Abstract
Although human CD56+CD3- natural killer (NK) cells participate in immune responses against microorganisms, their capacity to directly recognize and be activated by pathogens remains unclear. These cells encode members of the Toll-like receptor (TLR) family, involved in innate cell activation on recognition of pathogen-associated molecular patterns (PAMPs). We therefore evaluated whether the 2 bacterial protein PAMPs, the outer membrane protein A from Klebsiella pneumoniae (KpOmpA) and flagellin, which signal through TLR2 and TLR5, respectively, may directly stimulate human NK cells. These proteins induce interferon-γ (IFN-γ) production by NK cells and synergize with interleukin-2 (IL-2) and proinflammatory cytokines in PAMP-induced activation. Similar results were obtained using CD56+CD3+ (NKR-expressing) T cells. NK cells from TLR2-/- mice fail to respond to KpOmpA, demonstrating TLR involvement in this effect. Defensins are antimicrobial peptides expressed mainly by epithelial cells and neutrophils that disrupt the bacterial membrane, leading to pathogen death. We show that NK cells and NKR-expressing T cells constitutively express α-defensins and that KpOmpA and flagellin rapidly induce their release. These data demonstrate for the first time that highly purified NK cells directly recognize and respond to pathogen components through TLRs and evidence defensins as a novel and direct cytotoxic pathway involved in NK cell-mediated protection against microorganisms. (Blood. 2004;104:1778-1783)
Introduction
Natural killer (NK) cells are CD56+CD3- lymphocytes that spontaneously mediate cytotoxicity and are critical in the initial stages of immune defense.1 They kill transformed and infected cells characterized by modified, down-regulated, or absent host major histocompatibility complex (MHC) class 1 molecules.2 During microbial infections, they are activated by cytokines produced by surrounding innate cells and, in turn, synthesize a large panel of cytokines that tailor innate and adaptive immune responses. CD3+ T lymphocytes expressing the NK cell receptor CD56 (NKR-expressing T cells)3 are also involved in resistance to infected and transformed cells.4,5 CD56+ cells are primary producers of interferon-γ (IFN-γ), which has a protective role against many intracellular pathogens.6 Nevertheless, there is no evidence that these cells can in themselves recognize and be directly activated by bacterial components. In fact, NK cells are difficult to study; they are especially difficult to isolate when highly purified, but contaminated cells impede clear identification of NK cell functions. Numerous articles report a role for NK cell defense against bacterial infections, but never in a direct fashion because most purification strategies do not allow the isolation of pure NK cells. Consequently, whether NK cells are stimulated directly7,8 or in response to signals generated by activated bystander cells is still a matter of debate.9,10
Macrophages, dendritic cells, and some epithelial cells recognize pathogen-associated molecular patterns (PAMPs) expressed by microorganisms11 through cell surface receptors (called pattern-recognition receptors [PRRs]) involved in endocytosis or in cell activation (members of the Toll-like receptor [TLR] family).12 Most mammalian TLRs have a key role in initiating inflammatory responses.13 The TLR repertoire and the functional consequences of signaling through TLR have been mainly studied on antigen-presenting cells and epithelial cells.13 Recently, TLR mRNA expression has been reported in B and T lymphocytes9,14 and in NK cells.9 We analyzed their role in NK cell activation. As TLR ligands, we used 2 bacterial protein PAMPs, the outer membrane protein A from Klebsiella pneumoniae (KpOmpA), which activates DCs and macrophages through TLR2,15,16 and Escherichia coli flagellin, a major component of bacterial flagella that induces cytokine production by epithelial cells and monocytes through TLR5.17,18 In response to microbial invasion, macrophages, neutrophils, and epithelial cells produce mediators such as antimicrobial cationic peptides (called defensins) that directly kill or inactivate pathogens such as bacteria. We report that KpOmpA and flagellin act directly, and in synergy with proinflammatory cytokines, in inducing IFN-γ production by CD56+ cells. Moreover, we report that CD56+ cells constitutively express α-defensins19 and demonstrate that α-defensin synthesis and release are promoted after the activation of bacterial PAMP.
Materials and methods
Recombinant proteins
Endotoxin-free recombinant KpOmpA was expressed and purified as described.16 The gene fliC, specifying E coli flagellin, was isolated and cloned in the vector pET24 (Novagen, Madison, MI) in frame with the His6 tag sequence for purification. Recombinant flagellin was purified in apyrogenic conditions; endotoxin levels, determined through Limulus assay, were lower than 0.25 endotoxin units (EUs) per milligram protein. Tetanus toxoid (TT) and bovine serum albumin (BSA) were purchased from SBL Vaccine (Stockholm, Sweden) and Sigma (St Louis, MO), respectively.
Isolation and culture of cells
Murine DX5+ NK cells were isolated from nylon wool nonadherent splenocytes (isolated from C57BL/6 [Harlan, Gannat, France] and C57BL/6 TLR2-/- mice20 ) by positive selection using anti-DX5 monoclonal antibody (mAb)-coated magnetic beads (Miltenyi Biotec, Bergisch Gladbach, Germany). Then, DX5+CD3- cells were sorted using a FACSVantage cytofluorometer (Becton Dickinson, Erembodegem, Belgium) with a fluorescein isothiocyanate (FITC)-labeled rat anti-CD3 mAb (BD PharMingen, San Diego, CA). Purity of the DX5+CD3- NK cells, assessed by fluorescence-activated cell sorter (FACS) analysis using an anti-NK1.1 mAb (BD PharMingen), was higher than 99%. Human peripheral blood mononuclear cells (PBMCs) were isolated from freshly collected blood from healthy volunteers by centrifugation on Ficoll-Hypaque (Amersham Biosciences, Uppsala, Sweden). Neutrophils were isolated by sedimentation on 3% dextran (Amersham Biosciences). CD56+ cells were isolated from PBMCs by positive selection using mAb-coated magnetic beads (Miltenyi Biotech). CD56+CD3- and CD56+CD3+ cells were then sorted by FACS according to their CD3 expression, giving them 99% purity with no detectable contaminating cells. CD3+ T cells were enriched from PBMCs by rosetting with sheep red blood cells (RBCs; bioMérieux, Marcy l'Etoile, France). Naive CD4+CD45RA+ T cells were purified by FACS sorting using FITC-labeled anti-CD4 mAb and phycoerythrin (PE)-labeled anti-CD45RA mAb (both from DakoCytomation, Glostrup, Denmark). Cells were immediately cultured in RPMI 1640 medium supplemented with 10% fetal calf serum, 2 mM l-glutamine, 50 U/mL penicillin, 50 μg/mL streptomycin, 10 mM HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid), and 0.1 mM nonessential amino acids (all from Life Technologies, Cergy Pontoise, France) at a density of 1 to 2 × 106 cells/mL and were stimulated, as indicated, for 3 to 48 hours in 96-well plates with 0.12 to 1 mM KpOmpA,11 flagellin, BSA, or TT or with 10-7 M N-formyl-methionylleucyl-phenylalanine (fMLP) in the absence or the presence of 200 U/mL interleukin-2 (IL-2), 0.04 to 1 ng/mL IL-10, or 2 to 5 ng/mL IL-1β, IL-12, IL-15 (all from R&D Systems, Abingdon, United Kingdom) or IFN-α (PBL Biomedical Laboratories, Piscataway, NJ).
Cell surface labeling
Membrane CD69 expression was evaluated using FACS with FITC-labeled anti-CD69 mAb (Serotec, Kildington, United Kingdom). Results are expressed in mean fluorescence intensity (MFI) after subtracting the MFI obtained with the control antibody.
RT-PCR analysis
Expression of TLR1 to TLR10 and α-defensin 1 to 3 mRNA was analyzed using reverse transcription-polymerase chain reaction (RT-PCR) on FACS-sorted CD56+CD3-, CD56dim and CD56bright NK cells, and CD56+CD3+ T cells. Briefly, total cytoplasmic RNA was extracted using the RNeasy kit (Qiagen, Courtaboeuf, France) following the manufacturer's recommendations. Single-strand cDNA was synthesized using 2 μg total RNA by reverse transcription using an oligo-dT primer (Amersham Biosciences). PCR amplification was performed with an amount of cDNA corresponding to 25 ng starting total RNA (2 minutes at 94°C followed by 30 cycles consisting of 30 seconds at 94°C, 1 minute at 60°C, and 1 minute at 72°C, and then a final extension of 4 minutes at 72°C). Primer sequences have been previously reported for TLR21 and α-defensins.19 RNA integrity and cDNA synthesis were verified by amplifying GAPDH cDNA. Amplified fragments were size separated by electrophoresis and visualized by ethidium bromide.
Quantification of IFN-γ
Cells were incubated for 48 hours with various stimuli, as described above, and IFN-γ was measured in cell-free culture supernatants using enzyme-linked immunosorbent assay (ELISA) (murine IFN-γ, R&D Systems; human IFN-γ, Mabtech, Stockholm, Sweden) according to the manufacturers' recommendations (sensitivity of 2 ng/mL and 5 pg/mL, respectively).
Proliferation assays
NK cells (106 per milliliter) were seeded in quintuplicate in 96-well, flat-bottomed plates and were stimulated as described above for 2 days. Cells were then pulsed during the last 16 hours with 0.25 μCi (9.25 MBq)/well 3H-thymidine (Amersham Biosciences). Radioactive incorporation was measured by standard liquid scintillation counting, and results are expressed in cpm (mean of triplicate values).
Analysis of α-defensin expression and release
For analysis of α-defensins 1 to 3, CD56+CD3- and CD56+CD3+ cells were isolated from PBMCs by negative selection (StemCell Technologies, Meylan, France). FITC-labeled anti-CD3 and PE-labeled anti-CD56 mAbs (Coulter Immunotech, Marseille, France) were used to determine the purity of isolated populations by FACS (more than 95%) and to isolate CD56brightCD3- and CD56dimCD3- cell populations.1 Cells were first fixed for 20 minutes in 2% paraformaldehyde and were then labeled with anti-α-defensin 1 to 3 biotinylated mAbs (HBT, Uden, the Netherlands) at 5 μg/mL for FACS analysis or 20 μg/mL for confocal microscopy, in PBS containing 1 mM HEPES and 0.1% saponin. After 2 washes in PBS, 1 mM HEPES, and 0.01% saponin, NK cells were stained with biotinylated anti-α-defensin mAb followed by Alexa488-labeled streptavidin or streptavidin-Tri-Color (Caltag Laboratories, Burlingame, CA). Cells were then either directly analyzed by cytofluorometry or were adsorbed on poly-l-lysine-coated slides (Sigma), mounted in medium containing 0.2 μg/mL propidium iodide (Molecular Probes, Eugene, OR), and examined using an Axiovert 100M inverted confocal microscope (Zeiss, Oberkochen, Germany) using a C-Apochromat 40 × objective lens. Images were collected using an HAD 3CCD color video camera (Sony, Paris, France) and analyzed using Laser Scanning Microscope 510 software, version 2.02 (Zeiss). For α-defensin release measurements, supernatants of 2-hour-stimulated cells were quantified using a commercial kit (HBT) according to the manufacturer's recommendations.
Results
KpOmpA and flagellin directly activate human NK cells
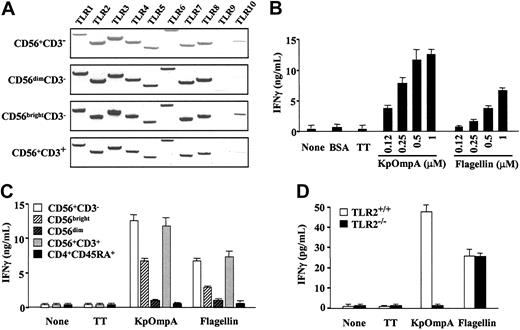
To analyze whether NK cells respond to protein PAMPs, we analyzed TLR1-10 mRNA expression using RT-PCR. In our experimental conditions, and using previously described primers,21 results showed that CD56+CD3- and CD56+CD3+ cells constitutively express TLR1-8 mRNA, TLR2, and TLR3 mRNA at the highest levels (Figure 1A).9,14 In contrast, TLR9 and TLR10 mRNA expression was low or undetectable in CD56+ cells (Figure 1A). Moreover, CD56bright and CD56dim cells exhibit similar profiles of TLR mRNA, though relative levels appeared higher in CD56bright than in CD56dim cells. Therefore, we analyzed the effects of KpOmpA and flagellin, which signal through TLR2 and TLR5, respectively, on CD56+ cell functions. Because previous studies demonstrated that the role of NK cells in resistance against many pathogens is principally mediated by IFN-γ production,6 we tested whether KpOmpA and flagellin may modulate IFN-γ production. KpOmpA and flagellin induce dose- and time-dependent production of IFN-γ by human CD56+CD3- NK cells, significant after 8 hours (Figure 1B) and maximal at 48 hours (data not shown). In contrast, NK cells exposed to the control proteins BSA and TT do not produce detectable levels of IFN-γ (Figure 1B). Two subsets of human NK cells have been described: the CD56bright subset produced cytokines, and the CD56dim subset had cytotoxic capacity against target cells once stimulated.1 Although the CD56bright subpopulation is mainly responsible for PAMP-mediated IFN-γ production, optimal IFN-γ production requires the presence of both subsets (Figure 1C).
KpOmpA and flagellin directly activate human CD56+ cells. (A) TLR1-10 mRNA expression was evaluated using RT-PCR in highly purified human CD56+CD3-NK cells, CD56dim and CD56bright NK cells, and CD56+CD3+ cells. Results are representative of 1 of 2 donors. (B) NK cells were or were not stimulated with 0.12 to 1 μM KpOmpA or flagellin or with 1 mM BSA or TT as negative controls. IFN-γ was quantified in the 48-hour supernatants using ELISA. Results are expressed in ng/mL as mean ± SD (n = 3). (C) Total CD56+CD3-NK cells, NK cell subpopulations CD56bright and CD56dim, IL-2-cultured CD56+CD3+ cells, and naive CD4+CD45RA+ T cells were or were not stimulated with 1 μM KpOmpA, flagellin, or TT. IFN-γ was quantified in the 48-hour supernatants using ELISA. Results are expressed in ng/mL as mean ± SD (n = 3). (D) DX5+CD3-NK cells from wild-type C57BL/6 and TLR2-/- C57BL/6 mice were or were not stimulated with 1 mM KpOmpA, flagellin, or TT in the presence of 200 U/mL IL-2. IFN-γ was quantified in the 48-hour supernatants using ELISA. Results are expressed in pg/mL as mean ± SD (n = 3).
KpOmpA and flagellin directly activate human CD56+ cells. (A) TLR1-10 mRNA expression was evaluated using RT-PCR in highly purified human CD56+CD3-NK cells, CD56dim and CD56bright NK cells, and CD56+CD3+ cells. Results are representative of 1 of 2 donors. (B) NK cells were or were not stimulated with 0.12 to 1 μM KpOmpA or flagellin or with 1 mM BSA or TT as negative controls. IFN-γ was quantified in the 48-hour supernatants using ELISA. Results are expressed in ng/mL as mean ± SD (n = 3). (C) Total CD56+CD3-NK cells, NK cell subpopulations CD56bright and CD56dim, IL-2-cultured CD56+CD3+ cells, and naive CD4+CD45RA+ T cells were or were not stimulated with 1 μM KpOmpA, flagellin, or TT. IFN-γ was quantified in the 48-hour supernatants using ELISA. Results are expressed in ng/mL as mean ± SD (n = 3). (D) DX5+CD3-NK cells from wild-type C57BL/6 and TLR2-/- C57BL/6 mice were or were not stimulated with 1 mM KpOmpA, flagellin, or TT in the presence of 200 U/mL IL-2. IFN-γ was quantified in the 48-hour supernatants using ELISA. Results are expressed in pg/mL as mean ± SD (n = 3).
Because KpOmpA activates macrophages through TLR2,16 we tested the involvement of TLR2 in KpOmpA-mediated NK cell activation. Although highly purified DX5+CD3- NK cells from TLR2+/+ and TLR2-/- produce comparable levels of IFN-γ in response to flagellin, NK cells from TLR2-/- mice fail to produce IFN-γ in response to KpOmpA, even in the presence of IL-2 (Figure 1D). These results show that KpOmpA-mediated activation of NK cells is dependent on functional TLR2. These data, added to the observation that polymixin B does not affect KpOmpA- or flagellin-induced IFN-γ production (data not shown), demonstrate that the effect of these protein PAMPs on NK cells is not mediated through contaminating endotoxins.16
CD3+CD56+ cells (Figure 1A) and peripheral blood T cells9,14 also express TLR2 and TLR5 mRNA. We thus evaluated their capacity to be stimulated by KpOmpA and flagellin. Results show that CD56+CD3+ cells produce IFN-γ in response to KpOmpA and flagellin, though to a lower extent (10-fold) than CD56+CD3- cells (data not shown). When CD56+CD3+ T cells were first maintained in culture for 24 hours with suboptimal doses of IL-2 or IL-12 (as described above), they produce levels of IFN-γ comparable to those of CD56+CD3- NK cells in response to KpOmpA and flagellin (Figure 1C). In contrast, naive CD4+CD45RA+ T cells do not respond to KpOmpA and flagellin (Figure 1C). Together these data show that highly purified NK cells recognize and are directly activated by KpOmpA and flagellin.
KpOmpA and flagellin synergize with cytokines in activating NK cells
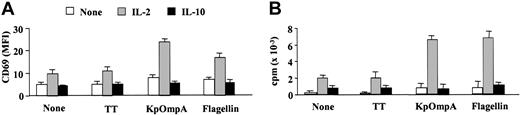
At the site of microorganism entry, innate cells (macrophages and epithelial cells) produce cytokines (eg, IL-1β, IL-12, IL-15, or IFN-α) that stimulate NK cells.4,6 Suboptimal concentrations of these cytokines (as mentioned in “Materials and methods”) induce low or undetectable levels of IFN-γ by CD56+CD3- NK cells (Figure 2A). However, they synergize with flagellin and KpOmpA in inducing IFN-γ, up to 5- and 7-fold increases, respectively (Figure 2A). Synergism is also observed with IL-2 (Figure 2B), a potent NK cell activation factor.6 The effect of the cytokines is dose dependent for all the cytokines tested (Figure 2B and data not shown). As controls, TT (Figure 2A-B) and BSA (data not shown) fail to stimulate NK cells, even in the presence of these cytokines. In contrast, IL-10, a late cytokine produced by T cells and macrophages that decrease cytokine production by NK cells,22 prevents KpOmpA- and flagellin-induced IFN-γ production (Figure 2C). As for the entire CD56+CD3- NK cell population, PAMPs activate the CD56bright subset in synergism with IL-2 for IFN-γ synthesis; conversely, IL-10 abrogates IFN-γ production (Figure 2D). PAMPs, without or with IL-2, do not up-regulate IFN-γ production by the CD56dim subset (Figure 2D). These results are in agreement with previous observations suggesting that this subpopulation is a poor cytokine producer.1 Results similar to those of the CD56bright subset are obtained with NKR-expressing T cells (Figure 2E). In parallel, in the presence of a suboptimal concentration of IL-2, KpOmpA and flagellin also up-regulate the expression of the activation marker CD69 on NK cells (Figure 3A) and induce their proliferation (Figure 3B) as opposed to IL-10, which prevents both effects (Figure 3A-B). These data suggest that cytokines, concomitantly produced by surrounding cells on bacterial invasion, tightly control NK cell sensitivity to PAMPs.
KpOmpA and flagellin synergize with cytokines in inducing IFN-γ production by NK cells. Freshly isolated CD56+ (A-C) and CD56dim and CD56bright NK cells (D) were unstimulated or were stimulated with 1 μM KpOmpA, flagellin, or TT in the absence or in the presence of 5 ng/mL IL-1β, IL-12, or IFN-α, 2 ng/mL IL-15, or 200 U/mL IL-2 (A); or 8 to 200 U/mL IL-2 (B), 0.04 to 1 pg/mL IL-10 (C), or 200 U/mL IL-2 or 1 pg/mL IL-10 (D). Results obtained from freshly isolated CD56+CD3+ cells are also presented (E). After 48-hour incubation, IFN-γ levels were determined using ELISA, and results are expressed in ng/mL as mean ± SD (n = 5).
KpOmpA and flagellin synergize with cytokines in inducing IFN-γ production by NK cells. Freshly isolated CD56+ (A-C) and CD56dim and CD56bright NK cells (D) were unstimulated or were stimulated with 1 μM KpOmpA, flagellin, or TT in the absence or in the presence of 5 ng/mL IL-1β, IL-12, or IFN-α, 2 ng/mL IL-15, or 200 U/mL IL-2 (A); or 8 to 200 U/mL IL-2 (B), 0.04 to 1 pg/mL IL-10 (C), or 200 U/mL IL-2 or 1 pg/mL IL-10 (D). Results obtained from freshly isolated CD56+CD3+ cells are also presented (E). After 48-hour incubation, IFN-γ levels were determined using ELISA, and results are expressed in ng/mL as mean ± SD (n = 5).
KpOmpA and flagellin induce CD69 expression and proliferation of NK cells. Freshly isolated CD56+CD3-NK cells were unstimulated or were stimulated with 1 μM KpOmpA, flagellin, or TT in the absence or in the presence of 200 U/mL IL-2 or 1 ng/mL IL-10. CD69 expression (A) and proliferation (B) were determined as described in “Materials and methods.” Results are expressed as mean ± SD (n = 5).
KpOmpA and flagellin induce CD69 expression and proliferation of NK cells. Freshly isolated CD56+CD3-NK cells were unstimulated or were stimulated with 1 μM KpOmpA, flagellin, or TT in the absence or in the presence of 200 U/mL IL-2 or 1 ng/mL IL-10. CD69 expression (A) and proliferation (B) were determined as described in “Materials and methods.” Results are expressed as mean ± SD (n = 5).
KpOmpA and flagellin directly trigger α-defensin synthesis and release by NK cells
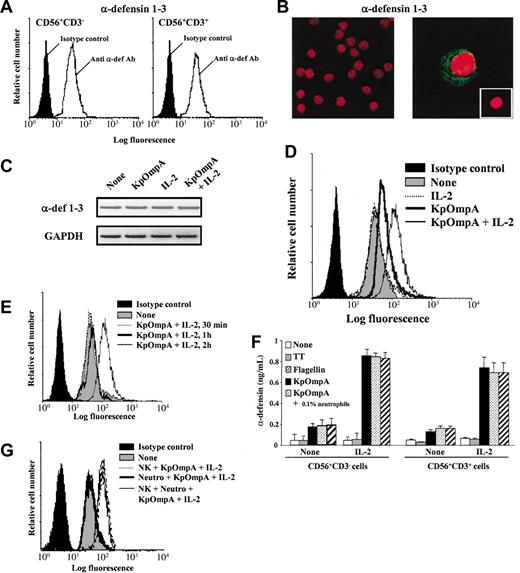
The bactericidal peptide α-defensins have an important role in innate host responses against microbes because they directly kill invaders by disrupting their membrane.23 FACS analysis on permeabilized cells shows that freshly isolated and highly purified CD56+CD3- and CD56+CD3+ cells constitutively express α-defensins 1 to 3 (Figure 4A). Confocal microscopy (Figure 4B) and RT-PCR analysis (Figure 4C) confirm the constitutive expression of α-defensins 1 to 3 by NK cells; similar observations are made with CD56+CD3+ cells (data not shown). These data are in agreement with previous studies reporting α-defensin 1 mRNA expression in CD56+ cells24 and CD56-α-defensin double staining in PBMCs.18 As for α-defensins retrieved in neutrophils,25 confocal analysis shows that α-defensins 1 to 3 are stocked in granules agglomerated in the vicinity of the cellular membrane (Figure 4B). These observations led us to hypothesize that CD56+ cells may respond to PAMPs by rapidly synthesizing and releasing antimicrobial peptides as a new form of toxicity. FACS analysis of PAMPs-activated NK cells pretreated with Brefeldin A shows an up-regulation of intracellular α-defensin MFI values (Figure 4D). This effect is potentiated by IL-2 (Figure 4D) and is significant as early as 2 hours after stimulation (Figure 4E). Identical results are obtained with CD56dim and CD56bright NK cell subsets and with CD56+CD3+ cells (data not shown). In addition, results show that 2-hour PAMP-activated CD56+CD3- and CD56+CD3+ cells readily release α-defensins and that this effect is potentiated by exogenous IL-2 (Figure 4F). It is important to note that α-defensin release is easily provoked by cell manipulation, underlying the importance of control proteins for each experiment. Interestingly, the time-dependent up-regulation of intracellular α-defensins is not associated with modification of α-defensin 1 to 3 mRNA expression, as assessed by RT-PCR (Figure 4C), as opposed to neutrophils.25 This observation suggests that the de novo synthesis responsible for intracellular α-defensin level up-regulation in CD56+ cells is independent of mRNA neosynthesis.
KpOmpA and flagellin trigger α-defensin synthesis and release by NK cells. Intracellular α-defensin expression was analyzed using (A) FACS in freshly and highly purified human NK cells and CD56+CD3+ T cells and using (B) confocal microscopy in NK cells (magnification: × 300 and × 1000 in left and right panels, respectively). (B, inset) Cells labeled with isotype control mAb (magnification: × 600). (C) α-defensin mRNA expression in NK cells either unstimulated or stimulated for 2 hours with KpOmpA in the absence or in the presence of IL-2 was analyzed using RT-PCR. RNA integrity and cDNA synthesis were verified by amplifying GAPDH cDNA. (D-E,G) Human CD56+CD3-NK cells were unstimulated or stimulated with 1 μM KpOmpA, flagellin, or TT in the absence or in the presence of 200 U/mL IL-2 or of 0.1% contaminating neutrophils, when indicated. After 2 hours, cells were stained for intracellular α-defensin expression analysis by cytometry. (F) α-Defensin released by NK cells and CD56+CD3+ cells activated by KpOmpA or flagellin in the absence or presence of IL-2 was quantified in cell culture supernatants using ELISA. Data are expressed in MFI (A,D,E,G) and in ng/mL, mean ± SD, n = 3 (F).
KpOmpA and flagellin trigger α-defensin synthesis and release by NK cells. Intracellular α-defensin expression was analyzed using (A) FACS in freshly and highly purified human NK cells and CD56+CD3+ T cells and using (B) confocal microscopy in NK cells (magnification: × 300 and × 1000 in left and right panels, respectively). (B, inset) Cells labeled with isotype control mAb (magnification: × 600). (C) α-defensin mRNA expression in NK cells either unstimulated or stimulated for 2 hours with KpOmpA in the absence or in the presence of IL-2 was analyzed using RT-PCR. RNA integrity and cDNA synthesis were verified by amplifying GAPDH cDNA. (D-E,G) Human CD56+CD3-NK cells were unstimulated or stimulated with 1 μM KpOmpA, flagellin, or TT in the absence or in the presence of 200 U/mL IL-2 or of 0.1% contaminating neutrophils, when indicated. After 2 hours, cells were stained for intracellular α-defensin expression analysis by cytometry. (F) α-Defensin released by NK cells and CD56+CD3+ cells activated by KpOmpA or flagellin in the absence or presence of IL-2 was quantified in cell culture supernatants using ELISA. Data are expressed in MFI (A,D,E,G) and in ng/mL, mean ± SD, n = 3 (F).
Because neutrophils produce high levels of α-defensins,25 it was important to exclude that, in our experimental conditions, α-defensin release resulted from contaminating neutrophils. Confocal microscopy (Figure 4B left) shows the absence of neutrophils in NK cell preparations. Moreover, FACS analysis of intracellular α-defensin staining was performed on cells gated first on forward scatter/side scatter (FSC/SSC) parameters, allowing the exclusion of granulocytes, and then on CD56+ cells. These data were confirmed by functional assays. Neutrophils do not produce α-defensin on stimulation with KpOmpA or flagellin, even in the presence of IL-2 (data not shown). After the addition of 0.1% neutrophils, NK cells were stimulated with KpOmpA or flagellin in the absence or the presence of IL-2. The presence of neutrophils does not lead to the up-regulation of α-defensin synthesis (Figure 4G) or release (Figure 4F). Taken together, these results demonstrate the specificity of analysis for the targeted cells.
Discussion
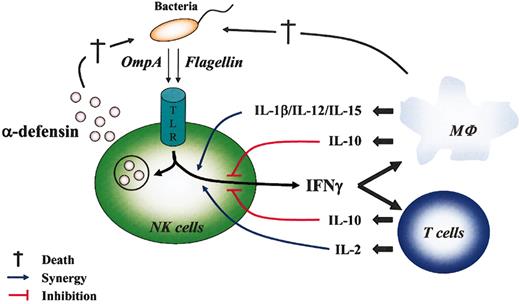
NK cells respond to cytokines synthesized by surrounding PAMP-activated innate cells,26,27 and their depletion causes susceptibility to several infections.6 Many studies suggest the importance of these cells in resistance to pathogen invasions. Nevertheless, whether NK cells directly recognize, and are subsequently activated by, the microorganism itself is still unclear. Constitutive expression of TLR mRNA by CD56+ cells suggests that they can be directly activated by bacterial PAMPs as foreign antigens. We report that the protein PAMPs KpOmpA and flagellin act alone and in synergy with different cytokines to stimulate some NK cell functions (Figure 5). This study is, to our knowledge, the first demonstration that highly purified CD56+ cells directly recognize bacterial proteins and are consequently directly stimulated in the absence of other cell types.
Direct and indirect NK cell responses during bacterial invasion. Direct NK cell activation by bacterial PAMPs was mediated through TLR and led to the secretion of cytokines (IFN-γ) and α-defensins that contribute, respectively, to activate surrounding cells (eg, macrophages, T lymphocytes) and to destroy (†) pathogens. In turn, surrounding cells that could be directly activated by PAMPs or environmental cytokines produced cytokines that synergized (IL-1β, IL-2, IL-12, IL-15; blue line) or inhibited (IL-10; red line) PAMP-mediated activation of NK cell functions.
Direct and indirect NK cell responses during bacterial invasion. Direct NK cell activation by bacterial PAMPs was mediated through TLR and led to the secretion of cytokines (IFN-γ) and α-defensins that contribute, respectively, to activate surrounding cells (eg, macrophages, T lymphocytes) and to destroy (†) pathogens. In turn, surrounding cells that could be directly activated by PAMPs or environmental cytokines produced cytokines that synergized (IL-1β, IL-2, IL-12, IL-15; blue line) or inhibited (IL-10; red line) PAMP-mediated activation of NK cell functions.
In this study, we demonstrate that the 2 bacteria-derived protein PAMPs, KpOmpA and flagellin, directly activate CD56+ cells. Most of the studies analyzing the activation of NK cells by PAMPs were focused on CpG oligonucleotides.7,9 More recently, 2 parasite-derived molecules, Leishmania lipophosphoglycan8 and LeIF,10 were shown to activate NK cells. Leishmania lipophosphoglycan activates NK cells through TLR2,8 but the activation pathway used by the protein LeIF is unknown.10 In this study, we demonstrate that KpOmpA and flagellin directly activate NK cells. Similarly, Leishmania lipophosphoglycan directly activates NK cells.8 In contrast, studies on CpG oligonucleotides and LeIF report that the PAMP activation of NK cells is indirect in that it is mediated by soluble mediators produced mainly by PAMP-activated antigen-presenting cells (APCs).9,10 These data suggest that the sensitivity of NK cells to PAMPs is dependent on their nature. Moreover, exogenous cytokines regulated KpOmpA- and flagellin-mediated activation of NK cells. In a similar manner, IL-12 and IL-15 synergize with LeIF to induce IFN-γ production.10 Collectively, these data suggest that priming with cytokines is required to render NK cells responsive to some PAMPs and that NK cell activation by PAMPs is tightly regulated by cytokines.
NK cells have an impressive arsenal of cytotoxic components permitting them to directly lyse infected cells, first by releasing their cytoplasmic granules containing, notably, perforin and granzymes and second by a nonsecretory pathway based on CD95L/CD95 interaction. They also contribute indirectly to pathogen elimination by secreting cytokines and chemokines, which activate innate cells and attract them to the infection site, respectively (Figure 5). The results presented here demonstrate clearly, despite the potentially contaminating presence of neutrophils, that CD56+ cell cytotoxic activity seems to be more complex than originally thought. In fact, by releasing preformed α-defensins upon contact with bacteria, these cells may participate in in vivo bacterial cytolysis (Figure 5). Because α-defensin release is rapid and highly sensitive, this fourth mechanism may permit CD56+ cells to act as soon as the bacteria are encountered, well before they have time to infect surrounding cells.27 Moreover, it was established that α-defensins exert chemotactic activity for monocytes, T lymphocytes, immature dendritic cells, and complement factors, thereby facilitating the rapid development of the immune response in addition to eliminating bacterial pathogens.23
Thus, expression of functional TLR by NK cells could allow them to develop specific and protective responses (cytokine synthesis, cytotoxic and bactericidal activities) to pathogen invasion. This hypothesis contributes to previous studies showing that CD56+ cell depletion has serious consequences in many infection models.1,6 Finally, in agreement with the hypothesis that NK cells efficiently adapt immune responses to the invader, it was demonstrated that NK cells directly recognize m157, an MHC 1-like glycoprotein encoded by murine cytomegalovirus (MCMV), allowing them to establish a resistant phenotype against the corresponding virus infection in vivo.28,29 In conclusion, these results highlight a new mechanism of direct pathogen recognition by NK cells, thereby enlarging their functional importance as first direct sensors and effectors of bacterial presence.
Prepublished online as Blood First Edition Paper, May 27, 2004; DOI 10.1182/blood-2003-08-2820.
Anick Chalifour is supported by a postdoctoral fellowship of the Le Fonds Québécois de la Recherche sur la Nature et les Technologies.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Nathalie Herbault-Bruguiere, Peggy Charbonnier, Béatrice Cavoret, and Sandrine Lauthier for their expert technical help. We also thank Dr Nathalie Corvaia for constant support and critical reading of the manuscript.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal