Abstract
Allogeneic hematopoietic stem cell transplantation can induce considerable tumor remissions in metastatic renal-cell carcinoma (RCC) patients. The precise effector mechanisms mediating these graft-versus-tumor reactions are unknown. We studied RCC-directed CD8+ T-cell responses in blood lymphocytes of healthy individuals matched with established RCC cell lines for HLA-class I. In 21 of 22 allogeneic mixed lymphocyte/tumor-cell cultures (MLTCs), RCC-reactive cytotoxic T-lymphocytes (CTLs) were readily obtained. From MLTCs, 121 CD8+ CTL clones with memory phenotype were isolated. Their anti–RCC reactivity was restricted by multiple classical HLA-Ia molecules, in particular by HLA-A2, -A3, -B7, -B44, -Cw7, and by a nonclassical HLA-Ib determinant. Extensive cross-reactivity analyses on a broad target panel identified CTLs that recognize antigens with expression restricted to renal tissue or to renal and colon tumors. Other CTLs were directed against antigens with broader tissue distribution being expressed in various epithelial and nonepithelial tumors or, additionally, in hematopoietic cells. With microcapillary liquid chromatography and matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF)/TOF mass spectrometry, we identified the HLA-A*0301-associated nonpolymorphic peptide KLPNSVLGR encoded by the ubiquitously expressed Eps15 homology domain–containing 2 gene as a CTL target. Defining human RCC antigens recognized by alloreactive CTLs may allow to improve the specificity and efficiency of allogeneic cell therapy (eg, specific donor-lymphocyte infusions or vaccination) in metastatic RCC patients.
Introduction
Metastatic renal-cell carcinoma (RCC) has an extremely poor prognosis, with a median survival of less than one year.1 The lack of effective chemotherapies for the treatment of this disease has spurred efforts to develop immunotherapies for patients with RCC. Such strategies are based on the assumption that RCC can activate immune cells to generate effective antitumor responses. The following observations support this view. First, RCC lesions are frequently infiltrated by natural killer (NK) cells and T lymphocytes.2-4 Cultivation of these lymphocytes in vitro yields T-cell responders that display HLA-restricted antitumor reactivity.5,6 Second, a few T-cell–defined RCC antigens have been identified by using clonal cytotoxic T lymphocytes (CTLs) generated from autologous patient blood and tumor-tissue samples.7,8 Third, systemic cytokine therapy using recombinant interleukin-2 and interferon-α can induce tumor remissions in metastatic RCC patients. The overall response rate to these agents, either alone or in combination, is less than 20%.9,10
In an attempt to increase antitumor response rates, pilot studies have been performed in which cytokine-refractory metastatic RCC patients were treated with allogeneic hematopoietic stem cell transplantation (allo-HSCT).11-15 The rationale for this approach is derived from observations in hematologic malignancies where allo-HSCT can induce curative graft-versus-leukemia responses.16 Accordingly, metastatic RCC patients received a conditioning regimen of reduced intensity, followed by the infusion of a peripheral blood stem cell allograft.11-15 Donors were siblings, either HLA-identical or with a mismatch of a single HLA antigen, or they were HLA-matched unrelated volunteers. Considerable graft-versus-RCC effects with complete or partial tumor remissions were observed in 30% to 50% of patients. In most cases, tumor regressions started 3 months to 6 months after transplantation and were preceded by acute graft-versus-host disease (GVHD) and tapering of cyclosporine immunosuppression. However, the precise immune response mechanisms mediating these graft-versus-RCC effects are completely unknown. Interestingly, a current report on a small number of patients observed an association between clinical RCC remissions and an expansion of interferon γ (IFN-γ)–producing CD8+ T cells in peripheral blood following allo-HSCT.17
In this study, we demonstrate for the first time that RCC-reactive CD8+ CTL clones can be isolated from peripheral blood mononuclear cells (PBMCs) of HLA class Ia–matched healthy donors. Alloreactive self-HLA–restricted CTLs seem to recognize a diverse set of antigens on RCC. One of these antigens was identified as an HLA-A*0301–associated peptide encoded by the Eps15 homology domain–containing 2 gene.
Materials and methods
Donors and cell lines
Donor-RCC pairs analyzed were matched for HLA class Ia in donor-versus-RCC direction according to serologic typing. To determine the HLA-Ia genotype of donor PBMC and RCC lines, individual alleles were amplified from genomic DNA with A/B/C allele-specific primers and were completely sequenced (HLA-class I GeneKits; Visible Genetics, Cambridge, United Kingdom). HLA-Ia genotypes are summarized in Table 1.
HLA class Ia genotyping results on healthy PBMC donors and RCC cell lines
. | HLA-A . | HLA-B . | HLA-Cw . |
|---|---|---|---|
| Donor 860 | *0201 *0301 | *0704 *4402 | *0501 *0702 |
| MZ1257-RCC | *0201 *0301 | *0702 *4402 | *0501 *0702 |
| Donor 880 | *0101 *0201 | *0702 *4402 | *0501 *0702 |
| MZ1851-RCC | *0101 *0201 | *0702 *4402 | *0702 *0702 |
. | HLA-A . | HLA-B . | HLA-Cw . |
|---|---|---|---|
| Donor 860 | *0201 *0301 | *0704 *4402 | *0501 *0702 |
| MZ1257-RCC | *0201 *0301 | *0702 *4402 | *0501 *0702 |
| Donor 880 | *0101 *0201 | *0702 *4402 | *0501 *0702 |
| MZ1851-RCC | *0101 *0201 | *0702 *4402 | *0702 *0702 |
PBMC donors were healthy individuals who participated in this study after informed consent in accordance with the Helsinki protocol. The study protocol was approved by the local ethics committee of the Landesaerztekammer Rheinland-Pfalz. MZ1257-RCC and MZ1851-RCC were clear-cell carcinoma cell lines previously established from RCC tissue of patients MZ1257 and MZ1851, respectively.18 RCC lines and primary nonmalignant kidney cells (NKCs) with the index “MZ” were kindly provided by Professor A. Knuth, University of Zuerich, and the Ludwig Institute for Cancer Research (LICR), Zuerich, Switzerland. RCC lines with the index “PB” were generous gifts of Professor W. Storkus, University of Pittsburgh, PA.
Mixed lymphocyte/tumor-cell culture (MLTC) and T-cell cloning
Donor PBMCs were separated from buffy-coat blood samples by Ficoll density centrifugation (Pharmacia, Uppsala, Sweden). With the use of CD8-Microbeads (Miltenyi Biotec, Bergisch Gladbach, Germany), CD8+ T cells were positively selected from PBMCs with a purity of more than 98%. They were cocultured in 24-well plates (Greiner, Nuertingen, Germany) at 106 cells/well with irradiated (100 Gy) allogeneic RCC stimulator cells at 105 cells/well in 2 mL AIM-V medium (GIBCO-BRL, Grand Island, NY) supplemented with 5% human serum (medium Ma). Interleukin 2 (IL-2) was added on day 3 at 250 IU/mL (Proleukin; Chiron, Emeryville, CA). Responder lymphocytes (106 cells/well) were restimulated weekly with 105 irradiated tumor cells in IL-2–containing medium Ma. MLTC responders (28 days old) were cloned by limiting dilution in round-bottomed 96-well plates (Greiner) preseeded with irradiated RCC stimulator (3 × 103/well) and allogeneic Epstein-Barr virus (EBV)–transformed B-lymphoblastoid cell line (LCL) feeder cells (4 × 104/well) in medium Ma supplemented with 250 IU/mL IL-2. After 3 weekly restimulations, growing clones were expanded in 24-well plates by addition of IL-2 (250 IU/mL), stimulator (5 × 104/well), and feeder (2 × 105/well) cells every 7 days.
Flow cytometric analysis
Cells were incubated for 15 minutes at 4° C with fluorescein isothiocyanate (FITC)– or phycoerythrin (PE)–conjugated monoclonal antibodies (mAbs). Abs were purchased from Beckman Coulter (Marseille, France), except for anti-CCR7 (R&D Systems, Minneapolis, MN). Flow cytometric analyses were performed on flow cytometer EPICS-ALTRA (Beckman Coulter).
IFN-γ ELISPOT assay
IFN-γ enzyme-linked immunospot (ELISPOT) assays were performed as previously described.19 Briefly, Multiscreen-HA plates (Millipore, Bedford, MA) were coated with 10 μg/mL mAb anti–hIFN-γ 1-D1K (Mabtech, Stockholm, Sweden). T cells were seeded at indicated numbers in AIM-V medium in triplicates. Stimulator cells at 5 × 104/well were added. After a 20-hour incubation at 37° C and 5% CO2, captured IFN-γ was detected by biotinylated mAb anti–hIFN-γ 7-B6-1 (Mabtech) at 2 μg/mL. Spots were developed by the sequential addition of Avidin-Peroxidase Complex (Vectastain Elite Kit; Vector, Burlingame, CA) and 3-amino-9-ethyl-carbazole (Sigma, St Louis, MO). Spot numbers were determined with computer-assisted video image analysis (Zeiss-Kontron, Jena, Germany).20
51Chromium-release assay
Target cells were incubated for one hour with 100 μCi (3.7 × 106 Bq) of Na512CrO4 (Amersham Buchler, Braunschweig, Germany). After washing, labeled targets (103/well) were plated in conical 96-well plates (Greiner). CTLs were added at indicated effector-to-target (E/T) ratios in a total volume of 180 μL medium Mb/well. Medium Mb was RPMI-1640 supplemented with 10% fetal calf serum (FCS; GIBCO-BRL). After a 4-hour incubation, 90 μL supernatant per well was collected for counting in a γ-counter. Percent specific 51Cr-release = (experimental release – spontaneous release) × 100 / (maximum release – spontaneous release).
Antibody-blocking test
To demonstrate HLA class I–restricted reactivity of CTLs, ELISPOT and cytotoxicity assays were performed in the presence of 100 μg/mL of the following murine mAbs: W6/32, an anti–HLA class I IgG2a21 ; MA2.1, an anti–HLA-A2 IgG122 ; GAP-A3, an anti–HLA-A3 IgG2a23 ; B1.23.2, an anti–HLA-B and –HLA-C IgG2a24 ; and L243, an anti–HLA-DR IgG2a.25
Cloning and transfection of cDNA coding for HLA class I alleles or EHD2
After synthesis of cDNA (Superscript First-Strand Synthesis; Invitrogen, Groningen, The Netherlands) from RCC-derived RNA (RNAeasy Mini-Kit; Qiagen, Hilden, Germany), full-length cDNAs were amplified using specific primers for HLA-A, HLA-B,26 and HLA-C alleles,27 or Eps15 homology domain–containing 2 (EHD2),28 respectively. cDNAs were inserted into the EcoRI and XhoI sites of the expression vector pcDNA3.1(+) (Invitrogen) and then completely sequenced. Authentic cDNA clones were transiently transfected into 293T cells29 seeded in precoated IFN-γ ELISPOT plates at 2 × 104 cells/well. Transfection was performed using PolyFect reagent (Qiagen). After a 24-hour incubation period, 293T transfectants were analyzed for spot production by RCC-reactive CTLs over 20 hours.
Extraction and HPLC separation of natural HLA-A*0301–associated RCC peptides
Naturally processed peptides were acid-eluted from HLA-A*0301 molecules purified from 2.5 × 1010 MZ1257-RCC cells using anti–HLA-A3 mAb GAP-A323 as previously described.30-32 Peptide ligands were fractionated on a microbore C18 column (2.1 × 150 mm, 5 μm particles, 300 Å pore size; Vydac, Hesperia, CA) on a reverse-phase (RP)–high-performance liquid chromatography (HPLC) instrument (HP1100; Agilent Technologies, Palo Alto, CA). The elution gradient used was 0% to 10% solvent B for 5 minutes, 10% to 75% solvent B in the next 65 minutes, and 75% to 100% solvent B in the next 5 minutes, whereas solvent A was 0.1% trifluoroacetic acid (TFA; Sigma) in water and solvent B was 0.085% TFA in 60% acetonitrile (Sigma) in water. Fractions were collected every 60 seconds at a flow rate of 250 μL/min.
Epitope reconstitution assay
Aliqouts of each HPLC fraction containing natural HLA-A*0301–associated peptides from 8 × 108 MZ1257-RCC cell equivalents were screened with CTL clone B5 in 6-hour-long 51Cr-release assays. As peptide-presenting cells, HLA-A*0301–transfected, transported associated with antigen processing (TAP)–deficient T2 (T2-A3) targets (provided by Dr van den Eynde, LICR, Brussels, Belgium) were used. To facilitate loading of exogenous HLA-A*0301–binding peptides, T2-A3 cells were pretreated with one mg/mL mAb W6/32 during 51Cr-labeling.33 After 4 washings, labeled T2-A3 at 1500 cells/well were incubated with natural HLA-A*0301 peptides for 90 minutes in 80 μL medium Mb supplemented with 5 μg/mL β2-microglobulin (β2m; Sigma). CTLs were then added in 80 μL medium Mb at an E/T ratio of 20:1.
Synthetic candidate peptides were screened with CTL clone B5 in an IFN-γ ELISPOT assay.20 W6/32-pretreated T2-A3 or HLA-A*0301–expressing LCLs at 7.5 × 104 cells/well served as peptide-presenting cells.
Screening and sequencing of candidate peptides by MALDI mass spectrometry (MS)
The peptide components of bioactive and inactive HPLC fractions were characterized by their molecular mass on a matrix-assisted laser desorption ionization time-of-flight (MALDITOF) instrument (Voyager-DE PRO Workstation; Applied Biosystems, Framingham, MA). Individual samples contained natural peptides obtained from 3.8 × 108 RCC cell equivalents and dissolved in 1% acetic acid/50% methanol (Sigma) in water.
Prior to sequencing, HPLC-purified peptides derived from 6 × 109 RCC cell equivalents were fractionated using a 75 μm × 150 mm microcapillary C18 column (PepMap; Dionex LC Packings, Amsterdam, Netherlands) at a flow rate of 200 nL/min (Ultimate Nano LC System; Dionex). Solvents A and B were identical to solvents used in first-dimension HPLC. Elution gradient increased 0.5% solvent B per minute. Liquid chromatography eluate (20-second subfractions; 67 nL) was mixed with 170 nL of α-Cyano-4-hydroxycinnamic acid-matrix solution before spotting onto the plate (Probot Microfraction Collector; Dionex). After drying, collision-induced dissociation (CID) mass spectra were recorded on selected candidate peptides using a MALDI-TOF/TOF instrument (4700 Proteomics Analyzer; Applied Biosystems). Resulting data were analyzed by GPS Explorer software (Applied Biosystems), which invoked a MASCOT database search (NCBInr) and a ProBLAST search engine, to determine protein source of candidate peptides.
Peptides were synthesized by solid-phase Fmoc chemistry on a Syro peptide synthesizer (Multisyntech, Witten, Germany). They were purified to more than 90% homogeneity by RP-HPLC and characterized by MS.
Results
RCC-reactive CD8+ T cells can be generated from HLA class I–matched healthy donors
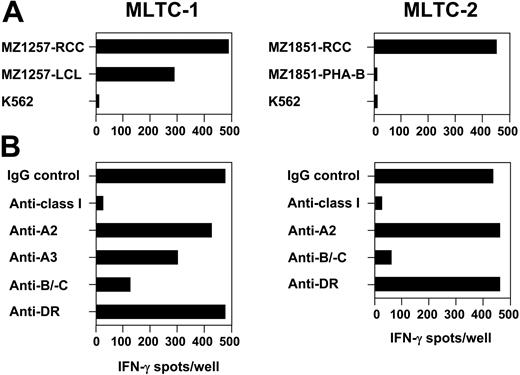
Allogeneic MLTCs were initiated by sensitizing CD8+ T lymphocytes purified from buffy coat–derived PBMCs of 5 different healthy donors against HLA class I–matched renal clear-cell carcinoma cell lines. In 22 independent MLTC experiments, responder lymphocytes showed a 2- to 6-fold expansion of cell numbers every week over a total culture period of 3 to 6 months. Continuous monitoring of responder-cell growth in the absence of RCC stimulators demonstrated that the proliferation of MLTC populations was strictly dependent on regular stimulations with tumor cells. In 21 of 22 MLTCs, allogeneic CD8+ T lymphocytes were obtained that showed HLA class I–restricted recognition of RCC stimulator cells in IFN-γ ELISPOT and 51Cr-release assays. We continued with detailed analyses on MLTCs sensitizing CD8+ T cells of healthy donors 860 and 880 against the tumor cell lines MZ1257-RCC and MZ1851-RCC, respectively (Table 1). In 2 representative MLTCs, clear reactivity was observed against the RCC stimulators, but not against NK target K562 (Figure 1A). However, both MLTCs differed in their ability to recognize hematopoietic cells available from the corresponding RCC patients: whereas MLTC-1 driven against MZ1257-RCC showed significant reactivity against an LCL line derived from patient MZ1257, MLTC-2 responding to MZ1851-RCC failed to recognize phytohemagglutinin (PHA)–activated PBMC blasts (PHA-Bs) obtained from patient MZ1851. Anti-RCC reactivity of MLTC-1 and MLTC-2 responders was completely blocked by an HLA class I–specific mAb demonstrating HLA I restriction (Figure 1B). Monoclonal Abs directed against single HLA-A or common HLA-B/-C determinants indicated the presence of RCC-reactive T cells restricted by HLA-A2, -A3, -B/C alleles in MLTC-1, and by HLA-B/C alleles in MLTC-2, respectively.
Generation of RCC-reactive CD8+ T cells from HLA-class Ia-matched healthy donors. In allogeneic MLTC-1 and MLTC-2, CD8+ T cells isolated from PBMCs of healthy donors 860 and 880 with a purity of more than 98% were stimulated weekly with HLA Ia–matched RCC lines MZ1257-RCC and MZ1851-RCC, respectively. HLA class Ia genotypes of donors and RCC lines are shown in Table 1. Both RCC lines were negative for HLA class II in flow cytometry. Specificity of MLTC responders was analyzed on day 25 of culture in 20-hour IFN-γ ELISPOT assays. (A) Targets were the RCC stimulator cell lines, the hematopoietic counterparts (ie, EBV-LCL or PHA-activated PBMC) derived from the same RCC patients, and NK target K562. PHA-activated PBMCs of patient MZ1257 and EBV-LCL of patient MZ1851 were not available. (B) HLA restriction of anti-RCC reactivity was determined by adding mAbs specific for total HLA class I, HLA-A2, HLA-A3, HLA-B/-C alleles, or HLA-DR. Each bar represents the mean spot number of triplicates with 10 000 MLTC responders seeded per well.
Generation of RCC-reactive CD8+ T cells from HLA-class Ia-matched healthy donors. In allogeneic MLTC-1 and MLTC-2, CD8+ T cells isolated from PBMCs of healthy donors 860 and 880 with a purity of more than 98% were stimulated weekly with HLA Ia–matched RCC lines MZ1257-RCC and MZ1851-RCC, respectively. HLA class Ia genotypes of donors and RCC lines are shown in Table 1. Both RCC lines were negative for HLA class II in flow cytometry. Specificity of MLTC responders was analyzed on day 25 of culture in 20-hour IFN-γ ELISPOT assays. (A) Targets were the RCC stimulator cell lines, the hematopoietic counterparts (ie, EBV-LCL or PHA-activated PBMC) derived from the same RCC patients, and NK target K562. PHA-activated PBMCs of patient MZ1257 and EBV-LCL of patient MZ1851 were not available. (B) HLA restriction of anti-RCC reactivity was determined by adding mAbs specific for total HLA class I, HLA-A2, HLA-A3, HLA-B/-C alleles, or HLA-DR. Each bar represents the mean spot number of triplicates with 10 000 MLTC responders seeded per well.
Allogeneic CTLs recognize antigens shared between tumor and hematopoietic cells
To analyze the specificity of allo-MLTC responders, MLTC-1 and MLTC-2 were cloned by limiting dilution. From 5600 wells initially seeded, 121 CD3+ CD8+ TCRαβ+ CTL clones were isolated that specifically recognized the RCC stimulator line in IFN-γ ELISPOT and 51Cr-release assays (MLTC-1: 69 clones; MLTC-2: 52 clones). In addition to IFN-γ, these CTLs demonstrated antigen-induced cytokine spot production for tumor necrosis factor α (TNF-α) and granzyme B, but not for TH2-associated cytokines IL-4, IL-5, and IL-10 (data not shown). In flow cytometry, CTLs were CD45RA– CD45RO+ CD25+ CD69+ and HLA-DR+, consistent with a phenotype of activated memory T cells (Table 2).
Flow cytometric analysis on RCC-reactive CTL clones B5, G16, K34, and J23
. | B5 . | G16 . | K34 . | J23 . |
|---|---|---|---|---|
| CD3 | 95 | 99 | 97 | 100 |
| CD8 | 95 | 99 | 97 | 100 |
| CD4dim | 2 | 3 | 73 | 0 |
| TCRαβ | 91 | 99 | 96 | 100 |
| TCRγδ | 2 | 1 | 0 | 0 |
| CD16 | 1 | 0 | 0 | 0 |
| CD56dim | 17 | 44 | 46 | 44 |
| CD57 | 1 | 1 | NT | 1 |
| CD45RA | 4 | 1 | 0 | 1 |
| CD45RO | 68 | 99 | 82 | 62 |
| CCR7dim | 26 | 7 | 18 | 10 |
| CD28dim | 46 | 48 | NT | 52 |
| CD94 | 13 | 33 | 17 | 1 |
| CD158a | 3 | 5 | 5 | 2 |
| CD158b | 5 | 14 | 4 | 2 |
| CD25 | 67 | 85 | NT | 95 |
| CD69 | 71 | 67 | NT | 93 |
| HLA-DR | 90 | 92 | NT | 79 |
. | B5 . | G16 . | K34 . | J23 . |
|---|---|---|---|---|
| CD3 | 95 | 99 | 97 | 100 |
| CD8 | 95 | 99 | 97 | 100 |
| CD4dim | 2 | 3 | 73 | 0 |
| TCRαβ | 91 | 99 | 96 | 100 |
| TCRγδ | 2 | 1 | 0 | 0 |
| CD16 | 1 | 0 | 0 | 0 |
| CD56dim | 17 | 44 | 46 | 44 |
| CD57 | 1 | 1 | NT | 1 |
| CD45RA | 4 | 1 | 0 | 1 |
| CD45RO | 68 | 99 | 82 | 62 |
| CCR7dim | 26 | 7 | 18 | 10 |
| CD28dim | 46 | 48 | NT | 52 |
| CD94 | 13 | 33 | 17 | 1 |
| CD158a | 3 | 5 | 5 | 2 |
| CD158b | 5 | 14 | 4 | 2 |
| CD25 | 67 | 85 | NT | 95 |
| CD69 | 71 | 67 | NT | 93 |
| HLA-DR | 90 | 92 | NT | 79 |
CTL clones were isolated from allogeneic MLTC-1 on blood-derived CD8+ T lymphocytes of healthy donor 860 and the HLA class la—matched RCC line MZ1257-RCC. T cells were stained with FITC- or PE-conjugated monoclonal antibodies directed against indicated cell surface markers and were then analyzed. Results shown represent percentage of cells with positive FITC- or PE-staining. Low staining intensity is marked as “dim.” NT indicates not tested.
Clonal CTLs were further analyzed for their HLA class I restriction and their cross-reactivity against K562, hematopoietic cell counterparts (ie, MZ1257-LCL, MZ1851-PHA-B, donor-derived LCL, and PHA-B), and a large panel of allogeneic targets matched for single HLA-Ia alleles with RCC stimulators. The latter included epithelial tumor cell lines originally established from renal, breast, colon, lung, pancreatic, hepatocellular, gallbladder, and cholangiocellular carcinomas as well as nonepithelial tumors such as melanomas and leukemias. If available, corresponding nonmalignant kidney cells or hematopoietic cells were also tested. Based on these results, CTLs were divided into 5 different groups.
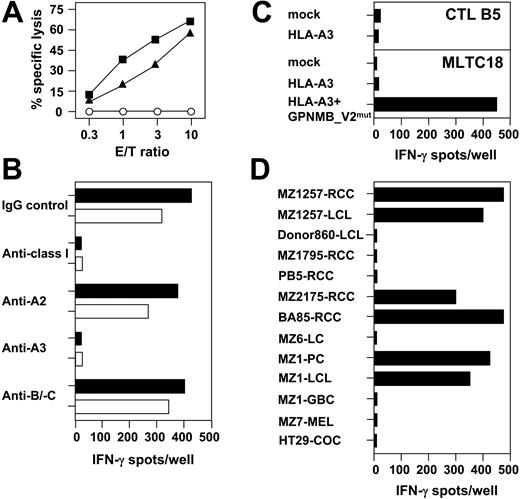
Group 1 CTLs comprising 48 individual clones simultaneously recognized RCC and hematopoietic cells isolated from the same patient, but not donor-derived hematopoietic cells or K562. This suggested that group 1 CTLs were directed against distinct peptide antigens shared between patient-derived RCC and hematopoietic cells and presented by self-HLA class I molecules. Alternatively, CTL could target HLA complexes by itself, provided that donors and RCC patients were mismatched for certain HLA alleles at the genomic level. As a representative example, results obtained with donor 860–derived CTL clone B5 are shown in Figure 2. This CTL recognized MZ1257-RCC and MZ1257-LCL, but not donor 860–derived LCL or K562 (Figure 2A,D). Antibody-blocking studies identified HLA-A3 as the CTL restriction element (Figure 2B). Indeed, HLA sequencing revealed HLA-A*0301 being identical between donor 860 and patient MZ1257 (Table 1). Consequently, HLA-A*0301 cDNA cloned from MZ1257-RCC and inserted into the pcDNA3.1(+) expression vector was not recognized by CTL B5 after transfection in 293T cells (Figure 2C). Accordingly, this allogeneic CTL clone was not directed against structural determinants of the HLA-A*0301 molecule independent of a distinct peptide antigen bound. Extensive cross-reactivity analysis on 25 different HLA-A3+ allogeneic target cell lines revealed that CTL B5 reacted with 4 of 7 RCC and one of 2 pancreatic carcinoma cell lines (Figure 2D). In addition to HLA-A*0301, HLA-A*0201 and HLA-B*4402 were also used as restriction elements by group 1 CTL clones (Table 3).
CTL clone B5 recognizes an HLA-A3–associated antigen expressed by renal and pancreatic carcinoma cell lines and their LCL counterparts. CTL B5 was cloned from allogeneic MLTC-1 that sensitized CD8+ T lymphocytes of healthy donor 860 against the HLA class Ia–matched RCC line MZ1257-RCC. (A) Lytic activity of CTL B5 was tested against MZ1257-RCC (▪), MZ1257-LCL (▴), and K562 (○) in a 4-hour 51Cr-release assay. Results shown are percentage of cytolysis at indicated effector-to-target (E/T) ratios. (B) Effect of different anti–HLA class I mAbs on recognition of MZ1257-RCC (▪) and MZ1257-LCL (□) by CTL B5 in 20-hour IFN-γ ELISPOT assay. (C) Full-length HLA-A*03011 cDNA cloned from MZ1257-RCC and inserted into pcDNA3.1(+) expression vector was transiently transfected in 293T cells. Transfectants were tested for recognition by CTL B5 in a 20-hour IFN-γ ELISPOT assay. In a control experiment, MZ1257-RCC–derived HLA-A*03011 cDNA was able to present a known HLA-A3–restricted peptide antigen derived from GPNMB_V2mut cDNA to antimelanoma CTL line MLTC18 (GPNMB_V2mut cDNA and MLTC18 provided by V.L. and T.W.). (D) Cross-reactivity of CTL B5 tested in 20-hour IFN-γ ELISPOT analyses against HLA-A3+ RCC, non-RCC tumor, and LCL cell lines. LCL indicates Epstein-Barr virus (EBV)–transformed B-lymphoblastoid cell line; MEL, melanoma cell line; LC, lung cancer cell line; COC, colon cancer cell line; PC, pancreatic-carcinoma cell line; and GBC, gallbladder-carcinoma cell line. Results shown are a representative part of 25 HLA-A3+ targets totally tested. All spot assays were performed with 5000 CTLs seeded per well. Each bar represents the mean spot number of triplicates.
CTL clone B5 recognizes an HLA-A3–associated antigen expressed by renal and pancreatic carcinoma cell lines and their LCL counterparts. CTL B5 was cloned from allogeneic MLTC-1 that sensitized CD8+ T lymphocytes of healthy donor 860 against the HLA class Ia–matched RCC line MZ1257-RCC. (A) Lytic activity of CTL B5 was tested against MZ1257-RCC (▪), MZ1257-LCL (▴), and K562 (○) in a 4-hour 51Cr-release assay. Results shown are percentage of cytolysis at indicated effector-to-target (E/T) ratios. (B) Effect of different anti–HLA class I mAbs on recognition of MZ1257-RCC (▪) and MZ1257-LCL (□) by CTL B5 in 20-hour IFN-γ ELISPOT assay. (C) Full-length HLA-A*03011 cDNA cloned from MZ1257-RCC and inserted into pcDNA3.1(+) expression vector was transiently transfected in 293T cells. Transfectants were tested for recognition by CTL B5 in a 20-hour IFN-γ ELISPOT assay. In a control experiment, MZ1257-RCC–derived HLA-A*03011 cDNA was able to present a known HLA-A3–restricted peptide antigen derived from GPNMB_V2mut cDNA to antimelanoma CTL line MLTC18 (GPNMB_V2mut cDNA and MLTC18 provided by V.L. and T.W.). (D) Cross-reactivity of CTL B5 tested in 20-hour IFN-γ ELISPOT analyses against HLA-A3+ RCC, non-RCC tumor, and LCL cell lines. LCL indicates Epstein-Barr virus (EBV)–transformed B-lymphoblastoid cell line; MEL, melanoma cell line; LC, lung cancer cell line; COC, colon cancer cell line; PC, pancreatic-carcinoma cell line; and GBC, gallbladder-carcinoma cell line. Results shown are a representative part of 25 HLA-A3+ targets totally tested. All spot assays were performed with 5000 CTLs seeded per well. Each bar represents the mean spot number of triplicates.
Results on cross-reactivity and HLA class I restriction of RCC-reactive CTL clones isolated from healthy donors 860 and 880
. | Patients . | . | Donors . | Other individuals . | . | . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTL group . | RCC . | LCL PHA-B . | LCL PHA-B . | LCL PHA-B . | NKC . | K562 . | Non-RCC . | HLA restriction . | ||||
| 1 | + | + | - | + | + | - | BC, LC, PC, LK, COC, HCC, MEL | A2, A3, B44 | ||||
| 2 | + | - | - | - | - | - | COC | A2, Cw7 | ||||
| 3 | + | - | - | - | + | - | - | A2, B7, Cw7 | ||||
| 4* | + | - | - | - | + | + | MEL | Cw7 | ||||
| 5* | + | - | - | - | NT | - | BC, LC, PC, COC | lb | ||||
. | Patients . | . | Donors . | Other individuals . | . | . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTL group . | RCC . | LCL PHA-B . | LCL PHA-B . | LCL PHA-B . | NKC . | K562 . | Non-RCC . | HLA restriction . | ||||
| 1 | + | + | - | + | + | - | BC, LC, PC, LK, COC, HCC, MEL | A2, A3, B44 | ||||
| 2 | + | - | - | - | - | - | COC | A2, Cw7 | ||||
| 3 | + | - | - | - | + | - | - | A2, B7, Cw7 | ||||
| 4* | + | - | - | - | + | + | MEL | Cw7 | ||||
| 5* | + | - | - | - | NT | - | BC, LC, PC, COC | lb | ||||
+ indicates targets that were recognized by specific CTL clones; - indicates targets that were not recognized by specific CTL clones; LCL, EBV B-lymphoblastoid cell line; PHA-B, phytohemagglutinin-activated PBMC blasts; NKC, primary nonmalignant kidney cells; BC, breast carcinoma; LC, lung cancer; PC, pancreatic carcinoma; LK, leukemia; COC, colon cancer; HCC, hepatocellular carcinoma; MEL, melanoma; NT, not tested.
Only observed in donor 860.
Mass spectrometry identification of the peptide epitope recognized by CTL clone B5
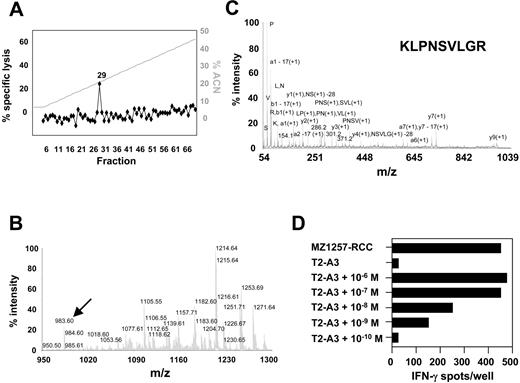
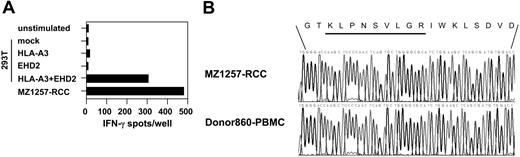
To identify the antigen recognized by CTL clone B5, HLA-A*0301–associated peptides were acid-eluted from purified HLA-A*0301 complexes of 2.5 × 1010 MZ1257-RCC cells and were fractionated by RP-HPLC. Individual fractions were screened for their ability to reconstitute CTL-mediated lysis using HLA-A*0301–transfected T2 cells (T2-A3) as targets and CTL B5 as effectors (Figure 3A). Bioactive fraction 29 and adjacent inactive fractions were screened by MALDI-TOF MS (Figure 3B). By comparing the abundance of peptide masses in spectra from bioactive and inactive fractions, several candidate peptides were identified that correlated with the bioactivity in the CTL epitope reconstitution assay. After microcapillary liquid chromatography separation of peptide material obtained from fraction 29, 7 of those candidates were sequenced by MALDI-TOF/TOF MS (Figure 3C). Candidate peptides were synthesized and tested for recognition by CTL clone B5. As peptide-presenting cells, T2-A3 or antigen-negative HLA-A*0301–expressing LCLs were used. Only target cells pulsed with peptide KLPNSVLGR deduced from candidate ion m/z 983.60 were recognized by CTL B5, with bioactivity seen at a peptide concentration of one nM or higher (Figure 3D). A search of protein sequence databases with peptide KLPNSVLGR generated a single perfect match with residues 480-488 of the Eps15 homology domain–containing 2 (EHD2) protein (accession no. NP_055 416). Furthermore, antigen-negative target cells cotransfected with full-length cDNAs of HLA-A*0301 and EHD2 were recognized by CTL B5 (Figure 4A). Comparison of genomic DNA isolated from MZ1257-RCC and PBMCs of donor 860 demonstrated identical nonpolymorphic sequences in the epitope-coding region of EHD2 (Figure 4B). Taken together, these results confirmed that the peptide KLPNSVLGR encoded by the nonpolymorphic EHD2 gene defines the HLA-A*0301–restricted, naturally processed CTL-B5 epitope. Further work excluded an artifactual expression of EHD2 in long-term cultured RCC lines by the observation that EHD2 cDNAwas detected in 4 of 5 ex vivo isolated primary RCC tumors (data not shown). In addition, the EHD2-specific CTL clone B5 recognized RCC cells obtained from fresh tumor tissue directly after the first in vitro culture passage.
Identification of the HLA-A*0301–associated peptide epitope KLPNSVLGR by mass spectrometry. (A) In vitro epitope reconstitution with naturally processed RCC peptides. Peptides were acid-eluted from immunoaffinity-purified HLA-A*0301 complexes of 2.5 × 1010 MZ1257-RCC cells and fractionated by RP-HPLC. Aliquots of each fraction corresponding to 8 × 108 cell equivalents were preincubated with HLA-A*0301–transfected T2 (T2-A3) cells and tested for recognition by HLA-A*0301–restricted CTL clone B5 at an E/T ratio of 20:1 in a 51Cr-release assay. Background lysis on T2-A3 by CTL B5 in the absence of peptides was 2%. Lysis on MZ1257-RCC by CTL B5 was 97% (not shown). ACN indicates acetonitrile. (B) MALDI-TOF mass spectrum from HPLC fraction 29 of the HLA-A*0301 peptide extract that contained the CTL B5 epitope. Data were obtained from 3.8 × 108 MZ1257-RCC cell equivalents and are shown in the 950 m/z to 1300 m/z range. The m/z 983.60 ion is indicated by arrow. (C) CID mass spectrum recorded on candidate peptide ions of m/z 983.60 observed in HPLC fraction 29. Peptide material from HPLC fraction 29 corresponding to 6 × 109 MZ1257-RCC cell equivalents was separated by microcapillary liquid chromatography prior to fragmentation by MALDI-TOF/TOF mass spectrometry. (D) Epitope reconstitution with synthetic peptide KLPNSVLGR. RCC-reactive CTL clone B5 was tested in an IFN-γ ELISPOT assay against T2-A3 cells pulsed with synthetic peptide KLPNSVLGR at indicated concentrations. Results obtained with 5000 CTLs/well are shown. Unpulsed T2-A3 cells and MZ1257-RCC cells were negative and positive controls, respectively. Results shown in panels A to D are representative of at least 2 experiments.
Identification of the HLA-A*0301–associated peptide epitope KLPNSVLGR by mass spectrometry. (A) In vitro epitope reconstitution with naturally processed RCC peptides. Peptides were acid-eluted from immunoaffinity-purified HLA-A*0301 complexes of 2.5 × 1010 MZ1257-RCC cells and fractionated by RP-HPLC. Aliquots of each fraction corresponding to 8 × 108 cell equivalents were preincubated with HLA-A*0301–transfected T2 (T2-A3) cells and tested for recognition by HLA-A*0301–restricted CTL clone B5 at an E/T ratio of 20:1 in a 51Cr-release assay. Background lysis on T2-A3 by CTL B5 in the absence of peptides was 2%. Lysis on MZ1257-RCC by CTL B5 was 97% (not shown). ACN indicates acetonitrile. (B) MALDI-TOF mass spectrum from HPLC fraction 29 of the HLA-A*0301 peptide extract that contained the CTL B5 epitope. Data were obtained from 3.8 × 108 MZ1257-RCC cell equivalents and are shown in the 950 m/z to 1300 m/z range. The m/z 983.60 ion is indicated by arrow. (C) CID mass spectrum recorded on candidate peptide ions of m/z 983.60 observed in HPLC fraction 29. Peptide material from HPLC fraction 29 corresponding to 6 × 109 MZ1257-RCC cell equivalents was separated by microcapillary liquid chromatography prior to fragmentation by MALDI-TOF/TOF mass spectrometry. (D) Epitope reconstitution with synthetic peptide KLPNSVLGR. RCC-reactive CTL clone B5 was tested in an IFN-γ ELISPOT assay against T2-A3 cells pulsed with synthetic peptide KLPNSVLGR at indicated concentrations. Results obtained with 5000 CTLs/well are shown. Unpulsed T2-A3 cells and MZ1257-RCC cells were negative and positive controls, respectively. Results shown in panels A to D are representative of at least 2 experiments.
Human EHD2 encodes for a nonpolymorphic antigen recognized by CTL B5 in association with HLA-A*0301. (A) From MZ1257-RCC cells, cDNAs coding for full-length HLA-A*0301 or EHD2 were isolated and inserted into pcDNA3.1(+) expression vectors. They were cotransfected into antigen-negative 293T cells and were analyzed for recognition by CTL clone B5 in a 20-hour IFN-γ ELISPOT assay. Controls included 293T cells transfected with either HLA-A*0301 or EHD2 cDNAs alone, and MZ1257-RCC cells. Each bar represents the mean spot number of triplicates with 5000 CTLs seeded per well. (B) Genomic DNA was isolated from MZ1257-RCC and PBMCs of donor 860. From each sample, an EHD2 fragment containing the KLPNSVLGR epitope-coding region was amplified using specific primers. PCR products were sequenced using standard procedures.
Human EHD2 encodes for a nonpolymorphic antigen recognized by CTL B5 in association with HLA-A*0301. (A) From MZ1257-RCC cells, cDNAs coding for full-length HLA-A*0301 or EHD2 were isolated and inserted into pcDNA3.1(+) expression vectors. They were cotransfected into antigen-negative 293T cells and were analyzed for recognition by CTL clone B5 in a 20-hour IFN-γ ELISPOT assay. Controls included 293T cells transfected with either HLA-A*0301 or EHD2 cDNAs alone, and MZ1257-RCC cells. Each bar represents the mean spot number of triplicates with 5000 CTLs seeded per well. (B) Genomic DNA was isolated from MZ1257-RCC and PBMCs of donor 860. From each sample, an EHD2 fragment containing the KLPNSVLGR epitope-coding region was amplified using specific primers. PCR products were sequenced using standard procedures.
Antigens restricted to certain epithelial tumors or to renal tissue serve as allo-CTL targets
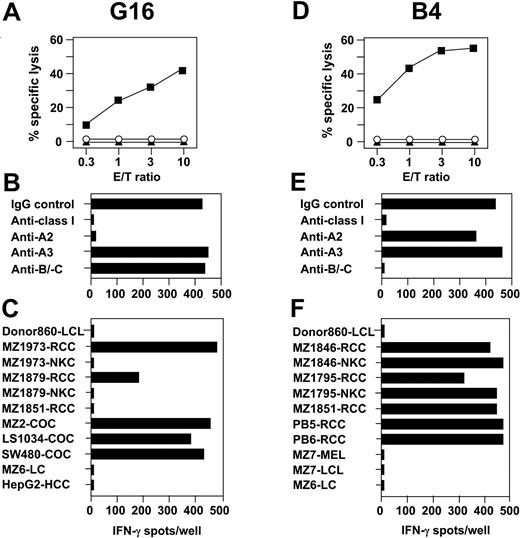
In addition to T-cell specificities simultaneously recognizing RCC and corresponding hematopoietic cells, 66 CTL clones were isolated that lysed RCC, but not LCL, PHA blasts, or K562. Of these CTL clones, two-thirds exhibited reactivity against allogeneic nonmalignant kidney cells (NKC) while one-third did not. CTLs representative of both specificities are shown in Figure 5. The HLA-A*0201–restricted CTL clone G16 recognized 3 of 6 HLA-A2+ allogeneic RCC lines, but not their corresponding NKC counterparts (Figure 5A-C). This lack of responsiveness against NKC defined G16 as a group 2 CTL. Of 21 other HLA-A2+ non-RCC tumor lines tested, CTL G16 cross-reacted exclusively with 5 of 5 colon cancers (COCs), suggesting an antigen restricted to malignant renal and colon cells as the target.
Allogeneic CTL clones define antigens restricted to renal cells or to renal and colon tumors. CTL G16 and B4 were cloned from MLTC-1 on CD8+ T lymphocytes of healthy donor 860 and the HLA class Ia–matched RCC line MZ1257-RCC. (A,D) CTL-mediated cytolysis was tested against MZ1257-RCC (▪), MZ1257-LCL (▴), and K562 (○) in 4-hour 51Cr-release assays. Results are given as percentage of specific lysis at indicated E/T ratios. (B,E) HLA class I restriction of CTLs reactive against MZ1257-RCC was identified by adding different anti-HLA mAbs in 20-hour IFN-γ ELISPOT assays. (C,F) Cross-reactivity pattern of CTLs observed in 20-hour IFN-γ spot assays against allogeneic target cell lines expressing HLA-A2 for CTL G16 and HLA-B7 for CTL B4, respectively. Results shown are representative of at least 20 different targets tested per CTL. Spot assays were plated with 5000 CTLs/well in triplicates.
Allogeneic CTL clones define antigens restricted to renal cells or to renal and colon tumors. CTL G16 and B4 were cloned from MLTC-1 on CD8+ T lymphocytes of healthy donor 860 and the HLA class Ia–matched RCC line MZ1257-RCC. (A,D) CTL-mediated cytolysis was tested against MZ1257-RCC (▪), MZ1257-LCL (▴), and K562 (○) in 4-hour 51Cr-release assays. Results are given as percentage of specific lysis at indicated E/T ratios. (B,E) HLA class I restriction of CTLs reactive against MZ1257-RCC was identified by adding different anti-HLA mAbs in 20-hour IFN-γ ELISPOT assays. (C,F) Cross-reactivity pattern of CTLs observed in 20-hour IFN-γ spot assays against allogeneic target cell lines expressing HLA-A2 for CTL G16 and HLA-B7 for CTL B4, respectively. Results shown are representative of at least 20 different targets tested per CTL. Spot assays were plated with 5000 CTLs/well in triplicates.
In contrast to group 2 specificities, CTL clones belonging to group 3 were characterized by their simultaneous recognition of allogeneic RCC lines and their NKC counterparts. As a representative example, CTL B4 restricted by HLA-B*0702 reacted with 5 of 5 HLA-B7+ RCC lines (Figure 5D-F). With comparable efficiency, this CTL clone recognized corresponding NKC cells that were available in 2 cases. The failure of CTL B4 to react with any other HLA-B7+ non-RCC tumor-cell lines pointed out that its target antigen might be exclusively expressed in renal tissue. In addition to HLA-A*0201 and HLA-B*0702, CTL clones of groups 2 and 3 used HLA-Cw*0702 as a restriction element (Table 3).
CD4+ CD8+ CTL exhibiting both HLA- and non-HLA–restricted cytolytic activity
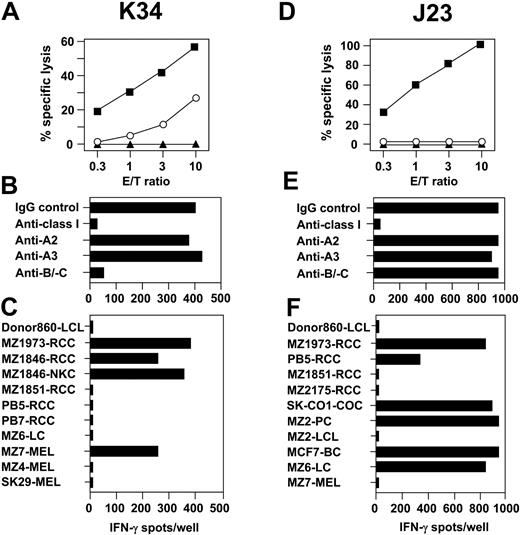
Specificity analysis on RCC-reactive CTLs identified clone K34 that exerted moderate but consistent cytolysis on NK target K562, but not on MZ1257-LCL (Figure 6A). This CTL recognized MZ1257-RCC in association with HLA-Cw7 as demonstrated by antibody-blocking and cross-reactivity assays (Figure 6B,C). Lysis of MZ1257-RCC could be blocked by mAbs directed against CD3 and CD8, indicating that the T-cell receptor (TCR) of clone K34 was involved in RCC recognition (data not shown). Consequently, CTL clone K34 defined a separate category (ie, group 4 CTL) among anti–MZ1257-RCC specificities containing CTL with simultaneous HLA- and non-HLA–restricted effector mechanisms. In flow cytometric analysis, CTL K34 showed coexpression of CD8 (bright) and CD4 (dim) not observed with other anti-RCC1257 CTL. Further phenotypic differences, in particular with regard to NK receptors CD94 and CD158a/b, were not found (Table 2).
Anti-RCC CTLs with HLA- and non–HLA-restricted cytotoxicity or with restriction by a nonclassical HLA-Ib molecule. CTL clones K34 and J23 were isolated from allo-MLTC-1 on CD8+ T lymphocytes of healthy donor 860 and the HLA class Ia–matched RCC line MZ1257-RCC. (A,D) Lysis on MZ1257-RCC (▪), MZ1257-LCL (▴), and K562 (○) by CTLs was determined in 4-hour 51Cr-release assays. Results are percentage of cytolysis at indicated E/T ratios. (B,E) IFN-γ ELISPOT assays were performed on MZ1257-RCC in the presence of anti–HLA class I mAbs to identify CTL restriction elements. (C) Reactivity of CTL K34 was tested against allogeneic HLA-Cw7+ target cell lines in IFN-γ spot assays. (F) IFN-γ spot production of CTL J23 was assayed against target cell lines matched or mismatched for single HLA-Ia alleles. ELISPOT results shown are representative of 50 different targets completely tested. They were obtained in 20-hour assays on 5000 CTLs/well in triplicates.
Anti-RCC CTLs with HLA- and non–HLA-restricted cytotoxicity or with restriction by a nonclassical HLA-Ib molecule. CTL clones K34 and J23 were isolated from allo-MLTC-1 on CD8+ T lymphocytes of healthy donor 860 and the HLA class Ia–matched RCC line MZ1257-RCC. (A,D) Lysis on MZ1257-RCC (▪), MZ1257-LCL (▴), and K562 (○) by CTLs was determined in 4-hour 51Cr-release assays. Results are percentage of cytolysis at indicated E/T ratios. (B,E) IFN-γ ELISPOT assays were performed on MZ1257-RCC in the presence of anti–HLA class I mAbs to identify CTL restriction elements. (C) Reactivity of CTL K34 was tested against allogeneic HLA-Cw7+ target cell lines in IFN-γ spot assays. (F) IFN-γ spot production of CTL J23 was assayed against target cell lines matched or mismatched for single HLA-Ia alleles. ELISPOT results shown are representative of 50 different targets completely tested. They were obtained in 20-hour assays on 5000 CTLs/well in triplicates.
Allogeneic RCC-reactive CTLs can be restricted by a nonclassical HLA-Ib molecule
From MLTC-1 several CTL clones were isolated that exhibited an extraordinary strong lytic activity against MZ1257-RCC, but did not recognize MZ1257-LCL or K562. Representative data on CTL clone J23 are shown in Figure 6D. Anti-RCC reactivity of these CTL clones was completely inhibited by anti–HLA class I mAb W6/32, but not by mAbs directed against HLA-A2, HLA-A3, or common HLA-B/-C determinants (Figure 6E). This result was confirmed in several independent antibody-blocking assays, including anti-RCC and anti-melanoma CTL clones with known HLA class Ia restriction as controls. From a broad panel of 50 different targets tested, CTL J23 cross-reacted with 3 additional RCC as well as single lung, colon, breast, and pancreatic carcinoma cell lines, but not with available LCL counterparts (Figure 6F). Tumor lines recognized were not matched for a distinct HLA class Ia allele. In summary, these results suggested that CTL J23 is restricted by a W6/32-detected nonclassical HLA-Ib molecule. CTL clones such as clone J23 were classified as group 5 CTL in our system. Anti–RCC reactivity mediated by these CTLs could be inhibited by anti-CD3 and anti-CD8 mAbs. Antibody typing of the TCR β-chain showed that these CTLs expressed either Vβ5.2 or Vβ1. No surface marker was found that allowed to discriminate them from group 1 to group 4 specificities (Table 2).
Discussion
The generation of RCC-reactive CTLs, followed by mass spectrometry identification of relevant tumor epitopes, requires RCC cell lines with a long-term in vitro growth characteristic. These cell lines are usually obtained only from a small minority of patients. In preparation of similar prospective studies on healthy sibling-donor RCC pairs, we initiated our project with clear-cell carcinoma lines previously established at our institution.18 In search of appropriate PBMC donors, we failed to find individuals matched with RCC lines for both HLA class I and II loci. Since the RCC cell lines used herein were negative for HLA class II in flow cytometry34 we applied PBMCs of healthy donors matched with these cell lines only for HLA class I. However, HLA class II expression below the flow cytometry level might be sufficient to lead to the induction of anti-allo HLA class II responses. To avoid the latter, we started allogeneic MLTCs with CD8+ T lymphocytes isolated from donor PBMCs with a purity of more than 98%. Following this experimental design, we never observed any anti-RCC CD4+ T-cell responses in vitro. With regard to HLA class I, a single HLA-B7 micromismatch between donor 860 (B*0704) and the MZ1257-RCC line (B*0702) was detected at the DNA level (Table 1). Small allelic disparities can certainly trigger potent anti-HLA mismatch immune responses both in vitro and in vivo.35,36 However, CTLs recognizing this HLA-B7 micromismatch by itself were never isolated within any clonal or nonclonal responder population. In the donor 880/MZ1851-RCC pair, the donor was heterozygous for HLA-Cw*0702 whereas the corresponding RCC was homozygous for this allele. In addition, donor 880 expressed HLA-Cw*0501, which was not detected within MZ1851-RCC DNA and, therefore, could not induce a T-cell response. As expected from the genomic HLA identity in the relevant donor-versus-RCC direction, anti-HLA CTLs were never obtained in this model.
Our study provides the first evidence that RCC-reactive CD8+ CTLs can be readily generated from PBMCs of HLA class I–matched healthy individuals. These CTLs secreted TH1-associated cytokines upon antigen contact and were restricted by various HLA class Ia and Ib molecules. Similar efficient generation and sustained propagation of a multitude of RCC-reactive T cells have not been described for autologous MLTC responders in patients MZ1257 and MZ185118,34 or in other RCC patients studied.4 This observation might reflect the superior ability to activate and expand RCC-directed T cells from PBMCs of allogeneic healthy donors compared with the autologous setting. Indeed, several reports demonstrated tumor-induced anergy or tolerance of autologous antitumor CTLs in RCC as well as other cancer types.37,38 Successful generation of RCC-reactive CD8+ T lymphocytes has also been achieved using PBMCs of allogeneic healthy donors who had only a single HLA class I allele match with their RCC stimulator cell lines.34,39 However, such an experimental set-up promotes dominant anti–HLA mismatch T-cell responses in vitro, potentially masking or even hindering the development of self-HLA–restricted antitumor CTLs.
Cloning of allogeneic MLTCs led to the isolation of RCC-reactive CTLs with phenotypic characteristics of pre–terminally differentiated memory T cells.40 Since healthy donors had no history of previous priming against RCC antigens, the question remains why these CTLs exhibited a memory phenotype. One explanation might be that originally naive CD45RA+ CD45RO– precursors converted to CD45RA– CD45RO+ memory CTLs during repeated antigen-specific stimulations in vitro.41 Considering the lack of costimulatory molecules CD80 and CD86 on MZ1257-RCC stimulators,34 this would require that T-cell priming against RCC antigens with subsequent differentiation into memory cells could occur in the absence of costimulation. Another possibility would be that anti-RCC CTLs developed from a memory precursor pool containing antigenic specificities similar to RCC antigens (“molecular mimicry”).42
Based on HLA class I restriction and cross-reactivity results, allogeneic CTL clones were divided into 5 different subgroups summarized in Table 3. Group 1 CTLs such as clone B5 recognized self-HLA Ia–associated peptide antigens that can be expressed by epithelial and nonepithelial tissues as well as hematopoietic cells. Accordingly, this cross-reactivity pattern suggested broadly expressed antigenic determinants such as ubiquitous unaltered or polymorphic minor histocompatibility antigens as their target structures.43 To define these antigens at the molecular level, we applied a combination of microcapillary liquid chromatography and MALDI-TOF/TOF mass spectrometry which previously has not been described for this purpose. Among several HLA-A*0301–binding, naturally processed RCC peptides successfully sequenced by this technique, peptide KLPNSVLGR was recognized by CTL clone B5. This epitope demonstrates a high binding score for HLA-A3 (data not shown). It is represented in a single known gene, Eps15 homology domain–containing 2 (EHD2), which is located on chromosome 19 and encodes a protein of 543 amino acid residues.28 The peptide is derived from residues 480-488 of the nonpolymorphic EHD2 gene product. Using a T-cell–independent approach, Weinschenk et al44 recently identified on solid RCC tumor tissue a natural HLA-A*02–associated peptide ligand encoded by residues 507-515 of EHD2. The precise function of the EHD2 gene is unknown. However, its strong homology to the Eps15 gene45 suggests that EHD2 might play a role in various cellular functions such as endocytosis of growth factor receptors, intracellular signal transduction, and actin remodeling.46
The EHD2 gene is broadly expressed in various malignant and nonmalignant tissues.28 Accordingly, raising an immune response against EHD2 might not only induce desired graft-versus-RCC effects but might also cause unwanted destruction of normal tissue. We observed EHD2 expression at the cDNA level in 4 of 5 ex vivo isolated primary RCC tumors and in all cell lines recognized by CTL clone B5 (data not shown). However, whereas only a few cell lines not recognized by this CTL (eg, donor 860-LCL, MZ7-MEL) were negative for EHD2 transcription, the majority of undetected targets did indeed demonstrate expression of EHD2. Representative examples were MZ1795-RCC, MZ6-LC, and HT29-COC (Figure 2D). The inability of CTLs to recognize EHD2-expressing targets might be explained by the failure of cells to process or present the appropriate CTL epitope. In fact, genomic HLA sequencing revealed that EHD2-positive cell lines not recognized by CTL B5 expressed an HLA-A3 allele different from the HLA-A*0301 originally used as the CTL restriction element. MZ1795-RCC and HT29-COC, for example, expressed HLA-A*0302, whereas MZ6-LC showed expression of HLA-A*0310. These findings suggest that CTL clone B5 can detect peptide epitope KLPNSVLGR only in cells expressing the HLA-A*0301 allele. Additional support for this hypothesis was derived from an experiment where 293T cells cotransfected with cDNAs coding for EHD2, and HLA-A*0302 were not recognized by CTL B5 (data not shown). This result might be explained by the failure of HLA-A*0302–expressing cells to properly present the EHD2 epitope, or, alternatively, by the lack of engagement between HLA-A*0302/peptide complex and the TCR of CTL B5. Similar allele-specific differences have been already described for viral and tumor epitopes presented by the various HLA-A2 subtypes.47,48
Clonal CTLs assigned to group 2 recognized multiple RCC and colon carcinoma lines, but not other tumors or NKC and LCL counterparts. Thus, group 2 CTLs might be directed against antigens specifically expressed or overexpressed in renal and colon carcinomas. In the case that antigens recognized by group 2 CTLs are in fact not expressed in nonmalignant tissue, they would be ideal candidates for tumor immunotherapy. Group 3 CTL clones exclusively reacted with RCC and NKC lines, indicating that the antigenic peptides are shared between normal and neoplastic renal cells. These CTLs might recognize renal lineage-specific antigens, as suggested by previous work on autologous antitumor T-lymphocyte responses in the MZ1257-RCC system18 and in other RCC49 or melanoma models.50 This observation indicates the possibility of overcoming the existing tolerance to kidney-specific self antigens in vivo. Raising specific CTLs against these antigens might ultimately lead to the development of autoimmune kidney disease, and, potentially, to regression of RCC lesions.
Our study further identified group 4 CTL clone K34 that exhibited both HLA- and non-HLA–restricted cytotoxicity. Indeed, CTLs with this kind of dual function have been isolated from the blood of healthy donors51 and ovarian carcinoma patients.52 They are potentially of great clinical importance as they have a backup mechanism that may act when tumor cells lose HLA class I expression. Phenotypically, CTL clone K34 showed simultaneous expression of CD4 and CD8. Although their precise function is poorly understood, CD4/CD8 double-positive T cells can be found among lymphocytes infiltrating autologous RCC tumors53 and rejected kidney allografts.54
We further isolated CTL clones of group 5 showing extraordinary strong recognition of various epithelial tumor cell lines. This reactivity could be completely blocked by pan–HLA class I antibody W6/32, but not by antibodies specific for HLA-A/-B or -C alleles. Antibody W6/32 detects not only classical HLA-Ia, but also certain nonclassical HLA-Ib molecules, such as HLA-E/-F and -G.55 HLA-E and -G molecules are thought to preferentially act as ligands for NK receptors.56 However, group 5 CTLs isolated in our study lacked expression of NK receptors CD94 and CD158 and exhibited TCR-mediated tumor recognition. While for most HLA-Ib molecules interaction with TCRα/β is still unclear, recent evidence suggests that HLA-E can function as a TCR ligand.57 If relevant in vivo, T-cell–mediated recognition of foreign nonclassical HLA-Ib antigens may contribute to the generation of alloreactivity in HLA-Ia–matched donor-recipient pairs, as has been suggested from animal studies.58
In conclusion, our results provide the first evidence that a heterogeneous panel of RCC-reactive CD8+ T cells can be isolated from PBMCs of HLA class I–matched healthy individuals. While we currently extend our studies on HLA-identical sibling-donor RCC pairs, our present observations are valuable for the increasing number of RCC patients who receive stem cell transplants from HLA-matched unrelated volunteers.14,15 CTLs generated from these donors were found to attack a diverse set of RCC antigens through multiple HLA-Ia/Ib restriction molecules. At this point, we cannot conclude whether these CTL-defined antigens are relevant target structures of anti-RCC immune responses occurring in vivo. Answering this question will certainly require us to analyze the expression of candidate antigens and the frequency of specific CTLs in RCC patients after allo-HSCT, and to correlate these findings with clinical graft-versus-tumor and graft-versus-host events.59-61 Nevertheless, RCC antigens recognized by group 2 CTLs and exclusively expressed in malignant tissue might be potential targets of single RCC regression responses that were observed in the absence of detectable GVHD.11 Thus, they could represent ideal candidates to develop specific immunotherapy, either by adoptive T-cell transfer or vaccination. In contrast, RCC antigens ubiquitously expressed in various epithelial and nonepithelial tissues (either of unaltered or polymorphic origin) are potential targets of graft-versus-host responses. The characterization of the latter antigen category will be crucial to generate specific strategies to avoid their immune attack. Finally, identification of molecular targets of graft-versus-host and graft-versus-tumor responses might provide the chance to improve the specificity and efficiency of allogeneic cell therapy in metastatic RCC.
Prepublished online as Blood First Edition Paper, July 1, 2004; DOI 10.1182/blood-2004-02-0459.
Supported by grant no. SFB432/A13 from the Deutsche Forschungsgemeinschaft (W.H. and A.S.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal