Abstract
Presence of the HLA-DR7 allele in patients who receive transplants has been proposed as a risk factor for human cytomegalovirus (HCMV)–associated complications; however, the precise mechanism of this increased risk remains unresolved. Here we show that HLA-DR7–restricted HCMV-specific CD4+ cytotoxic T lymphocytes (CTLs) can display an unusual dual specificity toward a glycoprotein-B (gB) epitope and the alloantigen HLA-DR4. However, no HLA-DR4–specific alloreactivity was observed when the gB-specific CTLs were generated from virus carriers expressing both HLA-DR7 and DR4 alleles. This most likely demonstrates the clonal inactivation of potentially self-reactive T cells in humans. Fine specificity analysis showed that gB-specific CTLs from HLA-DR7+/DR4- individuals displayed a distinct pattern of recognition when compared with CTLs from HLA-DR7+/DR4+ individuals, presumably evading an area of the epitope that mimics a structure presented on HLA-DR4. These data illustrate a possible mechanism for the clinical association between HCMV and graft-versus-host disease.
Introduction
Human cytomegalovirus (HCMV) has been recognized as one of the most important infectious causes of morbidity and mortality in immunocompromised transplant recipients1,2 Although evidence is far from conclusive, several studies have suggested that HCMV infections could trigger rejection in different transplantation settings. For instance, Inkinen et al3 have shown that HCMV infection can significantly increase the expression of collagen and the accumulation of myofibroblasts, which enhances the development of interstitial fibrosis in chronic renal allograft rejection. Additional studies have also suggested an association between HCMV infection and the immunologic mechanisms of rejection, as well as the role of HCMV in the development of bile duct damage in liver allografts.4 Indeed, studies carried out by Koskinen et al5 showed that HCMV-linked generalized immune activation and inflammation of the vascular structures might contribute to the initiation of allograft vasculopathy and to the pathogenesis of chronic heart allograft rejection. HCMV can also induce and enhance expression of major histocompatibility complex (MHC) class II antigens on various cells.6 You et al6 proposed that up-regulation in MHC class II expression may trigger or promote acute rejection by alloantigenic T-lymphocytes. Furthermore, the presence of certain HLA class II alleles (eg, DR7) in patients who receive transplants can significantly increase their risk of developing HCMV-associated complications.7-9 In the present study we show that HCMV-specific CD4+ T cells can display an unusual cross-reactivity toward alloantigens and may therefore have the potential to contribute toward transplant rejection or graft-versus-host disease.
Study design
Establishment and maintenance of cell lines
Epstein Barr virus–transformed lymphoblastoid cell lines (LCLs) transformed with B95.8 virus isolate were used in this study. In addition, murine L cell line expressing HLA-DR7 (referred to as DAP-DR7) were also used in this study.
Peptide synthesis
Overlapping peptides from HCMV glycoprotein-B (gB) and gH were synthesized by using the Merrifield solid-phase method10 and purchased from Chiron Mimotopes (Melbourne, Australia).
Generation of HCMV-specific CTL clones
Limiting dilution CTL microcultures and cytotoxicity assay
Peripheral blood mononuclear cells (PBMCs) were distributed in graded numbers (2-fold dilutions) from 6.25 × 103 to 5 × 104 cells per well in round-bottomed microtiter plates. Approximately 5 × 104 γ-irradiated (2000 rads) peptide-sensitized (1 μg/mL) autologous PBMCs were added to give a total volume of 100 μL. Twenty-four replicates were used at each concentration in each experiment. On day 10, each CTL microculture was split into 2 replicates and used as effectors in a standard 5-hour 51Cr-release assay11 against autologous DAP DR7 cells precoated with gB peptide or HLA-DR4+ LCLs.
Statistical analysis
The difference in the level of lysis from limiting dilution CTL microcultures were analyzed by Student t test.
Results and discussion
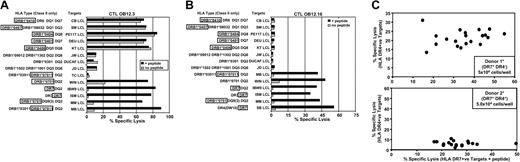
HCMV-specific CD4+ CTL clones were isolated from an HLA-DR7+, HCMV-seropositive individual (donor A) after in vitro stimulation of PBMCs with the HLA-DR7–restricted epitope DYSNTHSTRYV, derived from the gB antigen.13 In vitro analysis of these CTL clones revealed that all clones showing reactivity against the HCMV epitope DYSNTHSTRYV, in association with HLA-DR7, also lysed LCL target cells expressing the HLA-DR4 alloantigen. Figure 1A illustrates the CTL reactivity of one such clone from donor A (CTL OB12.3). Interestingly, this allorecognition was not constrained to 1 particular subtype of the HLA-DR4 antigen. LCLs expressing HLA-DR*0401, DR*0404, DR*0406, DR*0407, or DR*0410 were recognized with equal efficiency (Figure 1A). However, HLA-mismatched (HLA-DR7 without peptide or HLA-DR4-) LCLs were not recognized by these clones. A similar cross-reactivity was observed with all HLA-DR7–restricted gB-specific CTL clones (20 clones) expanded from donor A PBMCs (data not presented). Lysis of the HLA-DR4+ target cell line and the autologous LCL presensitized with peptide DYSNTHSTRYV by the CTL clone OB12.3 was significantly inhibited by unlabeled (no 51Cr) peptide-sensitized HLA-DR7 and unsensitized DR4+ target LCLs but was not inhibited by HLA-mismatched target cell lines (data not shown). The cross-reactivity was, therefore, clearly mediated by the same T-cell receptor (TCR) on a single population of CTLs.
CTL recognition of EBV-transformed LCLs by gB-specific CTL clones in the presence or absence of peptide epitope DYSNTHSTRYV. Data from 2 different clones isolated from HLA-DR7+, DR4- (CTL OB12.3) and HLA-DR7+, DR4+ (CTL OB12.16) individuals are presented in panels A and B, respectively. An effector-to-target ratio of 5:1 was used for both assays. These data are representative of 5 different experiments based on 4 different CTL clones. (C) Split-well analysis of CTL microcultures generated in limiting dilution analysis. Each point on the graph indicates the level of lysis for each CTL microculture of 2 target populations: HLA-DR4+ LCLs (y-axis) and DAP-DR7 cells, which express HLA-DR7, preincubated with peptide DYSNTHSTRYV (x-axis). DAP DR7 cells were also used as targets without peptide presensitization (data not shown); CTL microcultures lysed less than 5% of DAP DR7 targets without peptide presensitization. CTL microcultures from 2 different donors (donor 1, HLA-DR7+, DR4-; donor 2, HLA-DR7+, DR4+) were tested in these assays. Representative data from 24 replicates with 5 × 104 cells/well are presented. *P < .0001.
CTL recognition of EBV-transformed LCLs by gB-specific CTL clones in the presence or absence of peptide epitope DYSNTHSTRYV. Data from 2 different clones isolated from HLA-DR7+, DR4- (CTL OB12.3) and HLA-DR7+, DR4+ (CTL OB12.16) individuals are presented in panels A and B, respectively. An effector-to-target ratio of 5:1 was used for both assays. These data are representative of 5 different experiments based on 4 different CTL clones. (C) Split-well analysis of CTL microcultures generated in limiting dilution analysis. Each point on the graph indicates the level of lysis for each CTL microculture of 2 target populations: HLA-DR4+ LCLs (y-axis) and DAP-DR7 cells, which express HLA-DR7, preincubated with peptide DYSNTHSTRYV (x-axis). DAP DR7 cells were also used as targets without peptide presensitization (data not shown); CTL microcultures lysed less than 5% of DAP DR7 targets without peptide presensitization. CTL microcultures from 2 different donors (donor 1, HLA-DR7+, DR4-; donor 2, HLA-DR7+, DR4+) were tested in these assays. Representative data from 24 replicates with 5 × 104 cells/well are presented. *P < .0001.
The CTL recognition of alloantigen HLA-DR4 by gB-specific CTL clones raised the possibility that individuals who are both HLA-DR7+ and DR4+ may not respond to the gB epitope because of self-tolerance. To test this hypothesis, a series of CD4+ CTL clones were isolated from an HLA-DR4+/DR7+ individual, and their reactivity was tested against HCMV peptide-sensitized HLA-DR7+ and HLA-DR4+ targets (Figure 1B). As shown in Figure 1B, the gB-specific CTL clone could recognize the epitope presented in association with the HLA-DR7 molecule but did not lyse either peptide-sensitized or unsensitized HLA-DR4+ target cells. These observations suggest that the T-cell clonotype(s) that usually dominates the memory response for this gB epitope is probably inactivated by mechanisms of self-tolerance in HLA-DR4+ individuals.
To further confirm this observation, multiple CTL microcultures were raised by limiting dilution from 2 HLA-DR7+ individuals after in vitro stimulation with the gB epitope (Figure 1C). Donor 1 was HLA-DR7+/DR4-, while donor 2 was HLA-DR7+/DR4+. Each microculture was assayed separately for CTL activity against HLA-DR7+ target cells (DAP DR7) with and without the peptide DYSNTHSTRYV, as well as HLA-DR4+ LCLs (no peptide). All CTL microcultures from donor 1 (HLA-DR7+, DR4-) that recognized the gB peptide in association with HLA-DR7 cross-reacted with the alloantigen HLA-DR4 (Figure 1C). In contrast, the microcultures from donor 2 (HLA-DR7+ DR4+) showed reactivity toward the gB epitope in association with HLA-DR7 yet failed to recognize HLA-DR4+ targets. These observations were consistent with data obtained with individual CTL clones (Figure 1A-B) and demonstrate the clonal inactivation of potentially self-reactive T cells in HLA-DR7+ and DR4+ individuals. This clonal inactivation may be explained by thymic deletion of self-reactive T cells or induction of peripheral tolerance/anergy by regulatory T cells. It is possible that the peripherally tolerized T cells bind to HLA DR4 but fail to exert CTL lysis.
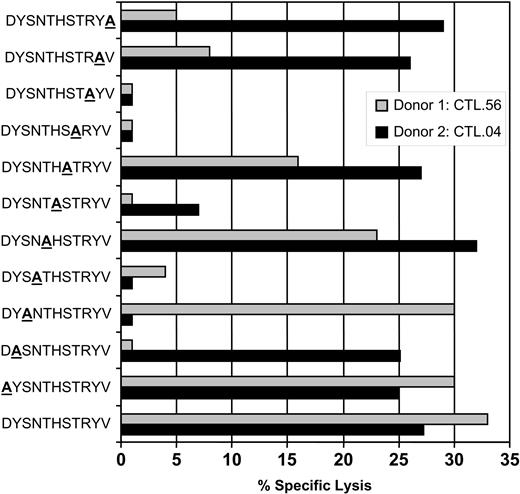
An overlapping alanine replacement net of peptides for the DYSNTHSTRYV epitope was used to further understand the possible mechanism of cross-recognition and delineate the interaction of TCR with HLA-peptide complex. Data presented in Figure 2 show that gB-specific CTL clones from donor 1 (HLA-DR7+/DR4-) bind amino acids toward the COOH-terminus of the peptide, while CTLs from donor 2 (HLA-DR7+/DR4+) did not seem to preferentially favor either NH2- or COOH-terminus, presumably evading an area that mimics a structure presented on the HLA-DR4 molecule. We tested 4 different clones from each donor, and all clones showed a similar pattern of CTL recognition (data not presented). This differential pattern of T-cell recognition of gB epitope by HLA-DR7+/DR4- and HLA-DR7+/DR4+ individuals may be due to the diversity in the expression of TcR on the clonotypes isolated from these individuals. This proposition is strongly supported by previous studies on cross-reactive Epstein-Barr virus (EBV)–specific CTLs which showed that background MHC can effectively diversify the TcR repertoire for a foreign epitope by deflecting the response away from a potentially self-reactive combination of TCR-binding residues.14
CTL recognition of alanine analogs of the peptide epitope DYSNTHSTRYV. HLA-DR7+ LCLs were presensitized with the individual peptides (0.1 μg/mL) and then exposed to DYSNTHSTRYV-specific CTL clones from either donor 1 (HLA-DR7+, DR4-; CTL.56) or donor 2 (HLA-DR7+, DR4+; CTL.04). An effector-to-target ratio of 5:1 was used in the assay. These data are representative of 3 different experiments based on 8 different CTL clones.
CTL recognition of alanine analogs of the peptide epitope DYSNTHSTRYV. HLA-DR7+ LCLs were presensitized with the individual peptides (0.1 μg/mL) and then exposed to DYSNTHSTRYV-specific CTL clones from either donor 1 (HLA-DR7+, DR4-; CTL.56) or donor 2 (HLA-DR7+, DR4+; CTL.04). An effector-to-target ratio of 5:1 was used in the assay. These data are representative of 3 different experiments based on 8 different CTL clones.
These data presented here are consistent with previous studies that suggest memory T cells play an important role in human alloresponses.15-17 It is now well established that healthy seropositive individuals generally display very high precursor frequencies of memory T cells specific for common herpes viruses like EBV and HCMV.12,18 The potency of the response to these viruses probably relates to their lifetime persistence and the repeated antigenic challenge with multiple viral epitopes. Previous studies have shown that up to 60% of MHC class I–restricted T cells cross-react with alloantigens.19,20 Thus, it is feasible that a significant proportion of memory T cells to viruses significantly influence human alloresponses. This study confirms the existence of cross-reactive memory T cells capable of recognizing both a HCMV gB-specific epitope and the HLA-DR4 molecule. Considering the fact that pretransplantation serologic studies have established that HCMV exposure is an important risk factor for various clinical complications in patients who receive solid organ or stem cell transplants,21 it will be interesting to explore the role of this cross-reactivity and the incidence of graft-versus-host disease (GvHD) in patients who receive transplants.
Prepublished online as Blood First Edition Paper, September 30, 2004; DOI 10.1182/blood-2004-07-2602.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal