Abstract
The t(12;21) (p13;q22) translocation resulting in ETV6/RUNX1 (previously named TEL/AML1) gene fusion is present in about 25% of children with precursor B-lineage acute lymphoblastic leukemia (B-ALL). We successfully tested 275 precursor B-ALL samples from children aged 1 to 17 years to determine the relation between t(12;21) and in vitro cellular drug resistance, measured by the fluorometric microculture cytotoxicity assay (FMCA). Samples from 83 patients (30%) were positive for t(12;21). The ETV6/RUNX1+ samples were significantly more sensitive than ETV6/RUNX1– samples to doxorubicin, etoposide, amsacrine, and dexamethasone, whereas the opposite was true for cytarabine. After matching for unevenly distributed patient characteristics, that is, excluding patients with high hyperdiploidy (> 51 chromosomes), t(9; 22), t(1;19), or 11q23 rearrangement, the ETV6/RUNX1+ samples remained significantly more sensitive to doxorubicin (P = .001) and etoposide (P = .001). For the other drugs tested (amsacrine, cytarabine, dexamethasone, prednisolone, vincristine, 6-thioguanine, and 4-hydroperoxy-cyclophosphamide), no significant difference in cellular drug sensitivity was found. In conclusion, we found that the presence of the t(12;21) translocation in childhood precursor B-ALL is associated with a high tumor cell sensitivity to doxorubicin and etoposide. High throughput techniques should now be used to elucidate the cellular mechanisms by which ETV6/RUNX1 gene fusion is linked to increased sensitivity to these drugs.
Introduction
Cytogenetic abnormalities of leukemic cells are known to be important independent prognostic factors in childhood acute lymphoblastic leukemia (ALL). The t(9;22)(q34;q11) translocation and 11q23/MLL rearrangements are associated with poor survival rates, whereas high hyperdiploidy correlates with a favorable outcome.1,2 The most common chromosomal aberration is translocation of t(12;21)(p13;q22), resulting in ETV6/RUNX1 gene fusion (previously named TEL/AML1), which occurs in 25% to 30% of newly diagnosed precursor B-lineage childhood ALL.3-7
The ETV6 gene, a member of the ETS family of transcription factors, is necessary for many developmental processes, for example, the establishment of bone marrow hematopoiesis (for reviews, see Rubnitz et al4 and Sawinska and Ladon8 ). The RUNX1 gene encodes a transcription factor and is normally expressed in cells of all hematopoietic lineages.9 Both ETV6 and RUNX1 are involved in chromosomal changes found in various forms of human hematologic malignancies.10,11 The exact mechanisms are not clear at present, but expression of the ETV6/RUNX1 fusion gene may influence the transcriptional activity of other genes and appears to interfere with RUNX1-dependent gene regulation.12
In childhood ALL, patients with ETV6/RUNX1 always display precursor B phenotype and most of them are between 1 and 10 years of age, factors associated with a favorable prognosis. High hyperdiploidy or presence of t(9;22), t(1;19)(q23;p13), or 11q23 rearrangements are very rarely found in ETV6/RUNX1+ patients. Some studies have indicated that patients with t(12;21) have an excellent prognosis.4,6,7,10,13 However, in patients treated with the Berlin-Frankfurt-Munster (BFM) group protocols, ETV6/RUNX1+ patients displayed better outcome in short-term follow-up, but appeared to have more late relapses.5,14,15 One possible explanation to these diverging results is that the prognostic impact of t(12;21) is dependent on therapy. This would be compatible with the hypothesis that the gene fusion results in a specific drug resistance profile. One study has indeed shown that ETV6/RUNX1 gene fusion is related to high in vitro drug sensitivity for l-asparaginase in childhood ALL.16
In the present study we have investigated whether the in vitro drug resistance profile of ETV6/RUNX1+ positive samples differs from other cases of precursor B-lineage ALL (B-ALL). Leukemic cells were tested by the fluorometric microculture cytotoxicity assay (FMCA), a rapid and reproducible method for determination of in vitro drug sensitivity.17,18
Patients, materials, and methods
Patients and samples
Leukemic cells from bone marrow or peripheral blood of children (ages 1-17 years) with newly diagnosed ALL were used in this study. Nordic centers for pediatric oncology participated and provided samples from 689 patients for test of in vitro drug resistance between 1992 and December 2002. The patients were representative of all children in this age group diagnosed with ALL in the Nordic countries during the study period, as described in detail elsewhere.19 Of the 689 ALL patients, 605 were diagnosed with precursor B-ALL. The diagnosis of ALL was established at a pediatric oncology center by analysis of bone marrow aspirates including morphology, immunophenotype, and cytogenetics of the leukemic cells. Patient characteristics and clinical follow-up data were obtained from annual reports submitted from the treating clinicians to the Nordic registry at the Childhood Cancer Research Unit in Stockholm. Precursor B immunophenotype was defined by positivity to TdT, HLA-DR, CD19, and usually also for CD22.20,21
The samples were collected in heparinized glass tubes, kept at room temperature, and sent by mail or through international express delivery companies. As a rule they reached the in vitro sensitivity laboratory in Uppsala, Sweden, for processing within 24 to 36 hours. Most of the samples (about 90%) were analyzed freshly, but for practical reasons some were cryopreserved in culture medium containing 10% dimethyl sulfoxide (DMSO) and 50% fetal calf serum (FCS) by initial freezing for 24 hours at –70° C followed by storage in liquid nitrogen. The cells were later thawed and analyzed. Previous studies showed that cryopreservation does not affect the in vitro sensitivity, and it has also been shown that the source of the leukemic cells (bone marrow or peripheral blood) does not affect the in vitro drug resistance measured.18,22,23
Leukemic cells were prepared by 1.077 g/mL Ficoll-Isopaque (Pharmacia, Uppsala, Sweden) density-gradient centrifugation. Viability was determined by the trypan blue exclusion test. The median viability was 95% and FMCA was performed only when the viability was 70% or higher. An independent hematologist estimated the proportion of leukemic cells on May-Grünwald-Giemsa–stained cytocentrifugate preparations, using light microscopy. The median proportion of lymphoblasts after separation was 90% and FMCA was performed only when this proportion was 70% or more.
Cytogenetic investigations
Chromosome banding analyses of bone marrow or peripheral blood samples were performed using standard methods in 15 cytogenetic laboratories in the Nordic countries. Since 1996 (Sweden) and 2000 (all 5 Nordic countries) the karyotypes have been centrally reviewed.24-27 All 375 patients in the study cohort were analyzed with fluorescence in situ hybridization (FISH) or reverse-transcriptase polymerase chain reaction (RT-PCR) or both, using standard methods for the presence of the cryptic translocation ETV6/RUNX1.2,5,8 The definition and description of clonal abnormalities followed the recommendations of the International System for Human Cytogenetic Nomenclature.28
FMCA procedure
FMCA is based on measurement of fluorescence generated from hydrolysis of fluorescein diacetate (FDA) to fluorescein by cells with intact plasma membranes and has been described in detail previously.17,29-31 One hundred thousand leukemic cells in 180 μL culture medium were seeded per well in 96-well microtiter plates prepared in advance with the different drugs to be tested. The culture plates were incubated at 37° C in a humidified atmosphere containing 95% air and 5% CO2 for 72 hours of continuous drug exposure. The plates were then centrifuged (200g, 5 minutes) and the medium removed by automatic pipetting. After one wash with phosphate-buffered saline (PBS), 200 μL/well PBS containing FDA (10 μg/mL) was added. Subsequently, the plates were incubated for 1 hour at 37° C and the fluorescence was then read by a scanning fluorometer (Fluoroscan 2; Labsystems, Helsinki, Finland). Drugs were tested in triplicate. Six wells without drugs served as controls and 6 wells containing culture medium only served as blanks. Quality criteria for a technically successful assay included a proportion of leukemic cells of 70% or more in control wells after 72 hours of incubation, a fluorescence signal in control wells of equal to or more than 5 times the mean blank value, and a mean coefficient of variation (CV) in control wells of less than 30%. The results are presented as survival index (SI), defined as fluorescence in test wells/fluorescence in control wells (blank values subtracted) × 100. Thus, a low numerical value indicates high sensitivity to the cytotoxic effect of the drug.
Reagents and drugs
FDA (Sigma, St Louis, MO) was dissolved in DMSO (Sigma) and kept frozen (–20° C) as a stock solution (10 mg/mL) protected from light. RPMI 1640 culture medium (Sigma) supplemented with 10% heat-inactivated FCS (HyClone, Cramlington, United Kingdom), 2 mM glutamine, 50 μg/mL streptomycin, and 60 μg/mL penicillin (HyClone) was used throughout.
Cytotoxic drugs were obtained from commercial sources and tested at the concentrations shown in Table 1. The active metabolite of cyclophosphamide, 4-hydroperoxy-cyclophosphamide (4-HC), was used. Experimental plates were prepared with 20 μL/well drug solution at 10 times the desired final concentration. The plates were stored at –70° C until further use. The desired final concentration of the drug was reached after addition of leukemic cells dissolved in 180 μL culture medium per well.
Drugs used for test of in vitro resistance
Drug . | Origin . | Solvent . | Concentration . |
|---|---|---|---|
| Doxorubicin | Pharmacia (Uppsala, Sweden) | PBS | 0.5 μg/mL |
| Etoposide | Bristol-Myers Squibb (Stockholm, Sweden) | PBS | 5 μg/mL |
| Amsacrine | Bristol-Myers Squibb | SW | 1 μg/mL |
| Dexamethasone | MSD (Stockholm, Sweden) | PBS | 1.4 μg/mL |
| Cytarabine | Sigma (St Louis, MO) | PBS | 0.5 μg/mL |
| Prednisolone | Organon (Gothenburg, Sweden) | PBS | 50 μg/mL |
| Vincristine | Lilly (Stockholm, Sweden) | PBS | 0.5 μg/mL |
| 6-thioguanine | Sigma | NaOH/SW | 10 μg/mL |
| 4-HC | Duke University (Durham, NC) | PBS | 2 μg/mL |
Drug . | Origin . | Solvent . | Concentration . |
|---|---|---|---|
| Doxorubicin | Pharmacia (Uppsala, Sweden) | PBS | 0.5 μg/mL |
| Etoposide | Bristol-Myers Squibb (Stockholm, Sweden) | PBS | 5 μg/mL |
| Amsacrine | Bristol-Myers Squibb | SW | 1 μg/mL |
| Dexamethasone | MSD (Stockholm, Sweden) | PBS | 1.4 μg/mL |
| Cytarabine | Sigma (St Louis, MO) | PBS | 0.5 μg/mL |
| Prednisolone | Organon (Gothenburg, Sweden) | PBS | 50 μg/mL |
| Vincristine | Lilly (Stockholm, Sweden) | PBS | 0.5 μg/mL |
| 6-thioguanine | Sigma | NaOH/SW | 10 μg/mL |
| 4-HC | Duke University (Durham, NC) | PBS | 2 μg/mL |
SW indicates sterile water.
The drugs tested were used at empirically derived cutoff concentrations, chosen to produce a large scatter of SI values among the samples. These concentrations were adopted from previous studies of leukemic cells.18
Statistical analysis
Nonparametric methods were used throughout. Differences in distribution of variables were tested with the Mann-Whitney U test, Kruskal-Wallis H test, or the χ2 test. The Spearman correlation coefficient was used to examine relationships between continuous variables. The SPSS (Chicago, IL) 11.5 software package was used for the calculations. All analyses were 2-tailed and the level of statistical significance was set at P < .05.
The patients and/or their guardians gave informed consent. The Ethics Committee of the Medical Faculty of Uppsala University approved the study.
Results
The presence of ETV6/RUNX1 gene fusion was explored in 375 (62%) of 605 samples from patients with precursor B-ALL sent to our laboratory for in vitro drug resistance analysis. In total, 101 ETV6/RUNX1+ and 274 ETV6/RUNX1– patients were identified, resulting in an overall prevalence of 27% in the precursor B-ALL samples. Clinical features and outcome for the patients not studied here due to lack of data on presence of ETV6/RUNX1 (n = 230) were comparable to those included in the study (data not shown).
Testing of cellular drug resistance by FMCA was accomplished successfully in 275 of the 375 samples where ETV6/RUNX1 data were available. Reasons for failure of the assay in the ETV6/RUNX1+ and ETV6/RUNX1– groups included insufficient cell numbers to test any drug (n = 1 and n = 17, respectively), transport problems (n = 1, n = 7), low proportion (< 70%) of lymphoblasts before the in vitro test (n = 1, n = 6), low proportion (< 70%) of lymphoblasts in control wells after 72 hours of incubation (n = 2, n = 12), low signal-to-noise ratio (n = 8, n = 31), and a coefficient of variation in controls of more than 30% (n = 5, n = 9). The technical success rate for in vitro testing of samples with sufficient number of cells was 84% and 77% in the ETV6/RUNX1+ and ETV6/RUNX1– groups, respectively (not significant).
The distribution of important clinical and biologic parameters within the ETV6/RUNX1+ and ETV6/RUNX1– groups with a successful in vitro test is summarized in Table 2. There was an expected difference in the modal number between the 2 groups since the hyperdiploid karyotype only rarely occurs in patients with the ETV6/RUNX1 gene fusion.
Characteristics of 275 children with ETV6/RUNX1+ or ETV6/RUNX1- precursor B-ALL tested for in vitro cellular drug resistance
ETV6/RUNX1 . | Positive . | Negative . | P . |
|---|---|---|---|
| No. of patients | 83 | 192 | |
| Median age, y (range) | 4.4 (1.3-15.6) | 5.1 (1.1-17.0) | .042 |
| Sex | .85 | ||
| Male, n (%) | 46 (55) | 104 (54) | |
| Female, n (%) | 37 (45) | 88 (46) | |
| WBC count | |||
| Median | 8 | 7 | .45 |
| Less than 10 × 109/L, n (%) | 46 (55) | 115 (60) | |
| Between 10 and 50 × 109/L, n (%) | 29 (35) | 57 (30) | |
| 50 × 109/L or more, n (%) | 8 (10) | 20 (10) | |
| Modal number | .001 | ||
| 45-46, n (%) | 41 (49) | 58 (30) | |
| 47-51, n (%) | 15 (18) | 21 (11) | |
| 52-60, n (%) | 0 (0) | 76 (40) | |
| More than 60, n (%) | 0 (0) | 5 (2) | |
| Unknown, n (%) | 27 (33) | 32 (17) |
ETV6/RUNX1 . | Positive . | Negative . | P . |
|---|---|---|---|
| No. of patients | 83 | 192 | |
| Median age, y (range) | 4.4 (1.3-15.6) | 5.1 (1.1-17.0) | .042 |
| Sex | .85 | ||
| Male, n (%) | 46 (55) | 104 (54) | |
| Female, n (%) | 37 (45) | 88 (46) | |
| WBC count | |||
| Median | 8 | 7 | .45 |
| Less than 10 × 109/L, n (%) | 46 (55) | 115 (60) | |
| Between 10 and 50 × 109/L, n (%) | 29 (35) | 57 (30) | |
| 50 × 109/L or more, n (%) | 8 (10) | 20 (10) | |
| Modal number | .001 | ||
| 45-46, n (%) | 41 (49) | 58 (30) | |
| 47-51, n (%) | 15 (18) | 21 (11) | |
| 52-60, n (%) | 0 (0) | 76 (40) | |
| More than 60, n (%) | 0 (0) | 5 (2) | |
| Unknown, n (%) | 27 (33) | 32 (17) |
P values were determined by χ2 and Mann-Whitney U test.
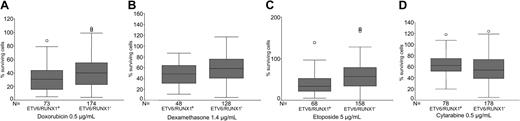
Table 3 and Figure 1 show the in vitro drug resistance in samples from patients with or without ETV6/RUNX1. Samples positive for ETV6/RUNX1 were significantly more sensitive to doxorubicin, etoposide, amsacrine, and dexamethasone, whereas the opposite was true for cytarabine. For prednisolone, vincristine, 6-thioguanine, and 4-HC no difference was observed.
In vitro drug resistance in 275 children with precursor B-ALL positive or negative for ETV6/RUNX1 gene fusion
. | ETV6/RUNX1+ . | ETV6/RUNX1- . | . |
|---|---|---|---|
| Drug . | Median (25th-75th) . | Median (25th-75th) . | P . |
| Doxorubicin | 32 (16-44) | 40 (23-56) | .003 |
| Etoposide | 35 (22-54) | 57 (35-80) | .001 |
| Amsacrine | 29 (17-45) | 37 (21-53) | .015 |
| Dexamethasone | 49 (31-66) | 60 (41-77) | .043 |
| Cytarabine | 63 (52-75) | 55 (39-74) | .016 |
| Prednisolone | 52 (32-73) | 54 (38-72) | .27 |
| Vincristine | 53 (41-67) | 57 (40-79) | .32 |
| 6-thioguanine | 42 (24-60) | 41 (24-58) | .90 |
| 4-HC | 20 (12-48) | 29 (13-42) | .56 |
. | ETV6/RUNX1+ . | ETV6/RUNX1- . | . |
|---|---|---|---|
| Drug . | Median (25th-75th) . | Median (25th-75th) . | P . |
| Doxorubicin | 32 (16-44) | 40 (23-56) | .003 |
| Etoposide | 35 (22-54) | 57 (35-80) | .001 |
| Amsacrine | 29 (17-45) | 37 (21-53) | .015 |
| Dexamethasone | 49 (31-66) | 60 (41-77) | .043 |
| Cytarabine | 63 (52-75) | 55 (39-74) | .016 |
| Prednisolone | 52 (32-73) | 54 (38-72) | .27 |
| Vincristine | 53 (41-67) | 57 (40-79) | .32 |
| 6-thioguanine | 42 (24-60) | 41 (24-58) | .90 |
| 4-HC | 20 (12-48) | 29 (13-42) | .56 |
The concentrations of the drugs used are denoted in Table 1.
All the values represent the percentage of surviving cells compared with control wells, presented as median or 25th to 75th percentile. P values were determined by the Mann-Whitney U test.
In vitro drug resistance in 275 children with precursor B-ALL positive or negative for ETV6/RUNX1 (TEL/AML1) gene fusion. ETV6/RUNX1+ patients were significantly more sensitive to (A) doxorubicin (P = .003), (B) dexamethasone (P = .043), and (C) etoposide (P = .001), but were more resistant to (D) cytarabine (P = .016). The box-and-whisker plot shows median, first, and third quartiles; whiskers extend to the highest and lowest value, excluding outliers, which are denoted by circles.
In vitro drug resistance in 275 children with precursor B-ALL positive or negative for ETV6/RUNX1 (TEL/AML1) gene fusion. ETV6/RUNX1+ patients were significantly more sensitive to (A) doxorubicin (P = .003), (B) dexamethasone (P = .043), and (C) etoposide (P = .001), but were more resistant to (D) cytarabine (P = .016). The box-and-whisker plot shows median, first, and third quartiles; whiskers extend to the highest and lowest value, excluding outliers, which are denoted by circles.
The next step in the data analysis was to exclude patients with high hyperploidy (> 51 chromosomes; n = 81). When comparing ETV6/RUNX1+ and ETV6/RUNX1– samples within this cohort, the positive samples remained significantly more sensitive to doxorubicin (P = .006) and etoposide (P = .005; data not shown). For the other drugs tested (amsacrine, cytarabine, dexamethasone, prednisolone, vincristine, 6-thioguanine, and 4-HC) no statistically significant difference was found.
Finally, we excluded patients with chromosomal aberrations known to be of prognostic significance and present almost exclusively in the ETV6/RUNX1– group: t(9;22), t(1;19), and 11q23 rearrangements.
As displayed in Table 4, the remaining patients in the ETV6/RUNX1+ and ETV6/RUNX1– groups showed very similar clinical and biologic characteristics, with the exception of age; ETV6/RUNX1+ children were significantly younger. Higher sensitivity to doxorubicin and etoposide was demonstrated in the ETV6/RUNX1+ group, with a trend in the same direction for dexamethasone (Table 5).
Characteristics of 182 children with ETV6/RUNX1+ or ETV6/RUNX1- precursor B-ALL excluding patients with high hyperdiploidy (> 51), t(9;22), t(1;19), and 11q23 rearrangement
ETV6/RUNX1 . | Positive . | Negative . | P . |
|---|---|---|---|
| No. of patients | 82 | 100 | |
| Median age, y (range) | 4.4 (1.3-15.6) | 6.3 (1.2-17.0) | .001 |
| Sex | .58 | ||
| Male, n (%) | 45 (55) | 59 (59) | |
| Female, n (%) | 37 (45) | 41 (41) | |
| WBC count | |||
| Median | 8 | 7 | .66 |
| Less than 10 × 109/L, n (%) | 45 (55) | 59 (59) | |
| Between 10 and 50 × 109/L, n (%) | 29 (35) | 29 (29) | |
| 50 × 109/L or more, n (%) | 8 (10) | 12 (12) |
ETV6/RUNX1 . | Positive . | Negative . | P . |
|---|---|---|---|
| No. of patients | 82 | 100 | |
| Median age, y (range) | 4.4 (1.3-15.6) | 6.3 (1.2-17.0) | .001 |
| Sex | .58 | ||
| Male, n (%) | 45 (55) | 59 (59) | |
| Female, n (%) | 37 (45) | 41 (41) | |
| WBC count | |||
| Median | 8 | 7 | .66 |
| Less than 10 × 109/L, n (%) | 45 (55) | 59 (59) | |
| Between 10 and 50 × 109/L, n (%) | 29 (35) | 29 (29) | |
| 50 × 109/L or more, n (%) | 8 (10) | 12 (12) |
Children with high hyperdiploidy (n = 81; data available in 78% of the cases), t(9;22) (n = 5; data available in 85% of the cases), t(1;19) (n = 6; data available in 50% of the cases), 11q23 rearrangement (n = 4; data available in 50% of the cases) were excluded from the cohort of 275 patients. Some patients had more than one chromosomal aberration.
P values were determined by χ2 and Mann-Whitney U test.
In vitro drug resistance in 182 children with ETV6/RUNX1+ or ETV6/RUNX1- precursor B-ALL excluding patients with high hyperdiploidy (> 51), t(9;22), t(1;19), and 11q23 rearrangement
. | ETV6/RUNX1+ . | ETV6/RUNX1- . | . |
|---|---|---|---|
| Drug . | Median (25th-75th) . | Median (25th-75th) . | P . |
| Doxorubicin | 32 (16-44) | 44 (27-57) | .001 |
| Etoposide | 35 (22-54) | 55 (33-73) | .001 |
| Amsacrine | 29 (17-45) | 34 (20-52) | .093 |
| Dexamethasone | 49 (31-67) | 60 (42-78) | .051 |
| Cytarabine | 63 (52-75) | 63 (45-75) | .51 |
| Prednisolone | 52 (31-73) | 56 (38-72) | .18 |
| Vincristine | 53 (41-67) | 53 (41-76) | .56 |
| 6-thioguanine | 42 (24-61) | 45 (29-62) | .30 |
| 4-HC | 20 (12-48) | 29 (15-44) | .53 |
. | ETV6/RUNX1+ . | ETV6/RUNX1- . | . |
|---|---|---|---|
| Drug . | Median (25th-75th) . | Median (25th-75th) . | P . |
| Doxorubicin | 32 (16-44) | 44 (27-57) | .001 |
| Etoposide | 35 (22-54) | 55 (33-73) | .001 |
| Amsacrine | 29 (17-45) | 34 (20-52) | .093 |
| Dexamethasone | 49 (31-67) | 60 (42-78) | .051 |
| Cytarabine | 63 (52-75) | 63 (45-75) | .51 |
| Prednisolone | 52 (31-73) | 56 (38-72) | .18 |
| Vincristine | 53 (41-67) | 53 (41-76) | .56 |
| 6-thioguanine | 42 (24-61) | 45 (29-62) | .30 |
| 4-HC | 20 (12-48) | 29 (15-44) | .53 |
The concentrations of the drugs used are denoted in Table 1. Children with high hyperdiploidy (n = 81; data available in 78% of the cases), t(9;22) (n = 5; data available in 85% of the cases), t(1;19) (n = 6; data available in 50% of the cases), 11q23 rearrangement (n = 4; data available in 50% of the cases) were excluded from the cohort of 275 patients. Some patients had more than one chromosomal aberration.
All the values represent the percentage of surviving cells compared with control wells, and are shown as median and 25th to 75th percentile. P values were determined by the Mann-Whitney U test.
Removal of patients 10 years old and older from both groups eliminated the difference in age (median 4.3 years in the positive group, n = 78, and 4.7 years in the negative group, n = 71; P = .50), but the ETV6/RUNX1+ samples were still more sensitive to doxorubicin (P = .004) and etoposide (P = .003; data not shown).
Discussion
We studied the relation between ETV6/RUNX1 gene fusion and in vitro drug resistance in precursor B-ALL patients within the framework of a Nordic multicenter study. As described in detail elsewhere,19 the samples sent to our laboratory for test of cellular drug resistance were representative of all children diagnosed with ALL in the Nordic countries during the study period, as demonstrated by similarities in sex, age, and white blood cell (WBC) count at diagnosis, immunophenotype, cytogenetics, and probability of disease-free survival (p-DFS). The ETV6/RUNX1 gene fusion was determined in 62% of the patients. The main reason for the low figure is that only few centers used methods for detection of this cryptic translocation before 1998. During the time period 1998 to 2002 (n = 409), when such methods were in common use, it was determined in 87% of the cases.
The prevalence of ETV6/RUNX1 gene fusion was 27% in the precursor B-ALL samples received, a figure very similar to what has been reported for pediatric precursor B-ALL patients in most Western populations studied.3-7 Biologic and clinical characteristics of the ETV6/RUNX1+ children included in the present study agree with those of other reports in the literature, in that they are exclusively of the precursor B phenotype, have nonhyperdiploid ALL, lack t(9;22), t(1;19), and 11q23 rearrangements, and are mostly between 1 and 10 years of age.2
For various reasons, the in vitro test was unsuccessful in a number of cases. For samples with enough cells on arrival at the laboratory, the overall success rate was 79%. Ramakers-van Woerden et al, using the 3-[4,5-dimethyl-thiazol-2,5-diphenyl] tetrazolium bromide (MTT) method, found a marked difference in success rates between ETV6/RUNX1+ (96%) and ETV6/RUNX1– (70%) patients, for which they had no explanation.16 In our material, the corresponding figures were 84% for the positive and 77% for the negative samples, a nonsignificant difference.
Our main finding was that the t(12;21) translocation in childhood ALL is associated with a specific drug-resistance profile. A crude analysis including all patients showed that ETV6/RUNX1+ children were significantly more sensitive than ETV6/RUNX1– children to doxorubicin, etoposide, amsacrine, and dexamethasone, but more resistant to cytarabine. To make the groups more comparable and focus on the effect of the ETV6/RUNX1 gene fusion, we excluded patients with high hyperdiploidy (> 51 chromosomes). Different dividing points between modal number groups have been proposed,34 but the 51/52 dividing point suggested by Mertens et al35 fits best with the findings in the Nordic population.25 In the present patient material 3 children had a modal number of 51, and one of them was t(12;21) positive. In children with more than 51 chromosomes, t(12;21) was not demonstrated.
The groups were further matched by excluding patients with t(9;22), t(1;19), and 11q23 rearrangements, all known to be of prognostic significance and only very rarely found in ETV6/RUNX1+ patients.2 We chose this approach as the best possible, aware that complete cytogenetic data were not available for all patients. In the remaining cohort of children, ETV6/RUNX1+ samples remained significantly more sensitive to doxorubicin and etoposide than ETV6/RUNX1– ones. Further matching of the groups by including only children ages 1 to 9 years gave similar results.
Ramakers-van Woerden et al studied the relation between ETV6/RUNX1 gene fusion and in vitro drug resistance in childhood ALL by the MTT method, a total cell kill assay similar to the FMCA used by us, and found that ETV6/RUNX1+ patients were significantly more sensitive to l-asparaginase.16 We also aimed at including l-asparaginase in our test panel of drugs, but unfortunately we chose a high drug concentration (10 IU/mL), which has later been shown to produce cytotoxicity by mechanisms that are not relevant in vivo.36 We now use considerably lower concentrations, but the number of patients correctly tested is too low as yet for a presentation of data. The other drugs tested by us were used at empirically derived cutoff levels, adopted from previous studies of leukemic cells, and chosen to produce a large scatter of survival rates among the samples tested. For some drugs these levels were 2 to 5 times higher than those achieved in pediatric patients. One should realize that the in vitro test does not aim at mimicking in vivo conditions and that this approach does not require drug concentrations for drug-induced cell kill in vitro to be the same as in vivo, provided that the mechanisms determining drug resistance are the same (for a review, see Larsson37 ). For all drugs tested, except l-asparaginase, the concentrations used in the present study were similar to the median LC50 value, that is, the drug concentration lethal to 50% of the cells or well within the 25th to 75th percentile interval for LC50 reported by Ramakers-van Woerden et al.16
In their crude analysis, Ramakers-van Woerden et al noted that ETV6/RUNX1+ patients tended to be more sensitive for glucocorticosteroids as well, but less sensitive for cytarabine, than ETV6/RUNX1– patients. We made similar observations. However, after matching for other prognostic factors, only the difference in sensitivity for l-asparaginase remained statistically significant in the study reported by Ramakers-van Woerden et al.16 One explanation for the diverging results might be that the number of patients was about half that in the present study. Other possible reasons are differences in patient selection and study design.
The main results of the present study, together with those of Ramakers-van Woerden et al, are that the ETV6/RUNX1+ patients show a selective sensitivity to a few drugs. However, it is not obvious how this knowledge should be translated into recommendations for the treatment of ETV6/RUNX1+ patients. It seems reasonable that demonstration of high activity in vitro speaks in favor of including a drug in relevant treatment protocols, but does high sensitivity in vitro mean that low doses are enough, or that high doses should be administered to maximize the effect? Only clinical trials can answer those questions. Loh et al found a very low frequency of ETV6/RUNX1+ patients at relapse in a group of patients treated on Dana-Farber Cancer Institute (DFCI) ALL Consortium protocols.13 The DFCI regimens, distinguished by early consolidation with intensive l-asparaginase for all patients and doxorubicin for higher risk patients (half of the ETV6/RUNX1+ patients were treated as high risk), have used fewer agents but at higher cumulative dosages than BFM-based protocols.15 It is conceivable that ETV6/RUNX1+ patients represent a biologically distinct subset of patients whose leukemia is more effectively treated by the agents used more intensively up-front by the DFCI group. However, this must be confirmed in long-term prospective studies including a substantial number of patients at diagnosis and relapse.
It will be of great interest to prospectively study whether sensitivity to doxorubicin, etoposide, or l-asparaginase is predictive of outcome in ETV6/RUNX1+ patients in a cohort undergoing uniform treatment. In the Nordic material, follow-up time is still too short because specific methods for detection of ETV6/RUNX1 gene fusion have only been in common use since 1998. Long follow-up times are of paramount importance because some reports indicate that ETV6/RUNX1+ patients relapse late; the majority of relapses (80%) occur off therapy (median, 3.8 years; range, 1.1-10.5 years).14,15 Awaiting such results, our data might generate new hypotheses. Doxorubicin and etoposide, the 2 drugs found here to be highly active against ETV6/RUNX1+ tumor cells, are both topoisomerase II inhibitors. The reason for this drug specificity is not clear because little is known about the function of the ETV6/RUNX1 fusion protein. Previous studies have shown that various inhibitors of topoisomerase II (also other than cytostatic drugs) may induce rearrangement of genes such as the MLL gene.38 Because the ETV6/RUNX1 gene fusion may also be induced by this mechanism,39 one can speculate that the observed sensitivity of ETV6/RUNX1+ samples to doxorubicin and etoposide reflects a cellular phenotype with aberrant or error-prone double-strand break repair. Modern high throughput techniques, for example, microarrays for gene expression profiling, should now be used to try to elucidate the mechanisms by which ETV6/RUNX1 gene fusion is linked to enhanced cytotoxic effects of doxorubicin, etoposide, and l-asparaginase.
Appendix
The following centers are members of the Nordic Society for Paediatric Haematology and Oncology and participated in the study: Department of Pediatrics, University of Umeå, Sweden; Department of Pediatric Hematology and Oncology, Uppsala University Children's Hospital, Sweden; Department of Pediatric Oncology, Karolinska Institute, Stockholm, Sweden; Department of Pediatric Oncology, University Hospital, Linköping, Sweden; Department of Pediatric Oncology, Queen Silvia Children's Hospital, Göteborg, Sweden; Department of Pediatric Oncology, University Hospital, Lund, Sweden; Department of Pediatrics, Landspitali University Hospital, Reykjavik, Iceland; Department of Pediatrics, Tromsö Hospital, Tromsö, Norway; Department of Pediatrics, St. Olav University Hospital, Trondheim, Norway; Department of Pediatric Oncology, Rikshospitalet, Oslo, Norway; Department of Pediatrics, Ullevål Hospital, Oslo, Norway; Department of Pediatric Oncology, Haukeland University Hospital, Bergen, Norway; Department of Pediatrics, Odense University Hospital, Denmark; Department of Pediatric Oncology, Skejby Hospital, Aarhus, Denmark; Department of Pediatrics, Aalborg Hospital, Aalborg, Denmark; Pediatric Clinic II, Rigshospitalet, Copenhagen, Denmark; Department of Paediatrics and Adolescents, University Central Hospital of Oulu, Finland; Kuopio University Hospital, Finland; University of Helsinki Hospital for Children and Adolescents, Finland; Department of Pediatrics, University Hospital TYKS, Turku, Finland; and Department of Pediatrics, Tampere University Hospital, Finland.
Prepublished online as Blood First Edition Paper, June 24, 2004; DOI 10.1182/blood-2003-12-4426.
Supported by the Lions' Cancer Research Fund, the Swedish Child Cancer Foundation, and the Nordic Cancer Union.
A complete list of the centers participating in the Nordic Society for Paediatric Haematology and Oncology appears in the “Appendix.”
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Ms Anna-Karin Lannergård, Ms Christina Leek, and Ms Lena Lenhammar for skillful technical assistance, and all colleagues in the Nordic Society for Paediatric Haematology and Oncology who provided the patient samples.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal