Abstract
Growth factor independence-1B (Gfi-1B) is a transcription factor with a highly conserved transcriptional repressor snail–Gfi-1 (SNAG) domain and 6 zinc-finger domains at the N- and C-terminus, respectively. Disruption of the Gfi-1B gene is lethal in the embryo with failure to produce definitive enucleated erythrocytes. In this study, we analyzed the role of Gfi-1B in human erythropoiesis. We observed an increase of Gfi-1B expression during erythroid maturation of human primary progenitor cells. We studied the consequences of variations in Gfi-1B expression in 2 transformed cell lines (K562 and UT7 cells), as well as in primary CD36+/GPA– progenitors. A knock-down of Gfi-1B delayed the terminal differentiation of K562 and primary cells. Forced expression of Gfi-1B in UT7 and K562 cells led to an arrest of proliferation and an induction of erythroid differentiation. Enforced expression of Gfi-1B in primary cells at the colony-forming units-erythroid (CFU-E) stage led to a partial glycophorin A (GPA) induction after erythropoietin (EPO) withdrawal but failed to protect cells from apoptosis. Deletion of the SNAG repressor domain abolished Gfi-1B–induced erythroid maturation, strongly suggesting that Gfi-1B acts in the late stage of erythroid differentiation as a transcriptional repressor.
Introduction
Recent studies have demonstrated that disruption of transcription factor function leads to either modifications in normal hematopoietic differentiation or leukemic transformation. Many genes coding for transcription factors have been reported to be mutated (Scl/Tal-1, Lmo2, Tel, E2A, Runx1)1 or their expression modified (Scl,2 Pu.1,3 and Ikaros4 ) in leukemic cells. Furthermore, important information about the role of these transcription factors in the regulation of hematopoiesis have been obtained from studies involving gene inactivation or overexpression (GATA,5 Fog,6 Tal,7 EKLF8 ).
Growth factor independence-1B (Gfi-1B) is a transcription factor that has been cloned by low-stringency hybridization to a probe derived from Gfi-1,9 the first described member of the family. Its expression is restricted to hematopoietic cells.9 Gfi-1 and Gfi-1B share high homology in 2 domains: an N-terminal repressor domain called Snail–Gfi-1 (SNAG) and 6 C2H2 zinc-fingers located at the C-terminus of the protein. The SNAG repressor domain also is shared by the orphan Hox gene, Gsh-1, the insulinoma-associated protein, IA-1, and the vertebrate members of the snail-slug protein family. Gfi-1 and Gfi-1B bind to a similar DNA sequence and encode both a position- and orientation-independent transcriptional repressor.9-12
While Gfi-1–deficient mice are neutropenic because of a blockage in myeloid maturation,13 gene targeting of Gfi-1B leads to deficient maturation of erythroid and megakaryocytic lineages.14 Gfi-1B–/– embryonic stem (ES) cells fail to contribute to red cells of adult chimeras, and Gfi-1B–/– embryos die by E15 with failure to produce definitive erythrocytes. Moreover, Gfi-1B seems to play a major role during human erythropoiesis, since its overexpression in early erythroid progenitors leads to their drastic expansion.15
Because erythroblasts from Gfi-1B–/– embryos exhibit a blockage of maturation at a c-kit+/ter119+ stage, we hypothesized that Gfi-1B could be implicated in the same way during the late step of human erythropoiesis. In this work, we analyze Gfi-1B function in human primary erythroid progenitors, as well as in 2 leukemic cell lines sharing an erythroid (and megakaryocytic) potential, K562 and UT7. Our results demonstrate that Gfi-1B is implicated in the induction of terminal erythroid maturation. Moreover, in contrast with its role in proliferation of early erythroblasts, Gfi-1B relies on its SNAG suppressor domain to achieve erythroid differentiation.
Materials and methods
K562 and UT7 cells culture
The K562 cell line was originally established from the pleural effusion of a patient with chronic myeloid leukemia16 and was maintained in RPMI medium (Invitrogen, Cergy Pontoise, France) supplemented with 10% fetal calf serum (FCS). The UT7 cell line was established from a patient with megakaryoblastic leukemia17 and maintained in α-minimum essential medium (MEM) (Invitrogen) supplemented with 10% FCS and 5 ng/mL recombinant human granulocyte-macrophage colony-stimulating factor (GM-CSF) (generously provided by Novartis Pharma S.A., Rueil Malmaison, France). Erythroid differentiation was induced in K562 cells by treatment with 0.1 μM aracytine (AraC) (Laboratoire Upjohn, Paris, France) and in UT7 cells by 2 UI/mL human recombinant erythropoietin (EPO) (a generous gift from OrthoBiotech, Paris, France) after 2 washes without growth factor.
Human primary cell culture
Human primary cells were obtained after informed consent according to the Declaration of Helsinki. Each patient, upon entering our institution, signed an agreement for the use of their samples for research. Study protocol was approved by the Institut Gustave Roussy ethical board. Cells from cord blood (CB) and peripheral blood (PB) were separated over a Ficollmetrizoate gradient (Biochrom, Berlin, Germany), and CD34+ cells were purified by an immunomagnetic selection (Miltenyi Biotec, Bergisch Gladbach, Germany). For erythroid differentiation, CD34+ cells were grown in Iscove modified Dulbecco medium (Invitrogen) containing 1.5% deionized bovine serum albumin (Cohn fraction V; Sigma, Saint-Quentin Fallavier, France), iron saturated human transferrin (Sigma), a mixture of sonicated lipids (20 μg/mL), 2 UI/mL EPO, 25 ng/mL human stem cell factor (SCF) (a generous gift from Amgen, Thousand Oaks, CA), 100 UI/mL interleukin 3 (IL3) (kindly provided by Novartis, France), and 10–6M dexamethasone (DXM) (Sigma). For colony assays, 1 000 to 5 000 cells were plated in H4100 medium (Stem Cell Technologies, Vancouver, Canada) supplemented with 10% FCS, 3 UI/mL EPO, 100 UI/mL IL3, 25 ng/mL SCF, and 10 ng/mL G-CSF (obtained from Bellon Laboratory, Neuilly-sur-Seine, France). Colony numbers were counted at day 7 for CFU-E and at day 14 for burst-forming unit-E (BFU-E) and CFU-GM.
Retroviral constructs, retrovirus production, and cell infection
The following human Gfi-1B cDNA were cloned upstream to the internal ribosomal entry site (IRES) linked to a green fluorescence protein (IRES-GFP) sequence in the MIGR (murine stem cell virus-internal ribosome entry site-green fluorescent protein) plasmid (a generous gift from J. Miller, Philadelphia, PA): full-length cDNA (Gfi-1B), full-length cDNA with a Flag sequence at the N-(NFlag) or at the C-terminus (CFlag), and SNAG deleted cDNA with or without Flag at the C terminus (Δ1 and Δ1Cflag, respectively). To obtain infectious retroviruses, 293 EBV-associated nuclear antigen (EBNA) cells (Invitrogen) were transfected by means of the Exgen reagent (Euromedex, Mundolsheim, France) with 3 different plasmids: pCMV-G (VSV-G coding sequence), pCMV-gag-pol (both kindly supplied by Dr J. Morgenstern, Millenium, Boston, MA), and a recombinant MIGR vector carrying the different Gfi-1B cDNAs. Supernatant containing infectious retroviral particles was recovered and concentrated by means of an Amicon membrane (Centricon, Millipore, Bedford, MA). Cells were incubated with the retroviral supernatant for 48 hours in the presence of hexadimethrine bromide (4 μg/mL, Sigma). Cells expressing GFP were sorted using a FacsVantage cytometer (BD Biosciences, Pont de Claix, France). Infection of human CD34+ cells was performed at day 4 and 5 of culture in the presence of EPO, SCF, IL3, and DXM as described above. At day 6, CD36+ and GPA– cells expressing GFP were sorted and plated in serum-free medium supplemented with 25 ng/mL SCF.
Immunolabeling for fluorescence microscopy and flow cytometric analysis of antigen expression
Cells were cytocentrifuged onto slides at 200g for 4 minutes, fixed in 0.2% paraformaldehyde (Serva, Heidelberg, Germany) for 5 minutes at room temperature, and permeabilized with 0.1% Triton X-100 (Sigma) prior to incubation with the M2 antibody (Sigma). After 3 washes, cells were incubated with a tetramethyl rhodamine [TRITC]–labeled antimouse antibody (Beckman Coulter, Villepinte, France). DNA was finally stained with 4′-6-diamidino-2-phenylindole-2HCl (DAPI) in Vectashield (Vector, Burlingame, CA). Phenotypic differentiation was monitored using the following antibodies: fluorescein isothiocyanate (FITC)–conjugated or allophycocyanin cell (APC)–conjugated CD36, FITC-conjugated CD34 (BD Biosciences), and a phycoerythrin (PE)–conjugated glycophorin A (Caltag, Burlingane, CA). Cells were analyzed using a FACSort or a FACScan cytometer (BD Biosciences).
Measurement of cell proliferation
DNA synthesis was estimated by measuring the [3H]-thymidine incorporation. In brief, 104 cells plated in 96-well dishes were incubated with [3H]-thymidine (0.5 μCi [0.0185 MBq] per well). Cells were collected after 6 hours, and the [3H]-thymidine incorporation into trichloroacetic acid–insoluble material was determined by liquid scintillation counting.
Northern Blot analysis
Total RNA were isolated by the guanidium isothiocyanate lysis procedure followed by centrifugation over a cesium-chloride cushion as described.18 Ten micrograms of total RNA was separated by electrophoresis on agarose-formaldehyde gels and transferred to nylon membranes (Gene-Screen; NEN Life Science Products, Boston, MA). Membranes were hybridized with fragments of human β-globin, δ-globin, ϵ-globin, and Gfi-1B cDNA. These fragments were labeled with α-32P-dCTP by random priming (Multiprime DNA-labeling system; Amersham, Orsay, France). After an overnight incubation at 42°C, membranes were washed and autoradiographed.
Quantitative reverse transcriptase–polymerase chain reaction (RT-PCR)
Total RNA were harvested using an acidic phenol extraction (RNAplus; Qbiogene, Carlsbad, CA). Samples were first treated with DNAase (Ambion, Huntington, United Kingdom), denatured, and reverse transcribed using a SuperScript II RNAse H- reverse transcriptase (Invitrogen). Real-time TaqMan PCR was carried out in the ABI Prism GeneAmp 5700 Sequence Detection System (Perkin-Elmer Applied Biosystems, Foster City, CA). In each experiment, Gfi-1B expression level was expressed relative to the housekeeping gene hypoxanthine-guanine-phosphoribosyltransferase (HPRT). Probes and primers were: Gfi-1B sense: 5′-AGCAAGAAGGCTCACACCTACC-3′, Gfi-1B antisense: 5′-GTTGCTTGGAGCCTGGTCT-3′, Gfi-1B probe: 5′-TGGGCACCGGGGTAAGGGCAT-3′, HPRT sense: 5′-GGCAGTATAATCCAAAGATGGTCAA-3′, HPRT antisense: 5′-TCAAATCCAACAAAGTCTGGCTTATAT-3′, HPRT probe: 5′-CTTGCTGGTGAAAAGGACCCCACGA-3′.
Transfection of K562 and primary erythroid progenitor cells with siRNA
Human Gfi-1B oligoribonucleotides were purchased from MWG-Biotech (Courtaboeuf, France). The sequence of the sense and the antisense strands are, respectively, GCCUAGCUUCUCCUGGGACdTdT and GUCCCAGGAGAAGCUAGGCdTdT. Oligonucleotides were deprotected as recommended by the supplier and were hybridized as previously described.19 For K562 cells, conditions for transduction of siRNA already have been published.20 CD34+ cells from CB or PB were cultured 5 days in serum-free medium in the presence of EPO, SCF, IL3, and DXM as described in “Human primary cell culture,” washed twice, and resuspended to a final concentration of 107 cells/mL in RPMI (Invitrogen). Cells were electroporated once or twice with 0.4 μg per 105 cells double-strand RNA (siRNA) or 0.2 μg per 105 cells antisense strand alone as control using a Biorad apparatus with the following conditions: voltage 280 V; capacitance 250 μF. After electroporation, cells were cultured in the presence of 2 UI/mL EPO and 25 ng/mL SCF. CD36 and glycophorin A (GPA) expression were evaluated by flow cytometry at 48 hours. Dead cells were excluded from the analysis using 7-amino–actinomycin D (7-AAD) staining (Sigma). Eg5 siRNA and a fluorescein-labeled scramble siRNA (Qiagen, Courtaboeuf, France) were used as transduction control. Sequences of Eg5 siRNA have been previously published.20 After electroporation with the fluorescent siRNA, transduction efficiency was evaluated the day after electroporation as the percentage of fluorescein-positive cells by flow cytometry.
Cell-cycle analysis
Cells were washed in phosphate-buffered saline (PBS) and labeled overnight in 400 μL of citrate buffer containing 25 μg/mL propidium iodine, 50 μg/mL RNAse A (Merck, Darmstadt, Germany), and 0.1% Nonidet P40 (Sigma) in the dark at 4°C. The samples were then analyzed with a FACSort cytometer, and the percentage of cells in the different phases of the cell cycle was calculated.
Statistical analysis
Student t test was used each time 3 experiments were performed, and P value was determined.
Results
Gfi-1B is up-regulated during erythroid differentiation of human primary cells
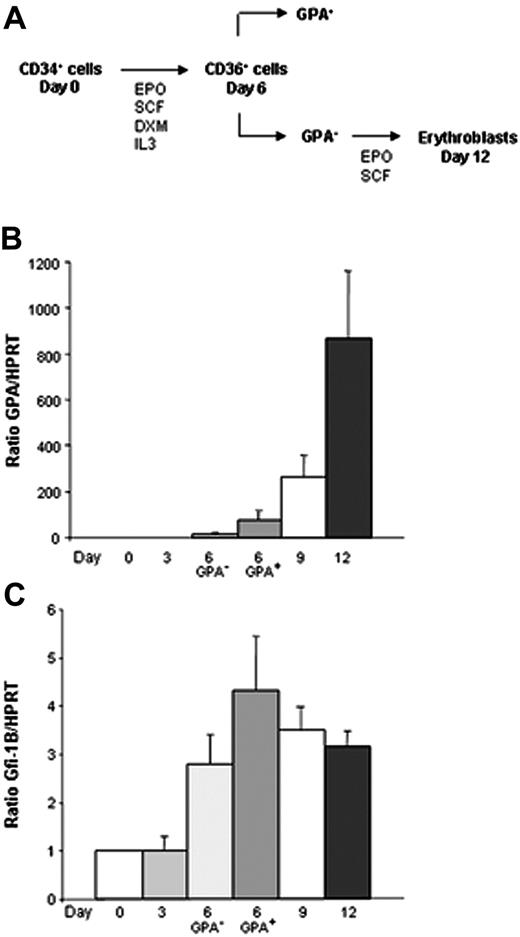
In order to approach the role of Gfi-1B in erythroid differentiation of human primary cells, we used a 2-step system of in vitro differentiation (Figure 1A). Purified CD34+ cells obtained from PB of patients treated with G-CSF were cultured in serum-free medium in the presence of EPO, SCF, IL3, and dexamethasone during 6 days, allowing the amplification of CD36+ erythroid progenitors. Cells were then sorted according to GPA expression. At last, CD36+/GPA– cells were induced into terminal differentiation in the presence of SCF and EPO during 6 additional days. Each 3 days of these kinetics, total RNA was prepared, real-time RT-PCR was performed, and the Gfi-1B/HPRT and the GPA/HPRT ratios were determined. As expected, GPA level was very low at day 3 of culture and increased dramatically during the late steps of differentiation from day 6 to day 12 (Figure 1B). As shown in Figure 1C, Gfi-1B expression was detectable in CD34+ cells, and increased during erythroid maturation. Of interest, Gfi-1B level was higher in the CD36+/GPA+ progenitor cells after 6 days of culture and did not increase furthermore. We concluded from these experiments that Gfi-1B is up-regulated during erythropoiesis at the moment where cells begin to enter into terminal differentiation.
Induction of Gfi-1B during erythroid maturation of human primary cells. Expression of Gfi-1B was determined by real-time PCR and normalized using HPRT as a housekeeping gene. (A) A 2-step system of erythroid differentiation: CD34+ cells from PB were cultured in a serum-free medium in the presence of EPO, SCF, IL3, and DXM. After 6 days of culture, CD34 expression was lost, and nearly all the cells expressed the CD36 antigen at their surface. Cells were sorted according to the GPA expression, and CD36+/GPA– cells were then induced into terminal maturation in the presence of SCF and EPO alone. (B) Up-regulation of GPA expression during erythroid differentiation. GPA level mainly increased during the late steps of maturation (RT-PCR: n = 3, each in triplicate). (C) High expression of Gfi-1B during erythroid maturation. Gfi-1B level increased at day 6 of erythroid differentiation and was higher in the CD36+/GPA+ fraction than in the CD36+/GPA– one (RT-PCR: n = 3 each in triplicate). Data are expressed as mean ± SD.
Induction of Gfi-1B during erythroid maturation of human primary cells. Expression of Gfi-1B was determined by real-time PCR and normalized using HPRT as a housekeeping gene. (A) A 2-step system of erythroid differentiation: CD34+ cells from PB were cultured in a serum-free medium in the presence of EPO, SCF, IL3, and DXM. After 6 days of culture, CD34 expression was lost, and nearly all the cells expressed the CD36 antigen at their surface. Cells were sorted according to the GPA expression, and CD36+/GPA– cells were then induced into terminal maturation in the presence of SCF and EPO alone. (B) Up-regulation of GPA expression during erythroid differentiation. GPA level mainly increased during the late steps of maturation (RT-PCR: n = 3, each in triplicate). (C) High expression of Gfi-1B during erythroid maturation. Gfi-1B level increased at day 6 of erythroid differentiation and was higher in the CD36+/GPA+ fraction than in the CD36+/GPA– one (RT-PCR: n = 3 each in triplicate). Data are expressed as mean ± SD.
Inhibition of Gfi-1B gene expression by small interfering RNA delays erythroid differentiation in K562 and CD36+ cells
K562 cells are able to differentiate toward erythropoiesis when exposed to different chemical inducers like AraC.21 After 4 days of 0.1 μM AraC treatment, more than 50% of cells synthesized hemoglobin and were benzidine (BZD) positive (data not shown). Because K562 cells constitutively expressed Gfi-1B, we investigated whether this transcription factor is absolutely required for the erythroid potential of these cells using a siRNA strategy. In a first step, we wanted to quantify the amount of short interfering RNA (siRNA) electroporated in the cells. Two approaches were used. First, siRNA targeting the mRNA of the kinesin Eg5 induces a mitotic arrest thus providing a convenient assay to quantify the penetration of siRNA.20 When 0.4 μg siRNA were transfected per 105 cells, nearly 80% of K562 cells accumulated in G2/M instead of 20% in the controls (Figure 2A). Second, we used a fluorescein-labeled scramble siRNA to quantify the transduction efficiency after electroporation. Flow cytometry analysis revealed that more than 60% of the K562 cells were fluorescein positive after 24 hours (Figure 2B). These results indicate that electroporation can enable the penetration of siRNA in a vast majority of the cells.
Effect of Gfi-1B knock-down by a RNAi strategy. (A) Antisense (left) or siRNA (right) Eg5 were electroporated in K562 cells. Percent of cells in G2/M phases of the cell cycle was determined 48 hours after electroporation. (B) A fluorescein-labeled nonrelevant siRNA was electroporated in K562 cells, and the percent of fluorescent cells was measured before (left) and the day after electroporation (right). (C) Gfi-1B expression determined by Northern blot analysis. The Gfi-1B–specific siRNA or the sense strand alone were electroporated in K562 cells, and Gfi-1B expression level was measured 7, 20, and 48 hours after electroporation. Ribosomal 18S RNA was used as control of RNA deposit. (D) siRNA delays AraC induced hemoglobinization of K562 cells. K562 cells were electroporated without (buffer, ♦) or with sense (▴), antisense (•), or siRNA (▪) and immediately exposed to aracytine. Benzidine staining was monitored each day.
Effect of Gfi-1B knock-down by a RNAi strategy. (A) Antisense (left) or siRNA (right) Eg5 were electroporated in K562 cells. Percent of cells in G2/M phases of the cell cycle was determined 48 hours after electroporation. (B) A fluorescein-labeled nonrelevant siRNA was electroporated in K562 cells, and the percent of fluorescent cells was measured before (left) and the day after electroporation (right). (C) Gfi-1B expression determined by Northern blot analysis. The Gfi-1B–specific siRNA or the sense strand alone were electroporated in K562 cells, and Gfi-1B expression level was measured 7, 20, and 48 hours after electroporation. Ribosomal 18S RNA was used as control of RNA deposit. (D) siRNA delays AraC induced hemoglobinization of K562 cells. K562 cells were electroporated without (buffer, ♦) or with sense (▴), antisense (•), or siRNA (▪) and immediately exposed to aracytine. Benzidine staining was monitored each day.
To determine the effects of the Gfi-1B knock-down, siRNA corresponding to a sequence of the Gfi-1B coding region was introduced in K562 cells using the same conditions of transfection. Sense or antisense strand alone was used as a negative control. After electroporation, cells were cultured in the presence of 0.1 μM aracytine, and erythroid differentiation was monitored each day by benzidine staining. As shown in Figure 2D, 3 and 4 days after introduction of Gfi-1B siRNA, hemoglobinization of K562 cells was reduced. The effect of the siRNA was transient, and the cells underwent the differentiation program 5 days after electroporation. The decrease in the level of Gfi-1B mRNA was confirmed by Northern blot analysis (Figure 2C). Seven and 20 hours after electroporation, Gfi-1B mRNA levels were lower in cells electroporated with the Gfi-1B siRNA compared to cells containing the sense strand alone. The residual Gfi-1B mRNA 7 and 20 hours after electroporation must have been due to a partial inhibition with the siRNA sequence we used and to the cells that were not transfected.
In order to test whether down-regulation of Gfi-1B had the same inhibitory effect on maturation of primary cells, we transfected Gfi-1B siRNA into erythroid progenitors. CD34+ cells were cultured and electroporated as described in “Material and methods.” As shown in Figure 3A, evaluation of transduction efficiency with the fluorescein-labeled siRNA revealed that only 24% of the cells were transfected. Eg5 siRNA could not be used as a second control in primary cells since mitotic arrest led to an early and massive mortality in transduced cells. Even though transduction of Gfi-1B siRNA was suboptimal, a delay in erythroid differentiation induced by EPO was observed. While 34.5% ± 4.5% of the cells transfected with a control siRNA expressed GPA 2 days after transfection, only 25% ± 5% expressed it after Gfi-1B siRNA transfection (Figure 3B-C). Because one-third reduction in GPA expression corresponds to the percentage of transduced cells, these results suggest that Gfi-1B is essential for erythroid maturation of primary erythroid progenitors.
Gfi-1B knock-down delays acquisition of GPA in CD36+ cells. (A) Control of transduction efficiency in primary cells. CD34+ from PB (n = 3) were cultured during 5 to 6 days in serum-free medium with EPO, SCF, IL3, and DXM. Fluorescein-labeled nonrelevant siRNA were electroporated day 6 after the beginning of the culture. The percentage of fluorescein-positive cells was determined before (left) and the day after (right) electroporation. (B) CD34+ from CB (n = 3) or PB (n = 1) were cultured in the presence of EPO, SCF, IL3, and DXM during 5 to 6 days, then electroporated with the Gfi-1B–specific siRNA (siRNA) or the antisense strand alone (AS) as control. Cells were then cultured in presence of EPO and SCF. After 48 hours, GPA expression level was determined by flow cytometry (antisense versus siRNA: P = .047, n = 4). Data are expressed as mean ± SD. (C) Flow cytometry analysis of one representative experiment, showing CD36/GPA expression (i) before electroporation, (ii) 48 hours after electroporation with the antisense strand alone, (iii) 48 hours after electroporation with the Gfi-1B–specific siRNA.
Gfi-1B knock-down delays acquisition of GPA in CD36+ cells. (A) Control of transduction efficiency in primary cells. CD34+ from PB (n = 3) were cultured during 5 to 6 days in serum-free medium with EPO, SCF, IL3, and DXM. Fluorescein-labeled nonrelevant siRNA were electroporated day 6 after the beginning of the culture. The percentage of fluorescein-positive cells was determined before (left) and the day after (right) electroporation. (B) CD34+ from CB (n = 3) or PB (n = 1) were cultured in the presence of EPO, SCF, IL3, and DXM during 5 to 6 days, then electroporated with the Gfi-1B–specific siRNA (siRNA) or the antisense strand alone (AS) as control. Cells were then cultured in presence of EPO and SCF. After 48 hours, GPA expression level was determined by flow cytometry (antisense versus siRNA: P = .047, n = 4). Data are expressed as mean ± SD. (C) Flow cytometry analysis of one representative experiment, showing CD36/GPA expression (i) before electroporation, (ii) 48 hours after electroporation with the antisense strand alone, (iii) 48 hours after electroporation with the Gfi-1B–specific siRNA.
Overexpression of Gfi-1B induces arrest of cell proliferation and promotes erythroid differentiation in K562 and UT7 cells
As described earlier,22 UT7 cells from a clone expressing Mpl (UT7/mpl, UT7 5.3) is able to differentiate toward erythropoiesis in the presence of EPO. Because both K562 and UT7 cells express Gfi-1B and share an erythroid potential, we studied the impact of Gfi-1B–forced expression in these 2 cell lines using a retroviral strategy. Complete human Gfi-1B cDNA was cloned into the retroviral vector MIGR and exogenous Gfi-1B expression was quantified by Northern blot in sorted GFP-positive cells. Figure 4A shows that, while endogenous Gfi-1B expression was low in K562 and UT7 cells, the exogenous retroviral RNA was highly expressed.
Forced expression of wild-type Gfi-1B cDNA induces erythroid differentiation in UT7 and K562 cell lines. (A) Northern blot analysis of mRNA from UT7 and K562 cells transduced with full-length Gfi-1B or the empty vector MIGR. Gfi-1B cDNA was used as a 32P-labeled probe. The expression level of the exogenous Gfi-1B is high (bicistronic mRNA of 5 Kb), while the endogenous expression level is low (2 Kb). (B) Measurement of cell proliferation by [3H]-thymidine incorporation. Two days after Gfi-1B transduction, the proportion of the cells in the S phase of the cell cycle was determined. Cells were incubated with [3H]-thymidine for 6 hours, and [3H]-thymidine incorporation was determined. Overexpression of Gfi-1B in K562 and UT7 cells inhibits cell proliferation in comparison with cells transduced with the empty vector MIGR. (C) Overexpression of Gfi-1B induces erythroid differentiation in both K562 and UT7 cells. GPA expression was evaluated by flow cytometry at day 4 after retroviral infection in UT7 cells (left). Hemoglobinization was determined by benzidine staining at day 6 in K562 cells (right). Full-length Gfi-1B expression induces expression of erythroid markers. Data are expressed as mean ± SD. (D) Globin expression analyzed by Northern blot using β, γ, and ϵ globin probes. Forced expression of full-length Gfi-1B induces expression of β and γ globins at a transcriptional level in UT7 cells in a same manner as EPO. In K562 cells, forced expression of Gfi-1B induces the transcription of the ϵ globin but not the β globin, in a same manner as AraC.
Forced expression of wild-type Gfi-1B cDNA induces erythroid differentiation in UT7 and K562 cell lines. (A) Northern blot analysis of mRNA from UT7 and K562 cells transduced with full-length Gfi-1B or the empty vector MIGR. Gfi-1B cDNA was used as a 32P-labeled probe. The expression level of the exogenous Gfi-1B is high (bicistronic mRNA of 5 Kb), while the endogenous expression level is low (2 Kb). (B) Measurement of cell proliferation by [3H]-thymidine incorporation. Two days after Gfi-1B transduction, the proportion of the cells in the S phase of the cell cycle was determined. Cells were incubated with [3H]-thymidine for 6 hours, and [3H]-thymidine incorporation was determined. Overexpression of Gfi-1B in K562 and UT7 cells inhibits cell proliferation in comparison with cells transduced with the empty vector MIGR. (C) Overexpression of Gfi-1B induces erythroid differentiation in both K562 and UT7 cells. GPA expression was evaluated by flow cytometry at day 4 after retroviral infection in UT7 cells (left). Hemoglobinization was determined by benzidine staining at day 6 in K562 cells (right). Full-length Gfi-1B expression induces expression of erythroid markers. Data are expressed as mean ± SD. (D) Globin expression analyzed by Northern blot using β, γ, and ϵ globin probes. Forced expression of full-length Gfi-1B induces expression of β and γ globins at a transcriptional level in UT7 cells in a same manner as EPO. In K562 cells, forced expression of Gfi-1B induces the transcription of the ϵ globin but not the β globin, in a same manner as AraC.
In order to study the effect of Gfi-1B forced expression on cell proliferation, we determined the proportion of cells in DNA synthesis by measuring [3H] thymidine incorporation. Figure 4B shows that proliferation of K562 and UT7 cells overexpressing Gfi-1B was strongly decreased in comparison with control cells infected with the empty retroviral vector MIGR.
Furthermore, in both cell lines, we observed that overexpression of Gfi-1B promoted erythroid differentiation in the absence of any inducer (ie, EPO for UT7 and AraC for K562) (Figure 4C). This was demonstrated by monitoring GPA and hemoglobin expression in UT7 and K562 cells at day 4 after transduction. Figure 4C shows that while 8% ± 3% of the UT7 cells infected with the empty vector MIGR expressed GPA, 81% ± 9% of the cells overexpressing Gfi-1B were GPA positive. 3% ± 3% of the control K562 cells were stained by benzidine 6 days after induction of differentiation, while 49% ± 11% of Gfi-1B–infected cells expressed hemoglobin. Induction of globin synthesis took place at mRNA level as shown by Northern blot analysis using β-, γ-, and ϵ-globin probes (Figure 4D). These results demonstrate that forced expression of Gfi-1B induces both arrest of cell proliferation and induction of differentiation in K562 and UT7 cells.
The SNAG domain of Gfi-1B is indispensable for induction of erythroid differentiation
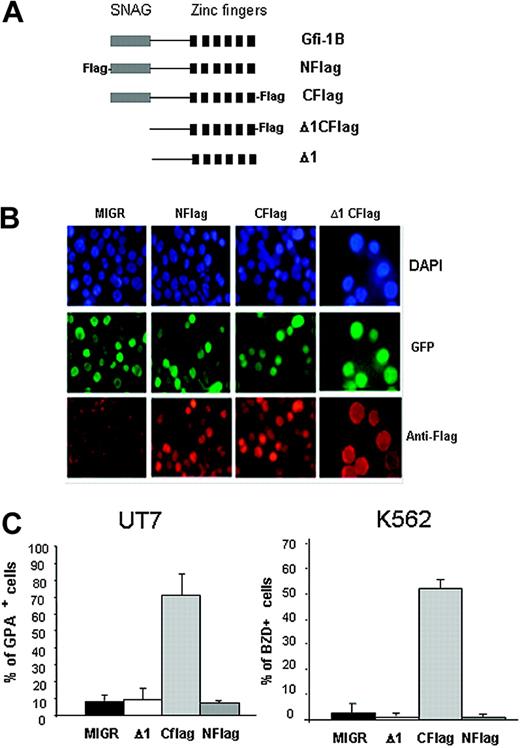
In order to study the localization of the transduced Gfi-1B, and because no antibody against Gfi-1B was available, we added a tag sequence to the N-terminus of Gfi-1B cDNA (Nflag, Figure 5A) and infected the cells with this modified retrovirus. No effect on differentiation was observed with this modified retrovirus, although N-Flag Gfi-1B was expressed in the nucleus of infected cells (Figure 5B). Because the N-terminus–tagged Gfi-1B was not able to induce arrest of proliferation or erythroid differentiation and because the SNAG domain is located at the N-terminus of the protein, we hypothesized that the Flag sequence prevented the function of the SNAG domain. For example, the Flag sequence could alter protein-protein interactions and thereby inhibit the SNAG function. We then cloned a C-terminus–tagged full-length Gfi-1B cDNA in the MIGR retroviral vector (Cflag) and overexpressed it in UT7 and K562 cell lines. Similar nuclear staining patterns were observed by immunofluorescence (Figure 5B). However, interestingly, the presence of the Flag sequence at the C-terminus did not abrogate Gfi-1B–induced erythroid differentiation of both cell lines (Figure 5C). To verify that the absence of effect observed with the N-tagged construct was due to the nonfunctionality of the SNAG domain, we deleted the SNAG domain of Gfi-1B and cloned the mutant cDNA with or without tag at the C-terminus in the retroviral vector MIGR (Δ1Cflag and Δ1, respectively). Despite a nuclear localization (as shown in Figure 5B, Δ1Cflag construct), the forced expression of the SNAG-deleted mutant was not able to induce GPA or hemoglobinization in K562 and UT7 cells (Figure 5C).
The SNAG domain of Gfi-1B is indispensable for induction of erythroid differentiation. (A) Schematic representation of the different constructs cloned into the retroviral vector MIGR. Full-length Gfi-1B (Gfi-1B), Flag-tagged at the N-terminus (NFlag) or at the C-terminus (CFlag), SNAG-deleted cDNA without (Δ1) or with (Δ1CFlag) at the C-terminus. (B) Immunofluorescence performed with an anti-Flag antibody showing a similar nuclear expression of all tagged proteins. DAPI staining reveals nucleus, all 4 populations are GFP positive, and anti-Flag antibody reveals Gfi-1B–infected cells. Cells were examined under a Nikon Eclipse E600 epifluorescence microscope equipped with an Apochromat Plan 60 ×/1.4 objective lens (Nikon, Tokyo, Japan). Images were acquired with a CoolSnap camera (Photometrics, Evry, France) and processed with Adobe Photoshop 7.0 software (Adobe, San Jose, CA). (C) Analysis of erythroid differentiation induced by Gfi-1B–forced expression. UT7 and K562 were infected with the empty MIGR or with the MIGR carrying the N-(Nflag) or the C-(Cflag) Flag or the deleted SNAG (Δ1) cDNA. GPA expression was determined by flow cytometry, 4 days after retroviral infection in UT7 cells (left). Hemoglobinization was determined by benzidine staining 6 days after infection of K562 cells (right). Data are expressed as mean ± SD.
The SNAG domain of Gfi-1B is indispensable for induction of erythroid differentiation. (A) Schematic representation of the different constructs cloned into the retroviral vector MIGR. Full-length Gfi-1B (Gfi-1B), Flag-tagged at the N-terminus (NFlag) or at the C-terminus (CFlag), SNAG-deleted cDNA without (Δ1) or with (Δ1CFlag) at the C-terminus. (B) Immunofluorescence performed with an anti-Flag antibody showing a similar nuclear expression of all tagged proteins. DAPI staining reveals nucleus, all 4 populations are GFP positive, and anti-Flag antibody reveals Gfi-1B–infected cells. Cells were examined under a Nikon Eclipse E600 epifluorescence microscope equipped with an Apochromat Plan 60 ×/1.4 objective lens (Nikon, Tokyo, Japan). Images were acquired with a CoolSnap camera (Photometrics, Evry, France) and processed with Adobe Photoshop 7.0 software (Adobe, San Jose, CA). (C) Analysis of erythroid differentiation induced by Gfi-1B–forced expression. UT7 and K562 were infected with the empty MIGR or with the MIGR carrying the N-(Nflag) or the C-(Cflag) Flag or the deleted SNAG (Δ1) cDNA. GPA expression was determined by flow cytometry, 4 days after retroviral infection in UT7 cells (left). Hemoglobinization was determined by benzidine staining 6 days after infection of K562 cells (right). Data are expressed as mean ± SD.
We therefore concluded that the presence of a functional SNAG domain of Gfi-1B is indispensable for promoting erythroid maturation in these 2 cell lines.
Gfi-1B overexpression in primary erythroid progenitors induces erythroid maturation without EPO
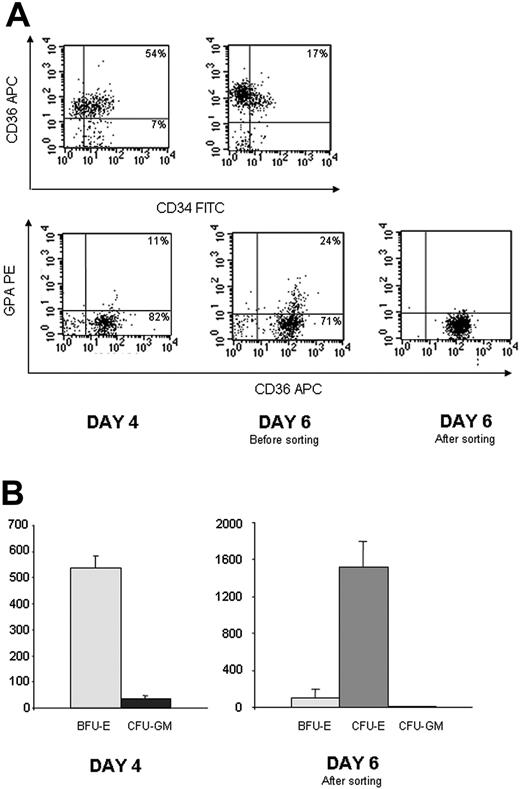
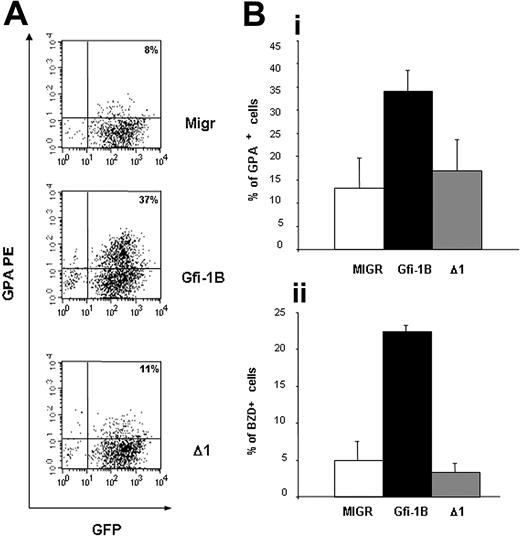
To verify whether Gfi-1B overexpression induces erythroid maturation in primary erythroid progenitors, we cultured CD34+ from PB in a serum-free medium in the presence of EPO, SCF, IL3, and DXM as described in “Materials and methods.” At days 4 and 5 of the culture, we infected these cells with retroviruses carrying the full-length or the SNAG-deleted Gfi-1B cDNA. Characterization of the cells after the first infection showed that most of them had an immature phenotype CD34+/CD36+/GPA– (Figure 6A). Colony assays revealed that they corresponded to the BFU-E stage (Figure 6B). When we sorted the infected cells at day 6, nearly all the cells had lost CD34 expression, and they cloned in colony assays as CFU-E (Figure 6A-B). Sorted cells CD36+/GPA–/GFP+ were plated in serum-free medium with 25 ng/mL of SCF alone. Two days after, 34% ± 5% of the cells overexpressing wild-type Gfi-1B were GPA positive versus only 13% ± 6% and 17% ± 7% after infection with the MIGR or Δ1 construct, respectively (Figure 7A-B). In the same way 23% ± 1% of the cells overexpressing wild-type Gfi-1B became benzidine positive versus 5% ± 3% and 3% ± 1% with the 2 other vectors (Figure 7B). Interestingly, Gfi-1B expression allowed neither cell proliferation nor cell survival in the absence of EPO. The cells transduced with MIGR, Gfi-1B, or Δ1 vectors started to die the third day after the cell sorting. These results demonstrate that Gfi-1B is involved in the erythroid differentiation process in the absence of antiapoptotic activity.
Characterization of CD34+ cells from PB cultured in serum-free medium in presence of EPO, SCF, DXM, and IL3 at day 4 (when the cells were retrovirally infected for the first time) and at day 6 (when the cells were sorted) (n = 3). (A) Flow cytometry analysis of one representative experiment showing that at day 4, most of the cells were still CD34 positive (71%), expressed CD36 but not GPA. At day 6, expression of CD34 was lost (17% positive), and cells were in majority CD36+/GPA–. (B) Colony-assay. 1 000 cells were plated in semisolid medium in presence of G-CSF, EPO, SCF, and IL3. Colonies were counted at day 7 for CFU-E and at day 14 for BFU-E and CFU-GM. Results are expressed as the number of colonies per 5 000 cells. Cells plated at day 4 are BFU-E (left). Infected CD36+/GPA– cells sorted after 6 days in culture gave rise to CFU-E (right). Data are expressed as mean ± SD.
Characterization of CD34+ cells from PB cultured in serum-free medium in presence of EPO, SCF, DXM, and IL3 at day 4 (when the cells were retrovirally infected for the first time) and at day 6 (when the cells were sorted) (n = 3). (A) Flow cytometry analysis of one representative experiment showing that at day 4, most of the cells were still CD34 positive (71%), expressed CD36 but not GPA. At day 6, expression of CD34 was lost (17% positive), and cells were in majority CD36+/GPA–. (B) Colony-assay. 1 000 cells were plated in semisolid medium in presence of G-CSF, EPO, SCF, and IL3. Colonies were counted at day 7 for CFU-E and at day 14 for BFU-E and CFU-GM. Results are expressed as the number of colonies per 5 000 cells. Cells plated at day 4 are BFU-E (left). Infected CD36+/GPA– cells sorted after 6 days in culture gave rise to CFU-E (right). Data are expressed as mean ± SD.
Overexpression of Gfi-1B in human CD36+/GPA– cells induces expression of GPA in the absence of EPO. CD34+ from PB were cultured in the presence of EPO, SCF, IL3, and DXM. The cells were then transduced twice with the different retroviral vector (MIGR, full-length Gfi-1B, or SNAG-deleted Gfi-1B, Δ1) at day 4 and day 5 of the culture. Infected CD36+/GPA– cells were sorted at day 6 of culture in the presence of IL3, EPO, SCF, and DXM and cultured in the presence of SCF, without EPO. GPA expression and BZD staining were evaluated 48 and 72 hours after EPO removal. (A) Flow cytometry analysis of the cells 48 hours after cell sorting (one representative experiment). In cells overexpressing full-length Gfi-1B, expression of GPA was higher than in cells transduced with the empty vector MIGR or with the SNAG-deleted Gfi-1B cDNA. (B) Induction 48 hours after cell sorting of erythroid markers determined by flow cytometry using antibody against GPA (i) (MIGR versus Gfi-1B: P = .01, n = 4; MIGR versus Δ1: P > .05) and by benzidine staining (ii) (MIGR versus Gfi-1B: P = .001, n = 4; MIGR versus Δ1: P > .05). Data are expressed as mean ± SD.
Overexpression of Gfi-1B in human CD36+/GPA– cells induces expression of GPA in the absence of EPO. CD34+ from PB were cultured in the presence of EPO, SCF, IL3, and DXM. The cells were then transduced twice with the different retroviral vector (MIGR, full-length Gfi-1B, or SNAG-deleted Gfi-1B, Δ1) at day 4 and day 5 of the culture. Infected CD36+/GPA– cells were sorted at day 6 of culture in the presence of IL3, EPO, SCF, and DXM and cultured in the presence of SCF, without EPO. GPA expression and BZD staining were evaluated 48 and 72 hours after EPO removal. (A) Flow cytometry analysis of the cells 48 hours after cell sorting (one representative experiment). In cells overexpressing full-length Gfi-1B, expression of GPA was higher than in cells transduced with the empty vector MIGR or with the SNAG-deleted Gfi-1B cDNA. (B) Induction 48 hours after cell sorting of erythroid markers determined by flow cytometry using antibody against GPA (i) (MIGR versus Gfi-1B: P = .01, n = 4; MIGR versus Δ1: P > .05) and by benzidine staining (ii) (MIGR versus Gfi-1B: P = .001, n = 4; MIGR versus Δ1: P > .05). Data are expressed as mean ± SD.
Discussion
Gfi-1 and Gfi-1B are 2 zinc finger repressors that play a major role in hematopoiesis. The role played by Gfi-1B on erythroid differentiation has been emphasized by several reports. First, Gfi-1B is highly expressed in the erythroid lineage. This has been shown in the chicken23 and in the mouse erythroid cell lines 32D-EPO-R and HCD 57.12 Gfi-1B expression also has been described in murine spleen and bone marrow9 as well as in human CD34+ cells undergoing erythroid differentiation.15 Second, Gfi-1B knock-out in mouse is lethal due to a major defect in the erythroid and megakaryocytic lineages. In the Gfi-1B–/– embryos, the absence of Gfi-1B does not alter erythroid commitment but totally blocks erythroid differentiation, leading to an accumulation of c-kit and Ter-119–positive cells in the fetal liver.14 In the present study, we show by quantitative reverse transcription polymerase chain reaction (RQ-PCR) that expression of Gfi-1B increases during erythroid differentiation at the moment cells enter into terminal differentiation. In agreement with this, we knocked down Gfi-1B by RNA interference in human cells undergoing erythroid differentiation. Decreased levels of Gfi-1B induced by siRNA transfection delayed acquisition of erythroid markers in K562 as well as in CD36-positive cells. Therefore, it appears that expression of Gfi-1B plays an important role in the maturation of committed erythroid progenitor cells.
The role of Gfi-1B in induction of erythroid maturation was further supported by the results of Gfi-1B overexpression experiments in K562 and UT7 cell lines. Indeed, Gfi-1B overexpression was sufficient to induce erythroid differentiation and a blockage of cell proliferation. This result contrasts with data reported by Jegalian and Wu12 in the murine erythroid cell lines 32D/EPO-R, where Gfi-1B overexpression increased the cell proliferation induced by EPO.12 However, 32D/EPO-R cells are different from UT7 cells with respect to their response to EPO. 32D/EPO-R cells are dependent on EPO for proliferation, while UT7 cells (cells from the UT7/Mpl 5.3 clone) respond to EPO by arrest of cell proliferation and induction of differentiation.
The arrest of cell proliferation observed in K562 and UT7 cells after Gfi-1B transduction may be the consequence of entry of the cells into a differentiation program. GATA-1, another transcription factor important for erythroid development, recently has been proposed to mediate proliferation arrest before induction of erythroid differentiation.24 The authors have demonstrated that, in a complementation assay based on synchronous inducible rescue of GATA-1– erythroblasts, GATA-1 both up-regulates p27 and represses the c-myc proto-oncogene. This could equally apply to Gfi-1B, which may repress c-myc transcription given the presence of Gfi-1B binding site in the myc promoter.11
Gfi-1B overexpression in human erythroid progenitor cells induced a partial erythroid maturation in the absence of EPO. However, these cells did not proliferate and could not survive after 3 days without EPO. This result is in sharp contrast with the previous work reported by Osawa et al,15 where overexpression of Gfi-1B led to the expansion of erythroid progenitor cells without terminal differentiation. This difference may be related to the stage of differentiation in which Gfi-1B overexpression was performed. Indeed, Osawa et al15 overexpressed Gfi-1B in immature CD34+ cells and observed an amplification at the BFU-E level, a cellular stage where cells were dependent on GM-CSF and not on EPO.15 In contrast, we studied the consequences of Gfi-1B overexpression in CD36+/GPA– cells. These cells were mainly CD34-negative, corresponded to the CFU-E stage, and were dependent on EPO and SCF for their proliferation and survival. When further maturing, they became dependent only on Epo, which induces the antiapoptotic protein Bcl-XL.25 This may explain why Gfi-1B overexpressing cells did not survive in the absence of EPO. Indeed, it has been shown that Gfi-1B did not induce Bcl-XL expression in erythroid cells.15 Thus Gfi-1B may play a crucial role during erythroid differentiation by permitting the exit of the cells from the BFU-E/CFU-E cell pool to the terminal differentiation.
The molecular mechanisms by which Gfi-1B acts on the erythroid program remains unknown. Several target genes of Gfi-1B have been identified. In the myelomonocytic M1 cells, Gfi-1B repressed the cell cycle CDKI p21Waf1,9 but relevance of p21Waf1 regulation in erythroid differentiation is questionable since there was no change in p21Waf1 expression levels in UT7 and K652 cells during erythroid differentiation (data not shown) or in Gfi-1B–/– embryos.14 Alternatively, in the murine erythroid cell lines 32D-EPO-R and HCD 57, Gfi-1B regulates SOCS1 and SOCS3 levels by a direct repression of their transcription.12 At the terminal stage of erythroid differentiation, Gfi-1B is expressed, while SOCS3 is not. Inactivation of the SOCS3 gene and upregulation of Gfi-1B would have the same implication: that is, induction of erythroid differentiation. In agreement with this hypothesis, it has been shown that SOCS3 inactivation induces a pathology characterized by a massive erythrocytosis throughout the embryo that can be due to an accelerated differentiation.26 In modulating SOCS expression, Gfi-1B may interfere with mitogenic signaling pathway activated by EPO, and this may promote the cells to enter into the terminal stages of erythroid differentiation.
Overexpression of a tagged form of Gfi-1B revealed that erythroid differentiation of UT7 and K562 cell lines occurred when the Flag-tag was at the C-terminus but not at the N-terminus end of the protein, despite a similar nuclear expression of both forms. This may be due to inhibition of the SNAG N-terminal repressor domain by the tag. In agreement with this hypothesis, a SNAG-deleted Gfi-1B did not induce erythroid differentiation in the 2 cell lines nor in CD36+ cells. This is consistent with the fact that all the target genes of Gfi-1B or of its homologs Gfi-1 identified to date appear to require a functional SNAG domain for transcriptional repression. For example, in the M1 cell line, repression of p21Waf1 relies on the presence of the SNAG domain,9 and a debilitating mutation of the SNAG domain abrogates repression of SOCS gene in the 32D-EPO-R cell line.12 By contrast, the SNAG domain is dispensable for inducing proliferation of immature human erythroblasts, and this effect is not dependent on transcriptional repression. Indeed, luciferase assays revealed in this work that in erythroid cells, a SNAG-deleted protein had a transactivating activity on a target promoter.15
Altogether our results and those from Osawa suggest that Gfi-1B could have distinct effects at the various cell stages during erythroid differentiation. In immature erythroid progenitors, through the zinc fingers domains, Gfi-1B would transactivate target genes implicated in cell proliferation. Later during the differentiation, Gfi-1B would regulate genes that have to be repressed for induction of terminal maturation. Of interest, the homologous protein Gfi-1 plays a major role during myeloid differentiation.27 Several target genes of Gfi-1 already have been identified.28 In particular, Gfi-1 represses the neutrophil elastase ELA2 gene. Inherited mutations of the Gfi-1 gene with loss of this repression capacity lead to congenital neutropenia.29
Therefore, characterization of the target genes of Gfi-1B in erythroblasts will provide important insights into the mechanisms of terminal erythropoiesis and may be relevant to understand at a molecular level pathologies that affect the erythroid lineage.
Prepublished online as Blood First Edition Paper, October 26, 2004; DOI 10.1182/blood-2003-11-4068.
Supported by grants from the Institut National de la Santé et de la Recherche Médicale and the Association pour la Recherche contre le Cancer, and the Institut Gustave Roussy (L.G.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We are grateful to Yann Lecluse and Frederic Larbret for cell sorting experiments, and to Adlen Foudi and Dr Nazanine Modjtahedi for their kind assistance.



![Figure 4. Forced expression of wild-type Gfi-1B cDNA induces erythroid differentiation in UT7 and K562 cell lines. (A) Northern blot analysis of mRNA from UT7 and K562 cells transduced with full-length Gfi-1B or the empty vector MIGR. Gfi-1B cDNA was used as a 32P-labeled probe. The expression level of the exogenous Gfi-1B is high (bicistronic mRNA of 5 Kb), while the endogenous expression level is low (2 Kb). (B) Measurement of cell proliferation by [3H]-thymidine incorporation. Two days after Gfi-1B transduction, the proportion of the cells in the S phase of the cell cycle was determined. Cells were incubated with [3H]-thymidine for 6 hours, and [3H]-thymidine incorporation was determined. Overexpression of Gfi-1B in K562 and UT7 cells inhibits cell proliferation in comparison with cells transduced with the empty vector MIGR. (C) Overexpression of Gfi-1B induces erythroid differentiation in both K562 and UT7 cells. GPA expression was evaluated by flow cytometry at day 4 after retroviral infection in UT7 cells (left). Hemoglobinization was determined by benzidine staining at day 6 in K562 cells (right). Full-length Gfi-1B expression induces expression of erythroid markers. Data are expressed as mean ± SD. (D) Globin expression analyzed by Northern blot using β, γ, and ϵ globin probes. Forced expression of full-length Gfi-1B induces expression of β and γ globins at a transcriptional level in UT7 cells in a same manner as EPO. In K562 cells, forced expression of Gfi-1B induces the transcription of the ϵ globin but not the β globin, in a same manner as AraC.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/105/4/10.1182_blood-2003-11-4068/6/m_zh80040573980004.jpeg?Expires=1765893200&Signature=SZ4U1i9Y1QIiUsqqNYnErpyLsmAOstoQuIPgulHL7~cds0oUiPODWxFOnanjKJwZHyNqE3CyUw90qivrA7UBwbDMw4s5NWXIDCdn4BZ5O1oLVC8LGTQ7d508RCHz3ah5PIf5DyRW5tL519HHq~pGHcpkWIwgg-SbzH6l0KZDfUoU~v0K-zHWAMqQWLMz2X4-VdNOH-bWSsE~cd2WxvuBsSH7M0nBzPGd3qDAQUbiGRN42JfVBNmMTjlodlbRM8iXzTAfJGhqUg8Xqs2RlhaEun-XKiSo76eAc0BCw1THE~X6kizOcWh8~7qMNZ-yG34Z6ZCpzYev4P8c3jG0hFd~UQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal