In their recent paper, Kunisato et al1 describe the role of stem cell leukemia gene (SCL) in regulating lineage fate in hematopoietic stem cells. Their experiments involve retroviral expression of SCL and a “dominant-negative” mutant of SCL (DN-SCL) in hematopoietic stem cells and their progeny. They propose that levels of SCL regulate lineage commitment: enforced expression of SCL favored myeloid differentiation, while expression of the DN-SCL favored lymphoid differentiation. We query the interpretation of the results obtained with the DN-SCL mutant, as its design and effects are not suggestive of a specific dominant-negative function. The authors cite Aplan et al2 and Krosl et al3 for the design of the dominant-negative SCL. In these papers the basic domain of SCL was deleted. This mutant is unable to bind to DNA, however, heterodimerization with E2A proteins remains intact through the presence of the helix-loop-helix (HLH) domain. The DN-SCL mutant used by Kunisato et al1 lacks both the basic and HLH domains. Such a mutant would be predicted to abrogate not only DNA binding, but also the ability to interact with E2A proteins. The remaining N- and C-terminal portions of SCL have no known function—indeed, a truncation mutant comprising only the basic and HLH domains could rescue hematopoiesis of SCL-null embryonic stem cells,4 suggesting that the N- and C-terminal amino acids are not essential. Since a dominant-negative mutant usually relies on deletion of specific functional domains while retaining vital protein interactions, it is difficult to understand how this mutant could act as a dominant negative. Moreover, enforced expression of the DN-SCL only mildly affects erythroid cell production in vitro or in vivo (Figures 3 and 7), whereas loss of SCL by conditional deletion has demonstrated that SCL is essential for erythroid burst-forming units (BFU-E) and production of red cells in vivo.5-7 Thus, there is no available data to positively suggest that the DN-SCL used by Kunisato and colleagues1 inhibits the function of SCL. Nonetheless, it is possible that the N- and C-terminal portions of SCL have an unknown function that causes the observed effects on lineage specification. However, without the correct controls, such as rescue of the DN-SCL effect with wild-type SCL, it is impossible to discriminate specific from nonspecific effects. In light of this and since the effects on myeloid and lymphoid lineage output are subtle and transient, it is important to regard with caution the assertion that the effects are due to a dominant-negative effect on SCL.
Dominant-negative activity of stem cell leukemia (SCL) lacking bHLH domain
Queries from Hall and Curtis on our paper1 in Blood include some important issues. As they argue, the construct of interest (ΔbHLH SCL) may not have an ability to interact with E2A proteins. Indeed, our experiment showed that it does not interact with wild-type (WT) stem cell leukemia (SCL) (data not shown). However, this does not imply that ΔbHLH SCL consisting only of the N- and C-terminal portions of SCL does not have any function. Contrary to the argument by Porcher et al,2 their results could indicate that the N- and C-terminal portions of SCL have some roles, since it appears that the bHLH domain alone does not completely rescue the SCL-null phenotype. In addition, as was described in our paper (Figure 7), we found maturation arrest in the erythroid progenitors by introducing ΔbHLH SCL. This observation is considered to be biologic evidence of dominant-negative effect of ΔbHLH SCL on wild-type SCL, given the phenotype of SCL conditional knockout mice.3 In this regard, we are afraid that the questioners may misunderstand our description in the paper.
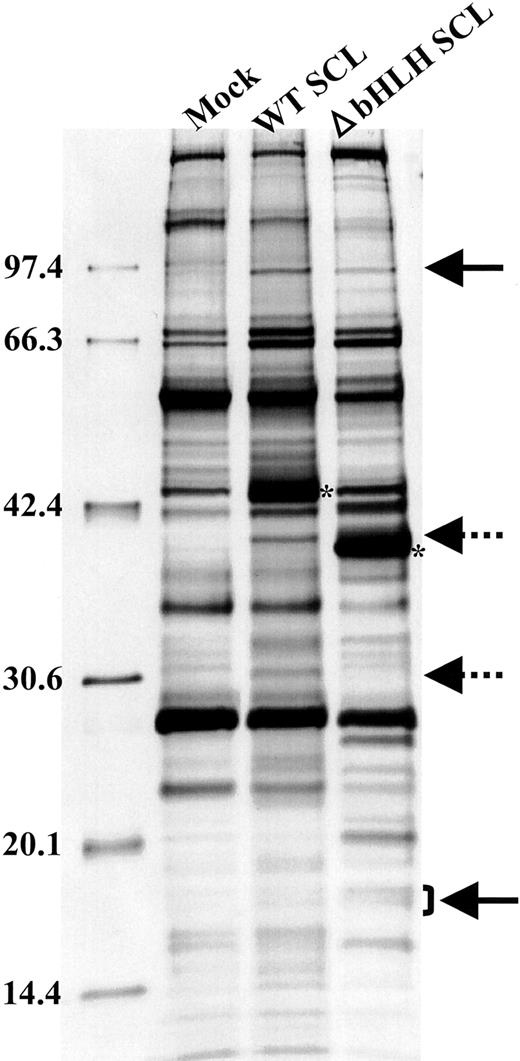
To explore the proteins that interact with ΔbHLH SCL, we have performed a coprecipitation analysis (Figure 1). We transfected HEK293 peak cells with plasmids containing FLAG-tagged WT SCL and ΔbHLH SCL under the cytomegalovirus (CMV) promoter. Two days after the transfection, lysates were prepared and immunoprecipitated with the anti-FLAG antibody-coated beads (Sigma, St Louis, MO). The samples then were resolved through sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), and the gel was silver-stained (Dai-ichi Kagaku, Tokyo, Japan). We found that some proteins coprecipitated commonly with WT SCL and ΔbHLH SCL (solid arrows), and others coprecipitated with WT SCL alone (dotted arrows). It is possible that the commonly precipitated proteins interact with the N- or C-terminal region of SCL, and thus, it is speculated that ΔbHLH SCL functions against WT SCL through the competition for binding to these proteins. To further explore the underlying biochemical mechanisms, we have sequenced some of these coprecipitated proteins, which we hope will be reported in the near future.
Proteins co-precipitated with WTSCL or ΔbHLH SCL. WT SCL and ΔbHLH SCL are indicated by asterisks. The bands indicated by arrows are reproducibly precipitated.
Proteins co-precipitated with WTSCL or ΔbHLH SCL. WT SCL and ΔbHLH SCL are indicated by asterisks. The bands indicated by arrows are reproducibly precipitated.
We disagree with the comment by the questioners that we should be able to show the rescue of the effect of ΔbHLH SCL with WT SCL. This is not an appropriate experiment to show the dominant-negative effect of ΔbHLH SCL.
Although the biochemical mechanisms need to be further disclosed, clear are our findings on the distinct biologic functions of WT SCL and ΔbHLH SCL on the commitment fate determination of hematopoietic stem cells. We hope that our ongoing study will give a clear answer to the mechanisms for how ΔbHLH SCL functions in a dominant-negative fashion against WT SCL.
Correspondence: Shigeru Chiba, Department of Cell Therapy and Transplantation Medicine, University of Tokyo Hospital 7-3-1, Hongo, Bunkyo-ku, Tokyo; e-mail: schiba-tky@umin.ac.jp.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal