteins interact with PBX1 through a highly conserved region just N-terminal to the DNA-binding homeodomain designated the PBX-interaction motif (PIM).16,17 Despite the observation that HOX and TALE proteins are co-overexpressed in a variety of leukemic cell lines and humanAMLs, it remains unclear if the activity of HOX proteins in hematopoiesis is dependent on direct interaction with PBX1.18,19 While some investigators have reported impaired HOX gene activity with mutation of the PIM, others have observed no difference or even enhanced HOX gene activity with disruption of the HOX-PBX1 interaction.20-24 Although HOX proteins of paralogs 1 to 10 are not thought to directly interact with MEIS proteins, they have been shown to form trimeric DNA-binding complexes with PBX proteins, and MEIS1 has been shown to accelerate AML onset when overexpressed with HOXA9, HOXA10, or HOXB3.8,25-28
HOXB6 appears to have a unique pattern of expression in hematopoietic cells. While other HOX genes are expressed only in primitive CD34+ cells, HOXB6 mRNA is undetectable in CD34+ cells and is transiently up-regulated in early granulocytic and monocytic differentiation.5,10 HOXB6 expression is also detectable in committed erythroid precursors (erythroid burst-forming units [BFU-Es] and erythroid colony-forming units [CFU-Es]).29 HOXB6 expression is present in human leukemic cells with erythroid features including K562, HEL, OCIM2, and MEG01 but not in other myeloid lines.30-32 HOXB6 is expressed in approximately 40% of human AMLs lacking major chromosomal translocations.5
Attempts to define the hematopoietic effects of HOXB6 have yielded conflicting results. Antisense knockdown, as well as retroviral overexpression of HOXB6, both result in impaired granulomonocytic differentiation.5,33,34 Antisense HOXB6 increases erythroid features in HEL cells, whereas the HOXB6 knockout mouse has increased erythroid progenitors without significant effects on other lineages.33,35 These data are consistent with a model in which HOXB6 expression promotes early erythroid and myeloid differentiation but sustained overexpression blocks late differentiation.
To conduct an analysis of the effect of HOXB6 on hematopoiesis, we overexpressed HOXB6 in murine bone marrow using murine stem cell virus (MSCV)–based vectors. Our findings indicate that HOXB6 increased self-renewal and proliferation of hematopoietic cells in vitro and resulted in immortalization of a myeloid progenitor with biphenotypic differentiation potential. In vivo, HOXB6 overexpression expanded hematopoietic stem cells (HSCs) and myeloid precursors while impairing erythropoiesis and lymphopoiesis. HOXB6 overexpression results in delayed AML, which shares many features, including recurring deletions of chromosome 2, with HOXA9-induced AML. These effects were independent of a functional PIM and appear largely, though not wholly, dependent on DNA binding. Finally, coexpression of MEIS1 dramatically reduced the latency of HOXB6-induced AML.
Materials and methods
Mice
B6.SJL-Ptprca Pep3b/BoyJ mice (8-12 weeks of age) from Jackson Labs (Bar Harbor, ME) were used for marrow donors. Recipients were 8- to 12-week-old female congenic C57BL/6 mice from Charles River Laboratories (Wilmington, MA). Donor and recipient mice are distinguishable based on allelic polymorphism at the Ly5 locus.
Retroviral vectors
The MIG vector is an MSCV-based vector containing an internal ribosomal reentry sequence (IRES) and enhanced green fluorescent protein (EGFP) cDNA, whereas the MIY vector contains the coding sequence for yellow fluorescent protein (YFP). The HOXB6 vector was constructed by inserting a cDNA encoding a full-length human HOXB6 protein fused to an N-terminal FLAG (fludarabine, cytarabine, and granulocyte colony-stimulating factor [G-CSF]) epitope.36 The MEIS1 vector was constructed by inserting the cDNA for human MEIS1 into MIY.37 To create the HOXB6WG and HOXB6NA vectors, site-directed mutagenesis of the HOXB6 cDNA resulting in a W130G substitution within the YPWM PBX-binding motif (HOXB6WG) and an N196A substitution within the DNA-binding region (HOXB6NA) was performed using the ExSite mutagenesis kit (Stratagene, La Jolla, CA). Mutations were confirmed by DNA sequencing. Functional inability to bind PBX1 (HOXB6WG) or the consensus HOXB6 DNA-binding sequence (HOXB6NA) was confirmed by electrophoretic mobility shift assay (EMSA) assay (data not shown).
Bone marrow transduction and generation of transplant chimeras
High-titer retrovirus was produced by transient transfection of 293-T cells with the appropriate retroviral vector and the pCL ecotropic packaging plasmid using calcium phosphate and used to infect murine bone marrow by spinoculation.38 Experimental details are described in the Supplemental Material, available on the Blood website (see the Supplemental Materials link at the top of the online article). Colony-forming cell (CFC) dose per animal was retrospectively calculated by performing CFC assays on freshly transduced marrow. Transduced CFC dose was also calculated by multiplying total CFC injected per animal by the transduction efficiency determined by fluorescence-activated cell sorter (FACS) analysis. In some experiments, CFC transduction was confirmed by direct examination of colonies with fluorescence microscopy.
CFC and serial replating assays
CFC assays were performed using freshly transduced marrow cells or single-cell suspensions of marrow and spleen from primary transplant chimeras killed 1 to 2 months after transplantation. Cells were plated in M3434 media (StemCell Technologies, Vancouver, BC, Canada) following manufacturer's protocol. For replating assays, contents of culture dishes were collected after 8 days incubation. Experimental details are described in the Supplemental Material.
Cell proliferation assays and generation of immortalized cell lines
Cell proliferation assays were performed in duplicate by plating freshly transduced cells in 24-well plates at a density of 1 × 105/mL in growth media (GM) (DME-H21 supplemented with 10% fetal calf serum [FCS], 100 ng/mL recombinant murine stem cell factor [rm SCF], 50 ng/mL rm interleukin 6 [rmIL-6], and 10 ng/mL rmIL-3). Immortalized cell lines were derived from transduced marrow cells grown continuously in liquid culture in GM or DME-H21 supplemented with 10% FCS and 10 ng/mL granulocyte-macrophage colony-stimulating factor (GM-CSF). Cell lines were also derived from transduced cells passaged 4 times in replating assays and subsequently grown in GM-CSF containing media.
Differentiation assays
Differentiation assays were performed as described by Calvo et al.39 Briefly, granulocytic differentiation was assessed by growth in the presence of 5 ng/mL rm G-CSF (StemCell Technologies) for 48 hours. Monocytic differentiation was assessed by growth in the presence of 10 ng/mL rm M-CSF (StemCell Technologies) for 7 days. Morphologic differentiation was assessed by performing 300-cell differentials on Wright-Giemsa–stained cytospins, and molecular evidence of differentiation was assessed by quantitative reverse transcriptase–polymerase chain reaction (Q RT-PCR). HOXB6 expression was measured by Western blotting and Q RT-PCR. All experiments were performed in duplicate using 2 independently derived cell lines per vector.
CFU-S12 assays
Marrow cells from primary transplant chimeras killed 1 to 2 months after transplantation were injected into lethally irradiated C57BL/6 recipients at a dose of 1 × 104 to 5 × 104 cells/mouse. Recipients were killed on day 12; their spleens fixed in 70% ethyl alcohol (EtOH), 2% acetic acid, and 2% formalin in a 40:2:2 ratio; and macroscopic colonies were counted.
Competitive repopulation assays
Competitive repopulation unit (CRU) assays were performed as previously described by Szilvassy et al.40 Experimental details are described in the Supplemental Material.
Real-time Q-RT-PCR analysis
RNA was extracted using Qiagen RNEasy kit (Valencia, CA) and treated with DNase (DNA-free; Ambion, Austin, TX). “No reverse transcriptase” controls were performed on all samples to confirm that genomic DNA was not present. RNA was reverse transcribed into cDNA with iScript (BioRad, Hercules, CA), 300 ng in a 20-μL volume. Q-PCR analysis was performed on an AB Prism 7900 system (Applied Biosystems, Foster City, CA), using assays on demand (AoD) from Applied Biosystems. The cDNA equivalent to 3 to 5 ng of RNA was measured in triplicate by real-time PCR in 20-μL volume in 384-well plates. For normalization, cDNA equivalent to 3 to 5 ng input RNA was measured for glyceraldehyde phosphate dehydrogenase (GAPDH) and 17L.
FACS analysis
Phycoerythrin (PE)–conjugated monoclonal antibodies directed against Mac-1, Gr-1, F4/80, Ter119, c-kit, B220, CD8, Ly5.1 as well as isotype controls were obtained from Caltag (Burlingame, CA). Peridinin chlorophyll A protein–cyanin 5.5 (PerCP-Cy5.5)–conjugated antibodies against Gr-1, Mac-1, B220, CD4, and Ly5.2 were obtained from PharMingen (San Diego, CA). Single-cell suspensions from bone marrow (BM), spleen, and thymus without red blood cell (RBC) lysis were prepared and 1 × 106 cells were incubated with antibody in a volume of 25 μL at 4°C for 30 minutes and washed once with phosphate-buffered saline (PBS), and data were acquired on a FACScan flow cytometer (Becton Dickinson, San Jose, CA). For distinguishing EGFP from YFP, 510/20 and 535/20 nM dichroic filters detected green or yellow fluorescence, respectively, using a FACSCalibur flow cytometer (Becton Dickinson). Data were analyzed using CellQuest (Becton Dickinson) or FlowJo (Tree Star, Stanford, CA) software.
Western blots
BM, spleen, or immortalized marrow cells (1 × 106) were lysed in urea cracking buffer, mixed with Laemmli sample buffer, and boiled for 10 minutes. Proteins were separated on 10% polyacrylamide sodium dodecyl sulfate (SDS) gels, transferred to Immobilon polyvinylidene fluoride (PVDF) filters (Millipore, Bedford, MA), and probed with polyclonal rabbit anti–human HOXB6 antiserum.41 Bound antibody was detected with horseradish peroxidase (HRP)–conjugated donkey antihuman secondary antibody and visualized using the enhanced chemiluminescence (ECL) kit (Amersham Biosciences, Piscataway, NJ).
Southern blots
Genomic DNA was isolated from 5 × 106 marrow, spleen, or immortalized marrow cells using Qiagen DNEasy columns (Qiagen) and digested with XhoI and ClaI to release the proviral fragment or with BamHI alone to create unique-sized fragments for individual proviral integrations. Blots were probed with EGFP cDNA. For demonstration of MEIS1 proviral integration, genomic DNA was digested with EcoR1 and probed with the EcoR1 fragment of MEIS1 retroviral plasmid DNA containing the MEIS1 cDNA sequence.
Spectral karyotyping analysis
Cytogenetic analysis was performed on cryopreserved spleen or bone marrow cells obtained from leukemic mice. The initiation of short-term (24-72 hours) cultures, metaphase cell preparations, and spectral karyotyping (SKY) were performed as described previously.42 A minimum of 10 metaphase cells were analyzed per leukemia.
Results
Overexpression of HOXB6 in marrow progenitors enhances self-renewal and cell growth in vitro
Retroviral vectors and experimental design are summarized in Figure 1. To assess in vitro effects of HOXB6 expression, serial replating and cell proliferation assays were performed on freshly transduced marrow cells. Cells overexpressing HOXB6 showed a dramatic increase in CFC frequency upon serial replating (Figure 2A). In contrast, CFC frequency rapidly declined after 1 to 2 replatings of cells transduced with the MIG control (Figure 2A). Cells overexpressing the PBX1-binding mutant HOXB6WG demonstrated increased CFC frequency with replating similar to the wild-type HOXB6 (Figure 2A). The DNA-binding mutant HOXB6NA, however, had a serial decline in CFC frequency similar to the MIG control (Figure 2A). Colonies from marrow transduced with HOXB6 or HOXB6WG had an increasing frequency of large, compact CFCs with little peripheral migration (Figure 2B). These colonies were composed of cells with blastlike morphology, denoted CFU-Bs, consistent with a HOXB6-induced differentiation block (Figure 2B). In contrast, CFCs from HOXB6NA and MIG control marrow produced no CFU-Bs (Figure 2C).
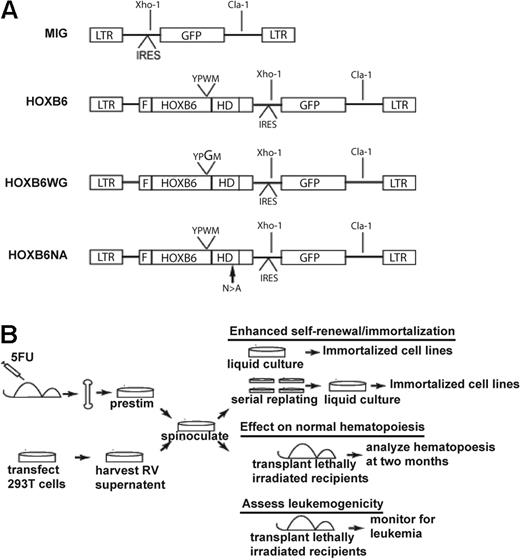
Retroviral vectors and experimental design. (A) The HOXB6 vector was constructed by cloning the cDNA encoding HOXB6, modified by the addition of an N-terminal FLAG epitope into the multiple cloning site of the MSCV-IRES-EGFP control vector. The HOXB6WG vector was constructed by introducing a single amino acid W → G substitution at position 130 within the conserved YPWM PBX-interaction motif (PIM). Similarly, the HOXB6NA vector includes one amino acid N → A substitution at position 196 within the homeodomain. The unique XhoI and ClaI restriction sights used for Southern blot analysis are shown. Digestion with both enzymes releases a 1.3-kb proviral fragment, whereas digestion with BamHI or XhoI alone cuts the provirus once, thus generating unique-sized fragments for individual viral integrations. LTR indicates long terminal repeat; F, N-terminal flag epitope; HD, homeodomain; 5FU, 5-fluorouracil; and RV, retroviral. (B) Schematic representation of experimental design.
Retroviral vectors and experimental design. (A) The HOXB6 vector was constructed by cloning the cDNA encoding HOXB6, modified by the addition of an N-terminal FLAG epitope into the multiple cloning site of the MSCV-IRES-EGFP control vector. The HOXB6WG vector was constructed by introducing a single amino acid W → G substitution at position 130 within the conserved YPWM PBX-interaction motif (PIM). Similarly, the HOXB6NA vector includes one amino acid N → A substitution at position 196 within the homeodomain. The unique XhoI and ClaI restriction sights used for Southern blot analysis are shown. Digestion with both enzymes releases a 1.3-kb proviral fragment, whereas digestion with BamHI or XhoI alone cuts the provirus once, thus generating unique-sized fragments for individual viral integrations. LTR indicates long terminal repeat; F, N-terminal flag epitope; HD, homeodomain; 5FU, 5-fluorouracil; and RV, retroviral. (B) Schematic representation of experimental design.
Overexpression of HOXB6 in bone marrow progenitors results in enhanced self-renewal and immortalization in vitro. (A) Results of 2 independent serial replating assays performed in duplicate demonstrated increased myeloid CFC frequency with successive replating for HOXB6 and HOXB6WG vectors, whereas CFC frequency rapidly declined in bone marrow transduced with HOXB6NA or MIG control. Results shown are means ± SD. P < .005 for comparison of HOXB6 or HOXB6WG with MIG in secondary and tertiary cultures. (B) HOXB6- and HOXB6WG-transduced marrow frequently produced dense colonies with little peripheral migration (left). Wright-Giemsa–stained cytospins of well-isolated dense colonies revealed these cells had blastlike morphology and thus the colonies were subclassified as CFU-blasts (right). CFC and cytospin images were obtained using an Axiovert 25 inverted microscope equipped with an AccuPlan 4 ×/0.10 objective lens (Carl Zeiss, Thornwood, NY) or a BH2 upright microscope equipped with a D Plan 100 ×/1.25 objective lens (Olympus America, Melville, NY). Images were captured with a Nikon Coolpix 4500 digital camera (Nikon USA, Torrence, CA) and enhanced using Nikon Photoimpression 4.0 software (Nikon USA). (C) Percentage of CFC composed of CFU-blasts increased with successive replatings in HOXB6- and HOXB6WG-transduced marrow, whereas HOXB6NA and MIG cultures had few or no CFU-blasts. P less than .005 for frequency of CFU-B in HOXB6 and HOXB6WG marrow versus MIG or HOXB6NA for all platings. Results shown are means ± SD for 2 independent experiments performed in duplicate. (D) Cell proliferation assays demonstrate exponential growth of HOXB6- and HOXB6WG-transduced bone marrow, whereas MIG-transduced cell cultures ceased to grow after approximately 4 weeks. The apparent intermediate growth of the HOXB6NA marrow was due to outgrowth of mast cells. (E) Western blot of lysates from HOXB6-immortalized cell lines showing robust expression of HOXB6. A HOXA9-immortalized cell line is used as a negative control. (F) Southern blot analysis of DNA isolated from immortalized cell lines using a fragment of EGFP cDNA as a probe. Digestion of genomic DNA with BamHI or XhoI alone cuts the provirus once liberating a unique-sized fragment for each proviral integration. One proviral integration was detected for all lines tested except HOXB6WG line 4, which may be oligoclonal or represent a single clone with multiple viral integrations. NS indicates nonspecific band. (G) FACS analysis of HOXB6-immortalized cell lines. In the top left panel, EGFP expression is shown on the x axis and FL-2 fluorescence in the unstained specimen is shown on the y axis. In the remaining 3 FACS histograms, Mac-1, B220, and c-kit staining are shown on the x axis (dark lines). The unstained cell line is overlaid as a control (light lines). Cell lines also had high Gr-1, intermediate F4/80, and negative CD8 expression (data not shown).
Overexpression of HOXB6 in bone marrow progenitors results in enhanced self-renewal and immortalization in vitro. (A) Results of 2 independent serial replating assays performed in duplicate demonstrated increased myeloid CFC frequency with successive replating for HOXB6 and HOXB6WG vectors, whereas CFC frequency rapidly declined in bone marrow transduced with HOXB6NA or MIG control. Results shown are means ± SD. P < .005 for comparison of HOXB6 or HOXB6WG with MIG in secondary and tertiary cultures. (B) HOXB6- and HOXB6WG-transduced marrow frequently produced dense colonies with little peripheral migration (left). Wright-Giemsa–stained cytospins of well-isolated dense colonies revealed these cells had blastlike morphology and thus the colonies were subclassified as CFU-blasts (right). CFC and cytospin images were obtained using an Axiovert 25 inverted microscope equipped with an AccuPlan 4 ×/0.10 objective lens (Carl Zeiss, Thornwood, NY) or a BH2 upright microscope equipped with a D Plan 100 ×/1.25 objective lens (Olympus America, Melville, NY). Images were captured with a Nikon Coolpix 4500 digital camera (Nikon USA, Torrence, CA) and enhanced using Nikon Photoimpression 4.0 software (Nikon USA). (C) Percentage of CFC composed of CFU-blasts increased with successive replatings in HOXB6- and HOXB6WG-transduced marrow, whereas HOXB6NA and MIG cultures had few or no CFU-blasts. P less than .005 for frequency of CFU-B in HOXB6 and HOXB6WG marrow versus MIG or HOXB6NA for all platings. Results shown are means ± SD for 2 independent experiments performed in duplicate. (D) Cell proliferation assays demonstrate exponential growth of HOXB6- and HOXB6WG-transduced bone marrow, whereas MIG-transduced cell cultures ceased to grow after approximately 4 weeks. The apparent intermediate growth of the HOXB6NA marrow was due to outgrowth of mast cells. (E) Western blot of lysates from HOXB6-immortalized cell lines showing robust expression of HOXB6. A HOXA9-immortalized cell line is used as a negative control. (F) Southern blot analysis of DNA isolated from immortalized cell lines using a fragment of EGFP cDNA as a probe. Digestion of genomic DNA with BamHI or XhoI alone cuts the provirus once liberating a unique-sized fragment for each proviral integration. One proviral integration was detected for all lines tested except HOXB6WG line 4, which may be oligoclonal or represent a single clone with multiple viral integrations. NS indicates nonspecific band. (G) FACS analysis of HOXB6-immortalized cell lines. In the top left panel, EGFP expression is shown on the x axis and FL-2 fluorescence in the unstained specimen is shown on the y axis. In the remaining 3 FACS histograms, Mac-1, B220, and c-kit staining are shown on the x axis (dark lines). The unstained cell line is overlaid as a control (light lines). Cell lines also had high Gr-1, intermediate F4/80, and negative CD8 expression (data not shown).
Proliferation assays performed in liquid culture demonstrated a similar phenomenon. Bone marrow cells transduced with HOXB6 grew exponentially, and cultures were rapidly overtaken by a uniform population of large, nonadherent cells (Figure 2D). In contrast, proliferation of marrow transduced with MIG decreased after 7 to 10 days and ceased within 4 weeks (Figure 2D). HOXB6WG-transduced marrow had a growth rate similar to wild-type HOXB6 (Figure 2D). In experiments performed with HOXB6NA-transduced marrow, proliferation decreased at 7 to 10 days, similar to control cultures; however, cultures were subsequently overgrown by mast cells, a phenomenon frequently seen when marrow cells are grown in the presence of SCF and IL-3 (Figure 2D).43 In contrast, marrow transduced with HOXB6NA and grown in media containing GM-CSF as the sole cytokine ceased proliferating within 2 to 4 weeks, whereas HOXB6- or HOXB6WG-transduced marrow continued to proliferate exponentially (data not shown). These experiments indicate that overexpression of HOXB6 increases self-renewal of myeloid precursors and imposes a differentiation block manifest by increasing frequency of CFU-Bs. This activity is dependent on DNA binding but independent of interaction with PBX1 via the PIM.
Overexpression of HOXB6 in bone marrow cells results in immortalization of a myelomonocytic progenitor with biphenotypic differentiation potential
In all experiments, retroviral transduction with HOXB6 or HOXB6WG resulted in outgrowth of immortalized myeloid cell lines (10/10 for HOXB6, 8/8 for HOXB6WG, 0/10 for HOXB6NA and MIG). Western blot analysis confirmed sustained expression of HOXB6 protein (Figure 2E). Southern blot analysis of DNA from independently derived cell lines demonstrated the lines were clonal and arose from retroviral integrations at several unique sites within the mouse genome (Figure 2F; data not shown). HOXB6- and HOXB6WG-immortalized cell lines had blastlike morphology by light microscopy and an indistinguishable myelomonocytic immunophenotype by FACS analysis, with high levels of Mac-1 and Gr-1, moderate c-kit, and no B220 (Figure 2G). All cell lines were initially factor dependent, requiring GM-CSF for growth. However, after 2 to 6 months in culture, several HOXB6 and HOXB6WG cell lines became independent of GM-CSF, suggesting the lines gained the ability to produce cytokines, a phenomena observed in HoxA9-transduced murine marrow.44 Alternatively, the cells may have acquired secondary mutations rendering them factor independent. These factor-independent HOXB6 cell lines continued to express high levels of HOXB6 protein and had an unchanged morphologic appearance and immunophenotype (data not shown).
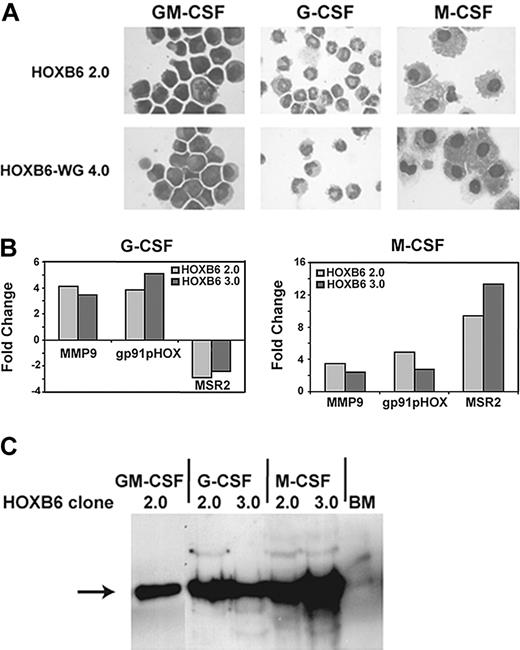
Previously, Calvo et al39 demonstrated that HOXA9-immortalized myeloid cell lines differentiate into granulocytes and monocytes under appropriate cytokine conditions. Similarly, HOXB6 and HOXB6WG cells differentiated into mature appearing neutrophils or monocytes when grown in the presence of G-CSF or M-CSF, respectively (Figure 3A; Table 1). Morphologic differentiation was accompanied by characteristic changes in myeloid gene transcription assessed by Q RT-PCR (Figure 3B). G-CSF–induced differentiation was accompanied by up-regulation of genes associated with neutrophil differentiation, including gp91pHOX and matrix metalloproteinase (MMP9; gelatinase b), whereas expression of macrophage scavenger receptor 2 (MSR2), a marker of monocytic differentiation, was down-regulated.39,45 Conversely, M-CSF–induced differentiation was accompanied by up-regulation of MSR2. Differentiation in response to G-CSF or M-CSF occurred in the presence of sustained HOXB6 protein expression as assayed by Western blot (Figure 3C).
HOXB6-immortalized cell lines are able to differentiate into neutrophils and monocytes when grown in the presence of G-CSF or M-CSF, respectively. (A) Representative photomicrographs from HOXB6 and HOXB6WG cell lines grown in the presence of GM-CSF, G-CSF, or M-CSF. Images were acquired as described for Figure 1. (B) Morphologic evidence of differentiation is accompanied by characteristic changes in gene transcription measured by real-time Q RT-PCR. Granulocytic differentiation is accompanied by up-regulation of MMP9 and gp91pHOX, whereas expression of MSR2, a gene normally expressed late in monocytic differentiation, is down-regulated. M-CSF–induced monocytic differentiation, in contrast, is accompanied by up-regulation of MSR2 in addition to less pronounced increases in MMP9 and gp91pHOX. Results shown are for 2 independently derived HOXB6 cell lines. Q RT-PCR assays were performed in triplicate. (C) Differentiation in response to G-CSF or M-CSF occurs without significant change in HOXB6 expression as assayed by Western blot. Cells grown in GM-CSF containing media and whole unfractionated bone marrow were used as positive and negative controls, respectively.
HOXB6-immortalized cell lines are able to differentiate into neutrophils and monocytes when grown in the presence of G-CSF or M-CSF, respectively. (A) Representative photomicrographs from HOXB6 and HOXB6WG cell lines grown in the presence of GM-CSF, G-CSF, or M-CSF. Images were acquired as described for Figure 1. (B) Morphologic evidence of differentiation is accompanied by characteristic changes in gene transcription measured by real-time Q RT-PCR. Granulocytic differentiation is accompanied by up-regulation of MMP9 and gp91pHOX, whereas expression of MSR2, a gene normally expressed late in monocytic differentiation, is down-regulated. M-CSF–induced monocytic differentiation, in contrast, is accompanied by up-regulation of MSR2 in addition to less pronounced increases in MMP9 and gp91pHOX. Results shown are for 2 independently derived HOXB6 cell lines. Q RT-PCR assays were performed in triplicate. (C) Differentiation in response to G-CSF or M-CSF occurs without significant change in HOXB6 expression as assayed by Western blot. Cells grown in GM-CSF containing media and whole unfractionated bone marrow were used as positive and negative controls, respectively.
Cytokine-induced differentiation of HOXB6-immortalized myeloid cell lines
. | Morphology, no. . | . | . | . | . | . | . | . | . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | GM-CSF . | . | . | G-CSF . | . | . | M-CSF . | . | . | ||||||||
| Cell line . | Blast . | Neut . | Mono . | Blast . | Neut . | Mono . | Blast . | Neut . | Mono . | ||||||||
| B6 2.0 | 75 | 12 | 13 | 6 | 86 | 8 | 6 | 6 | 87 | ||||||||
| B6 3.0 | 92 | 1 | 7 | 2 | 86 | 12 | 26 | 4 | 70 | ||||||||
| B6WG 2.0 | 83 | 11 | 6 | 5 | 94 | 1 | 83 | 2 | 15 | ||||||||
| B6WG 4.0 | 97 | 1 | 2 | 18 | 73 | 9 | 2 | 1 | 97 | ||||||||
. | Morphology, no. . | . | . | . | . | . | . | . | . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | GM-CSF . | . | . | G-CSF . | . | . | M-CSF . | . | . | ||||||||
| Cell line . | Blast . | Neut . | Mono . | Blast . | Neut . | Mono . | Blast . | Neut . | Mono . | ||||||||
| B6 2.0 | 75 | 12 | 13 | 6 | 86 | 8 | 6 | 6 | 87 | ||||||||
| B6 3.0 | 92 | 1 | 7 | 2 | 86 | 12 | 26 | 4 | 70 | ||||||||
| B6WG 2.0 | 83 | 11 | 6 | 5 | 94 | 1 | 83 | 2 | 15 | ||||||||
| B6WG 4.0 | 97 | 1 | 2 | 18 | 73 | 9 | 2 | 1 | 97 | ||||||||
Three hundred cell differentials were performed on Wright-Giemsa-stained cytospins from 2 independently derived HOXB6 and HOXB6WG cell lines grown in GM-CSF, G-CSF, or M-CSF as the sole cytokine.
Neut indicates neutrophils; and Mono, monocytes.
In vivo, HOXB6 confers a proliferative advantage to myeloid progenitors and impairs erythropoiesis and lymphopoiesis
To assess the effect of HOXB6 overexpression on hematopoiesis in vivo, lethally irradiated congenic mice received transplants of HOXB6-infected marrow cells. Primary recipients were chimeras, as they received a mixture of transduced and nontransduced marrow. Transduction efficiencies were similar for all vectors, as were total CFCs and transduced CFCs transplanted per mouse (Table 2). Mice were bled between 4 and 8 weeks after transplantation, and a subset was killed at 4 to 8 weeks after transplantation for analysis of hematopoietic tissues.
Retroviral transduction efficiency and CFCs transplanted per mouse
. | Transduction efficiency, % ± SD . | CFCs transplanted, × 103 . | . | |
|---|---|---|---|---|
| Vector . | . | Total . | Transduced . | |
| MIG | 35 ± 14 | 8.1 | 2.8 | |
| HOXB6 | 32 ± 21 | 7.8 | 2.5 | |
| HOXB6WG | 55 ± 15 | 12 | 6.6 | |
| HOXB6NA | 42 ± 34 | 13 | 5.5 | |
. | Transduction efficiency, % ± SD . | CFCs transplanted, × 103 . | . | |
|---|---|---|---|---|
| Vector . | . | Total . | Transduced . | |
| MIG | 35 ± 14 | 8.1 | 2.8 | |
| HOXB6 | 32 ± 21 | 7.8 | 2.5 | |
| HOXB6WG | 55 ± 15 | 12 | 6.6 | |
| HOXB6NA | 42 ± 34 | 13 | 5.5 | |
For each vector, n = 15.
Transduction efficiency was defined as the percent GFP+ bone marrow cells approximately 24 hours following retroviral transduction. Transduced CFC dose was calculated as CFC transplanted × transduction efficiency.
Transduction efficiency calculated in this manner agreed with direct examination of CFC assays by fluorescent microscopy (data not shown). Results for each vector reflect 3 independent experiments (5 mice per experiment).
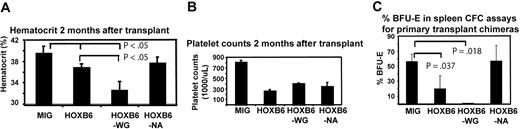
HOXB6 and HOXB6WG recipients had mild splenic enlargement, whereas HOXB6NA chimeras had spleen weights similar to MIG controls (Table 3). Thymus weights were similar in all groups (Table 3). Analysis of complete blood counts revealed HOXB6 and HOXB6WG transplant chimeras were anemic, whereas HOXB6NA mice had hematocrits similar to MIG chimeras (Figure 4A). In addition, HOXB6, HOXB6WG, and HOXB6NA recipients had moderate thrombocytopenia compared with controls (Figure 4B). White blood cell counts in HOXB6 and HOXB6NA mice were unchanged with respect to controls; however, HOXB6WG mice had mild leukopenia (data not shown). The defect in erythropoiesis was dependent on DNA binding but independent of a functional PIM. Surprisingly, the defect in megakaryopoiesis appeared to be independent of both DNA binding and interaction with PBX1 via the PIM.
Organ weights (mean ± SD) of primary transplant chimeras 2 months after transplantation
. | Organ weights, mg (P vs MIG) . | . | |
|---|---|---|---|
| Vector . | Spleen . | Thymus . | |
| MIG | 87 ± 11 (NA) | 41 ± 20 (NA) | |
| HOXB6 | 138 ± 11 (.001) | 44 ± 14 (NS) | |
| HOXB6WG | 117 ± 10 (.09) | 62 ± 20 (NS) | |
| HOXB6NA | 78 ± 10 (NS) | 48 ± 3 (NS) | |
. | Organ weights, mg (P vs MIG) . | . | |
|---|---|---|---|
| Vector . | Spleen . | Thymus . | |
| MIG | 87 ± 11 (NA) | 41 ± 20 (NA) | |
| HOXB6 | 138 ± 11 (.001) | 44 ± 14 (NS) | |
| HOXB6WG | 117 ± 10 (.09) | 62 ± 20 (NS) | |
| HOXB6NA | 78 ± 10 (NS) | 48 ± 3 (NS) | |
NA indicates not applicable; NS indicates P value not significant.
n = 5 for MIG, 4 for HOXB6, and 3 for HOXB6WG and HOXB6NA.
HOXB6 primary transplant chimeras have mild splenomegaly and impaired erythropoiesis and thrombopoiesis. (A) HOXB6 and HOXB6WG transplant chimeras were anemic compared with MIG controls and HOXB6WG mice had a greater degree of anemia than HOXB6 mice. Results shown are means ± SD. n = 17 for HOXB6, 16 for HOXB6WG, 14 for HOXB6NA, and 13 for MIG cohorts. (B) HOXB6, HOXB6WG, and HOXB6NA primary transplant chimeras all had moderate thrombocytopenia compared with MIG controls. P less than .005 for all vectors. n for each cohort as in panel A.. Results shown are means ± SEM. (C) CFC assays performed with spleen suspensions from primary transplant chimeras demonstrate significantly reduced BFU-E frequency in mice that received transplants of HOXB6- and HOXB6WG-overexpressing bone marrow. Results represent means ± SEM from 4 individual experiments for MIG, 3 for HOXB6, and 2 each for HOXB6WG and HOXB6NA.
HOXB6 primary transplant chimeras have mild splenomegaly and impaired erythropoiesis and thrombopoiesis. (A) HOXB6 and HOXB6WG transplant chimeras were anemic compared with MIG controls and HOXB6WG mice had a greater degree of anemia than HOXB6 mice. Results shown are means ± SD. n = 17 for HOXB6, 16 for HOXB6WG, 14 for HOXB6NA, and 13 for MIG cohorts. (B) HOXB6, HOXB6WG, and HOXB6NA primary transplant chimeras all had moderate thrombocytopenia compared with MIG controls. P less than .005 for all vectors. n for each cohort as in panel A.. Results shown are means ± SEM. (C) CFC assays performed with spleen suspensions from primary transplant chimeras demonstrate significantly reduced BFU-E frequency in mice that received transplants of HOXB6- and HOXB6WG-overexpressing bone marrow. Results represent means ± SEM from 4 individual experiments for MIG, 3 for HOXB6, and 2 each for HOXB6WG and HOXB6NA.
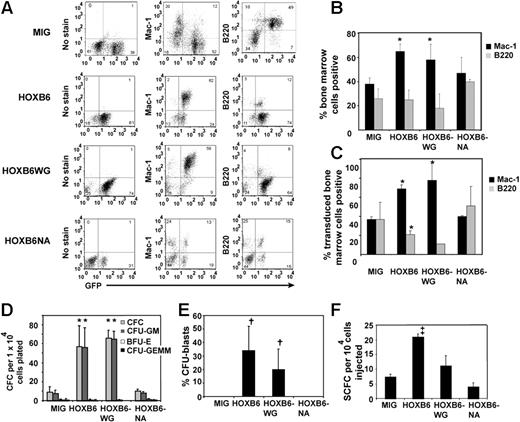
FACS analysis of marrow, spleen, and thymus from primary recipients using a panel of lineage-specific fluorescent antibodies and GFP expression as a marker for transduction confirmed that HOXB6-transduced hematopoietic progenitors contribute to myeloid and lymphoid lineages (Figure 5A; and Figure S1). In marrow of HOXB6 and HOXB6WG recipients there was an increase in frequency of myeloid cells (Mac-1+) compared with controls (mean ± standard deviation 65% ± 6%, 68% ± 13%, 38% ± 5%, respectively; Figure 5B). This increase was not seen in HOXB6NA chimeras (47% ± 13% Mac-1+). The increase in Mac-1+ cells in HOXB6 and HOXB6WG transplant chimeras was due to enhanced contribution of transduced (EGFP+) cells to the myeloid compartment (71% ± 2% and 78% ± 2% vs 38% ± 1% for HOXB6, HOXB6WG, and MIG, respectively; Figure 5C). In contrast, the frequency of B lymphocytes (B220+) produced by HOXB6- and HOXB6WG-transduced marrow was decreased compared with MIG controls (19% ± 5%, 11% ± 0%, and 37% ± 18%, respectively; P < .05 for HOXB6, P = .08 for HOXB6WG; Figure 5C). The distribution of Mac-1+ and B220+ cells did not differ between MIG and HOXB6NA chimeras (Figure 5C). FACS analysis also revealed a defect in thymocyte development. In HOXB6 mice there was a moderate decrease in single-positive CD4+CD8– and CD4–CD8+ cells, whereas MIG controls had the expected distribution of thymocytes (Figure S2). Finally, HOXB6 marrow had a mild increase in c-kit+ cells (Figure S1).
HOXB6 overexpression expands myeloid precursors and impairs B lymphopoiesis. (A) Representative FACS plots from bone marrow suspensions of primary transplant chimeras killed 1 to 2 months after transplantation. For all plots, EGFP expression (a marker for transduced cells) is represented on the x-axis, and the y-axis represents staining with Mac-1, B220, or no antibody (also see Figure S1). (B) HOXB6 and HOXB6WG transplant chimeras have an increase in frequency of myeloid cells (Mac-1+) in the bone marrow. n = 5 for MIG and HOXB6, 3 for HOXB6WG, and 2 for HOXB6NA. *P less than .05 versus MIG. (C) The increase in myeloid progenitors was due to a predominance of myeloid forms among HOXB6- and HOXB6WG-transduced cells. Additionally, HOXB6-transduced marrow had a decrease in B220+ cells, whereas a similar trend existed for HOXB6WG. *P < .05 versus MIG. Results from panels B and C represent mean percentages ± SD. (D) HOXB6 and HOXB6WG overexpression produced an approximately 5-fold increase in CFC production. Results shown are mean CFC production per 10 000 cells plated ± SEM. n = 4 mice for MIG and 3 for HOXB6, HOXB6WG, and HOXB6NA. The increase in CFC was due almost entirely to expansion of CFU-GMs. *P = .03 versus MIG. (E) HOXB6 and HOXB6WG marrow had significantly increased CFU-Bs when compared with MIG controls. Results represent mean percentages ± SD. n as for panel D. †P = .001 and .01, respectively, for HOXB6 and HOXB6WG versus MIG. (F) Bone marrow from HOXB6 and HOXB6WG chimeras had an increase in spleen colony-forming cells (SCFCs) as measured by the CFU-S12 assay. Results represent mean SCFCs per 1 × 104 cells injected ± SEM for 3 (MIG and HOXB6WG) or 2 (HOXB6 and HOXB6NA) individual experiments. n = 20, 12, 19, and 10 recipients for MIG, HOXB6, HOXB6WG, and HOXB6NA, respectively. ‡P less than .005 versus MIG.
HOXB6 overexpression expands myeloid precursors and impairs B lymphopoiesis. (A) Representative FACS plots from bone marrow suspensions of primary transplant chimeras killed 1 to 2 months after transplantation. For all plots, EGFP expression (a marker for transduced cells) is represented on the x-axis, and the y-axis represents staining with Mac-1, B220, or no antibody (also see Figure S1). (B) HOXB6 and HOXB6WG transplant chimeras have an increase in frequency of myeloid cells (Mac-1+) in the bone marrow. n = 5 for MIG and HOXB6, 3 for HOXB6WG, and 2 for HOXB6NA. *P less than .05 versus MIG. (C) The increase in myeloid progenitors was due to a predominance of myeloid forms among HOXB6- and HOXB6WG-transduced cells. Additionally, HOXB6-transduced marrow had a decrease in B220+ cells, whereas a similar trend existed for HOXB6WG. *P < .05 versus MIG. Results from panels B and C represent mean percentages ± SD. (D) HOXB6 and HOXB6WG overexpression produced an approximately 5-fold increase in CFC production. Results shown are mean CFC production per 10 000 cells plated ± SEM. n = 4 mice for MIG and 3 for HOXB6, HOXB6WG, and HOXB6NA. The increase in CFC was due almost entirely to expansion of CFU-GMs. *P = .03 versus MIG. (E) HOXB6 and HOXB6WG marrow had significantly increased CFU-Bs when compared with MIG controls. Results represent mean percentages ± SD. n as for panel D. †P = .001 and .01, respectively, for HOXB6 and HOXB6WG versus MIG. (F) Bone marrow from HOXB6 and HOXB6WG chimeras had an increase in spleen colony-forming cells (SCFCs) as measured by the CFU-S12 assay. Results represent mean SCFCs per 1 × 104 cells injected ± SEM for 3 (MIG and HOXB6WG) or 2 (HOXB6 and HOXB6NA) individual experiments. n = 20, 12, 19, and 10 recipients for MIG, HOXB6, HOXB6WG, and HOXB6NA, respectively. ‡P less than .005 versus MIG.
We previously reported that HOXB6 expression inhibits erythroid maturation in an in vitro culture model.33 Consistent with those findings, we observed a decrease in marrow TER-119+ cells in HOXB6 mice compared with MIG controls (22% ± 5% vs 48% ± 7% for HOXB6 vs MIG; P < .005; Figure S1). We also performed methylcellulose progenitor assays on splenic suspensions from transplant recipients. Whereas spleen cells from MIG chimeras contained the expected high percentage of BFU-Es, those from HOXB6 and HOXB6WG mice had markedly reduced BFU-E frequency. BFU-E frequency in HOXB6NA chimeras was similar to MIG controls (Figure 4D).
HOXB6 overexpression results in expansion of committed and primitive myeloid progenitors
CFC and spleen CFU (CFU-S) assays were performed using marrow from primary recipients 1 to 2 months after transplantation. Overexpression of HOXB6 or HOXB6WG produced a 5-fold increase in CFC frequency compared with controls, whereas HOXB6NA overexpression had no detectable effect (Figure 5D). This increase was almost entirely due to an increase in CFU-GMs (Figure 5D). Similar to assays performed on freshly transduced marrow, 30% to 50% of CFU-GMs from HOXB6 and HOXB6WG chimeras could be subclassified as CFU-Bs. No CFU-Bs were present in HOXB6NA and MIG CFC assays (Figure 5E).
More primitive progenitors were assessed with day-12 CFU-S assays. HOXB6 transplant chimeras had an approximately 4-fold increase in spleen colony-forming cells (SCFCs) compared with controls. While there was a trend toward increased SCFC activity in the HOXB6WG mice, this did not reach statistical significance. As with CFC assays, HOXB6NA overexpression did not have an appreciable effect on SCFC activity (Figure 5F).
HOXB6 overexpression enhances repopulating ability
Standard CRU assays were performed using limiting dilutions of donor marrow derived from primary recipients 1 to 2 months after transplantation.40 Overexpression of HOXB6 or HOXB6WG produced an approximately 30- to 160-fold expansion in HSCs compared with MIG controls (Table 4). HOXB6NA chimeras had a modest but statistically significant increase in HSC frequency. Of note, the HSC frequency observed in HOXB6 primary recipients of 1 in 4.0 × 104 approximates that of untransplanted normal marrow. This number may represent the maximal HSC number permissible by the hematopoietic stem cell “niche” and approximates the frequency observed in mice that received transplants of bone marrow overexpressing HOXB4.7,46 These results indicate that HOXB6 has a dramatic effect on HSC expansion independent of interaction with PBX1 via the PIM. Interestingly, the HOXB6NA chimeras also had a significant increase in HSC frequency, suggesting a possible DNA-binding–independent effect.
HOXB6 increases CRU frequency
Vector . | CRU frequency . | 95% CI . | Fold expansion . |
|---|---|---|---|
| MIG | 1 in 6.3 × 106 | 1 in 3.1-13 × 106 | — |
| HOXB6 | 1 in 4.0 × 104 | 1 in 3.0-5.3 × 104 | 160 |
| HOXB6WG | 1 in 2.1 × 105 | 1 in 1.4-3.1 × 105 | 30 |
| MIGB6NA | 1 in 5.1 × 105 | 1 in 3.7-7.0 × 105 | 12 |
Vector . | CRU frequency . | 95% CI . | Fold expansion . |
|---|---|---|---|
| MIG | 1 in 6.3 × 106 | 1 in 3.1-13 × 106 | — |
| HOXB6 | 1 in 4.0 × 104 | 1 in 3.0-5.3 × 104 | 160 |
| HOXB6WG | 1 in 2.1 × 105 | 1 in 1.4-3.1 × 105 | 30 |
| MIGB6NA | 1 in 5.1 × 105 | 1 in 3.7-7.0 × 105 | 12 |
For each vector, 2 primary transplant chimeras were killed for CRU donors (except MIG, n = 4) and 3 to 4 dilutions analyzed per experiment (secondary recipient n = 32, 36, 22, and 29 for MIG, HOXB6, HOXB6WG, and HOXB6NA, respectively). Fold expansion is expressed relative to MIG control.
— indicates that MIG is the control to which other vectors are compared.
HOXB6 overexpression results in delayed AML
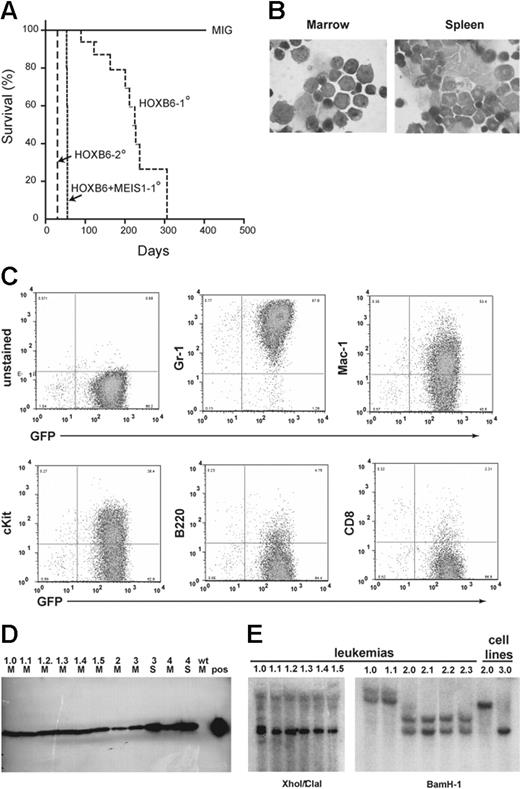
To assess the leukemogenic potential of HOXB6 overexpression, primary recipients were monitored for up to 1 year after transplantation. Nine of 9 evaluable HOXB6 transplant chimeras developed fatal AML with a median onset of 223 ± 19 days (Figure 6A). Although anemia, thrombocytopenia, and circulating dysplastic myelomonocytic forms were present for months preceding AML, there was no myeloproliferative prodrome (Figure S3). Leukemias were readily transplantable and resulted in rapid death of secondary recipients (Figure 6A). According to the Bethesda classification of hematopoietic neoplasms in mice, HOXB6-induced AMLs have phenotypes consistent with acute myelomonocytic leukemia and acute monocytic leukemia (Figure 6B).47 Leukemic blasts expressed Mac-1, Gr-1, and less intense F4/80 and c-kit (Figure 6C; data not shown). Lymphoid antigens were absent except for variable low-level B220 (Figure 6C). Leukemias were highly invasive, involving spleen, liver, kidneys, lung, bone, bowel, and thymus (data not shown). HOXB6 protein expression was easily detectable in marrow and spleen suspensions from leukemic animals (Figure 6D). Southern blot analysis demonstrated that leukemias were clonal or oligoclonal (Figure 6E).
HOXB6 overexpression results in delayed AML. (A) HOXB6 primary transplant chimeras developed fatal acute myeloid leukemia with a latency of approximately 8 months (median, 223 days; range, 83-261 days). Leukemias were transplantable with a rapid disease onset. (B) Leukemic blasts had immature morphologic appearance on Wright-Giemsa–stained cytospins; however, there was a variable expression of monocytic features such as increased cytoplasm, cytoplasmic vacuolization, and nuclear indentation. Leukemic blasts infiltrated all tissues examined including spleen, thymus, liver, kidneys, and lung (data not shown). Image acquisition was performed as described for Figure 1. (C) FACS analysis of leukemic cells, with EGFP expression on the x-axis and lineage markers on the y-axis, demonstrates the blasts express myeloid markers Gr-1 and Mac-1 as well as c-kit. The blasts do not stain for lymphoid antigens B220 or CD8. (D) HOXB6 protein was easily detected in bone marrow (M) and spleen (S) lysates from both primary and secondary recipients. Wild-type (wt), unfractionated BM was used as a negative control, and HOXB6-transfected 293-T packaging cells were used as positive control (pos). (E) Southern blot analysis of 2 HOXB6-induced primary leukemias and secondary recipients. XhoI/ClaI double digest confirms proviral integration in all leukemias (left). BamHI single digest indicates the primary leukemias were clonal (#1) or oligoclonal versus clonal with 2 proviral integrations (#2) and were transmitted to secondary recipients.
HOXB6 overexpression results in delayed AML. (A) HOXB6 primary transplant chimeras developed fatal acute myeloid leukemia with a latency of approximately 8 months (median, 223 days; range, 83-261 days). Leukemias were transplantable with a rapid disease onset. (B) Leukemic blasts had immature morphologic appearance on Wright-Giemsa–stained cytospins; however, there was a variable expression of monocytic features such as increased cytoplasm, cytoplasmic vacuolization, and nuclear indentation. Leukemic blasts infiltrated all tissues examined including spleen, thymus, liver, kidneys, and lung (data not shown). Image acquisition was performed as described for Figure 1. (C) FACS analysis of leukemic cells, with EGFP expression on the x-axis and lineage markers on the y-axis, demonstrates the blasts express myeloid markers Gr-1 and Mac-1 as well as c-kit. The blasts do not stain for lymphoid antigens B220 or CD8. (D) HOXB6 protein was easily detected in bone marrow (M) and spleen (S) lysates from both primary and secondary recipients. Wild-type (wt), unfractionated BM was used as a negative control, and HOXB6-transfected 293-T packaging cells were used as positive control (pos). (E) Southern blot analysis of 2 HOXB6-induced primary leukemias and secondary recipients. XhoI/ClaI double digest confirms proviral integration in all leukemias (left). BamHI single digest indicates the primary leukemias were clonal (#1) or oligoclonal versus clonal with 2 proviral integrations (#2) and were transmitted to secondary recipients.
To date, AML has been observed in 4 of 15 HOXB6WG primary recipients with an onset of 118 days (median observation of 115 days, range 74-186 days) and in 0 of 15 HOXB6NA primary transplant chimeras. However, 2 of 15 HOXB6NA mice have been killed due to poor health. Both were pancytopenic. However, evaluation of bone marrow by FACS analysis and light microscopy revealed a mixture of transduced and nontransduced cells without objective evidence ofAML.
MEIS1 accelerates HOXB6-induced AML
Antennapedia-like HOX proteins are not thought to interact directly with MEIS1, though an indirect interaction may occur through PBX proteins.26-28 Since MEIS1 has recently been reported to convert HOXB4 stem cell expansion to overt leukemia,25 we investigated the potential cooperativity between HOXB6 and MEIS1 in leukemogenesis. Cohorts of mice received transplants of marrow overexpressing HOXB6 alone, MEIS1 alone, and both HOXB6 and MEIS1 (HOXB6/MEIS1). At 5 weeks, the earliest time point evaluated, all 5 HOXB6/MEIS1 mice were killed on the basis of having severe anemia, moderate thrombocytopenia, and circulating blast forms (Figure 6). Analysis of bone marrow and spleen from these mice confirmed the presence of AML with a similar morphology and immunophenotype as HOXB6-induced AML (Figure S4). Proviral integration of MEIS1 in leukemic blasts was confirmed by Southern blot, and coexpression of HOXB6 and MEIS1 was confirmed by FACS analysis for EGFP and YFP as well as Western blot for HOXB6 (Figure S4). In addition, lack of rearrangement of the MEIS1 locus in HOXB6-alone AMLs was confirmed by Southern blot (data not shown). Similar to HOXB6-induced leukemias, HOXB6/MEIS1-induced AMLs were readily transplantable with a latency of approximately 30 days (data not shown). Consistent with prior reports, mice that received transplants of marrow transduced with MEIS1 alone remained healthy and had blood counts and blood morphology similar to MIY controls.8,25
HOXB6-induced AMLs have recurring cytogenetic abnormalities consistent with those observed in HOXA9-induced AMLs
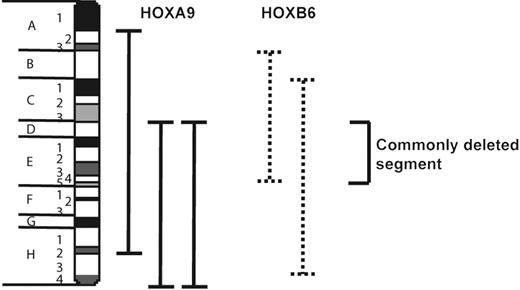
HOXB6-induced AML occurs with substantial latency, suggesting that additional genetic insults are required to develop the full leukemic phenotype. To explore potential cooperative mutational events in HOXB6-induced leukemogenesis, we performed SKY analysis of 5 HOXB6-induced AMLs from 3 separate experiments (Table 5). The sole cytogenetic abnormality observed in 2 of 5 individual leukemias was an interstitial deletion of chromosome 2. Deletions of chromosome 2 were also found as the sole cytogenetic abnormality in 3 of 6 HOXA9-induced AMLs. Mapping the deletion breakpoints revealed that the smallest commonly deleted segment that was deleted in all of the HOXB6- and HOXA9-induced AMLs consisted of bands 2D-E4 (Figure 7).
Cytogenetic abnormalities encountered in HOXB6- and HOXA9-induced AMLs
Vector and ID . | Karyotype . |
|---|---|
| HOXB6 | |
| 29847 | 40,XY,del(2)(BE4)[6]/41,idem,+del(2)(BE4)[4] |
| 29848 | 40,XX,del(2)(C1H3)/[9]/40,XX,del(2)(C2F3)[1] |
| 29849 | 40,XX[10] |
| 29850 | 40,XY[10] |
| 29851 | 40,XX[10] |
| HOXA9 | |
| 29495 | 40,XY[10] |
| 29497 | 41,XX,+X,del(2)(A2H2)[10] |
| 29498 | 40,XX[10] |
| 29779 | 40,XY[10] |
| 28299 | 40,XY,del(2)(DH4)[9]/39, idem, -10, -17, +dic mar[1] |
| 28298 | 40,XY,del(2)(DH4)[10] |
Vector and ID . | Karyotype . |
|---|---|
| HOXB6 | |
| 29847 | 40,XY,del(2)(BE4)[6]/41,idem,+del(2)(BE4)[4] |
| 29848 | 40,XX,del(2)(C1H3)/[9]/40,XX,del(2)(C2F3)[1] |
| 29849 | 40,XX[10] |
| 29850 | 40,XY[10] |
| 29851 | 40,XX[10] |
| HOXA9 | |
| 29495 | 40,XY[10] |
| 29497 | 41,XX,+X,del(2)(A2H2)[10] |
| 29498 | 40,XX[10] |
| 29779 | 40,XY[10] |
| 28299 | 40,XY,del(2)(DH4)[9]/39, idem, -10, -17, +dic mar[1] |
| 28298 | 40,XY,del(2)(DH4)[10] |
Spectral karyotyping performed on 5 HOXB6- and 5 HOXA9-induced AMLs reveals recurrent deletions of chromosome 2 in 3 HOXA9-induced and 2 HOXB6-induced AMLs.
HOXB6-induced AMLs have recurrent cytogenetic abnormalities that span a common chromosomal region with HOXA9-induced AML. Schematic diagram of murine chromosome 2 indicating region of overlap in deletions observed in HOXB6- and HOXA9-induced AMLs.
HOXB6-induced AMLs have recurrent cytogenetic abnormalities that span a common chromosomal region with HOXA9-induced AML. Schematic diagram of murine chromosome 2 indicating region of overlap in deletions observed in HOXB6- and HOXA9-induced AMLs.
Discussion
Here we demonstrate that HOXB6 overexpression results in increased self-renewal and proliferation of granulomonocytic precursors. Together with the observed impairment in erythropoiesis, megakaryopoiesis, and B lymphopoiesis, our data support a model in which HOXB6 expression promotes expansion of granulocytes and monocytes at the expense of other blood elements. Within the scope of these experiments it was not possible to determine if these effects were due to altered lineage commitment or altered proliferation of committed progenitors. Other investigators have demonstrated similar lineage-specific defects with overexpression of HOX genes, including impaired erythropoiesis with HOXA5, impaired B lymphopoiesis with Hoxa9 and HOXA10, and impaired macrophage production and increased megakaryoblastic precursors with HOXA10.3,4,48 Together with these reports, our data support a model in which stage- and lineage-specific down-regulation of certain HOX gene expression is crucial for normal hematopoietic differentiation.
HOXB6 protein overexpression has a striking effect on HSC self-renewal. In its ability to expand HSCs, HOXB6 is similar to its 3′ relative HOXB4 as well as Hoxa9.3,46,49 However, unlike HOXB4, HOXB6 markedly affects the production and differentiation of the various blood lineages. While HOXB4 is likely involved in the normal regulation of the HSC pool, it is unclear what physiologic role HOXB6 or other HOX proteins may play.50,51
Overexpression of HOXB6 in murine bone marrow results in AML with a latency of approximately 8 months, similar to that for HOXB3, Hoxb8, Hoxa9, and HOXA10. The ability of HOXB6 to immortalize murine marrow, expand HSCs and other myeloid precursors, and cause AML was independent of direct interaction with PBX1 via the PIM. This is the first demonstration of PIM-independent immortalization of murine marrow by a member of the Antennapedia-like HOX proteins, proteins that preferentially heterodimerize with members of the PBX family as opposed to MEIS. The hypothesis that HOX genes rely on interaction with PBX1 to exert biologic activity is based on their cooperative DNA binding, their coexpression in leukemic cell lines, and synergistic transforming activity in Rat-1 fibroblast cell lines.19,52 However, there is mounting evidence that members of the Antennapedia-like HOX genes and the Abd-B–like HOX genes retain biologic activity without direct interaction with PBX1. In fact, the HSC expanding capacity of the HOXB4 protein is increased upon down-regulation of PBX1, and direct or indirect interaction with PBX1 is not required for induction of AML by HOXA9.23,53,54
Interestingly, the latent period for HOXB6 to induce overt leukemia was dramatically reduced by co-overexpression of MEIS1 protein. While this may seem improbable given the lack of direct association between MEIS1 and Antennapedia-like HOX proteins, cooperative leukemogenic interactions have been described between MEIS1, HOXB3, and HOXB4.8,25 In both of the aforementioned cases, however, the latency of AML was approximately 150 days, suggesting additional genetic events were required. In contrast, the short latency of HOXB6/MEIS1-induced AML suggests that coexpression of HOXB6 and MEIS1 is sufficient to produce AML. Although DNA-binding studies with purified proteins have shown that MEIS does not form direct cooperative DNA-binding complexes with the paralog 1-10 HOX proteins,55 trimeric complexes for HOXB1 and HOXD4 have been described in which the HOX and MEIS proteins are bridged by the PBX protein.26,28 Our data show that a HOXB6WG mutant, which does not appear to form DNA-binding complexes with PBX, mimics wild-type HOXB6 in its in vitro and in vivo activities. Taken together, these data suggest that additional cellular proteins may mediate HOXB6-MEIS interactions to accelerate leukemia in vivo. Alternatively, MEIS1 may modulate HOX activity by titrating PBX1 and preventing its binding to HOX proteins. This later hypothesis is supported by the observations that knockdown of PBX1 expression accentuates the proliferative effects of HOXB4 on HSC proliferation and co-overexpression of a mutant MEIS1 protein unable to bind PBX1 together with HOXA9 did not accelerate HOXA9-induced leukemogenesis like its wild-type counterpart.23,53 Future studies will be directed toward elucidating these possible mechanisms.
As a screen for cooperating genetic events in HOXB6-induced leukemogenesis, we have performed SKY analysis on a limited set of HOXB6- and Hoxa9-induced AMLs. Notably, the most common cytogenetic abnormality observed in both HOXB6 and HOXA9 AMLs was a recurring interstitial deletion of chromosome 2 with a common region of deletion spanning 2D-E4. Zimonjic et al56 have observed similar recurrent loss of the same region of chromosome 2 in a mouse model of acute promyelocytic leukemia. In that system, chromosome 2 deletions were highly associated with the acquisition of additional cytogenetic abnormalities, suggesting genes deleted in this region may play a role in maintaining genomic integrity. Additionally, Sauvageau et al57 have reported that overexpression of HOX genes, HOXA9 in particular, can predispose to genomic instability. These findings suggest that HOXB6 and other leukemogenic HOX proteins promote leukemia in part due to nonrandom loss of the 2D-E4 region. Interestingly, we did not observe additional cytogenetic abnormalities associated with interstitial deletions of chromosome 2.
In summary, our results show that HOXB6 enhanced self-renewal of myeloid precursors consistent with its normal expression in the early stages of myeloid differentiation. Constitutive HOXB6 expression blocks myeloid differentiation and impairs erythropoiesis, megakaryopoiesis, and lymphopoiesis. Finally, when aberrantly expressed, HOXB6 exhibits many cardinal features of a potent oncoprotein, inducing enhanced proliferation of early hematopoietic precursors, inhibition of differentiation, and perhaps loss of critical loci on chromosome 2, findings all consistent with the frequent overexpression of HOXB6 in human AML.
Prepublished online as Blood First Edition Paper, November 2, 2004; DOI 10.1182/blood-2004-04-1583.
Supported by grants from the Research Service of the Department of Veterans Affairs (N.A.F., H.J.L., C.L.); the National Institutes of Health DK48642 (H.J.L.), CA08009 (C.L.), CA91245 (H.J.L.), CA84221 (S.C.K., M.M.L.); and the American Cancer Society via an Institutional Research Grant through the University of California San Francisco (UCSF) Comprehensive Cancer Center (N.A.F.). N.A.F. is a Veterans Affairs (VA) Career Development Award Recipient.
Presented in abstract form at the 45th annual meeting of the American Society of Hematology, San Diego, CA, December 9, 2003.
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The authors gratefully acknowledge Dr Robert Hawley for providing the MSCV retroviral vector, Dr Andrei Goga for providing the pCL Ecotropic packaging plasmid, Dr Mark Kamps for providing the B16 GM-CSF–producing cell line, and Dr Keith Humphries for the MIG and MIY vectors and technical assistance.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal