Abstract
Activating FLT3 mutations are the most common genetic aberrations in acute myeloid leukemia (AML), resulting in the constitutive activation of this receptor tyrosine kinase (RTK), but such mutations are rarely found in acute lymphoblastic leukemia (ALL). Here we describe a unique subset of de novo adult T-cell ALL (T-ALL) cases that coexpress CD117/KIT and cytoplasmic CD3 (CD117/KIT+ ALL). Activating mutations in the FLT3 RTK gene were found in each of 3 CD117/KIT+ cases that were analyzed, but not in 52 other adult T-ALL samples from the same series that lacked CD117/KIT expression. Our results indicate the need for clinical trials to test the efficacy of drugs that inhibit the FLT3 RTK in this subset of patients with T-ALL. (Blood. 2004;104:558-560)
Introduction
Early hematopoietic stem and progenitor cells express CD117/KIT, the stem cell factor receptor.1 Detection of CD117/KIT expression by lymphoblasts in acute lymphoblastic leukemia (ALL) is rare and is restricted to T-lineage disease.1-3 During normal lymphopoiesis, CD117/KIT is expressed by a fraction of CD34+CD3-CD4-CD8- (triple-negative) thymocytes, which have not yet rearranged their T-cell receptor (TCR) genes.4 In these thymocytes,5 as in normal bone marrow progenitors,6 expression of CD117/KIT coincides with that of CD135, the FLT3 receptor tyrosine kinase (RTK) that is activated by the FLT3 ligand. FLT3 and CD117/KIT share extensive structural homology.7 Treatment with FLT3 ligand and stem cell factor elicits in vitro differentiation of CD117/KIT+CD135/FLT3+ T-cell progenitors toward the myeloid lineage, whereas stem cell factor together with interleukin-7 induces development toward a mature T-cell phenotype.5
Activating mutations of FLT3 are the most common genetic abnormality found in acute myeloid leukemia (AML) and encode a protein with constitutive RTK activity in the absence of ligand.6 While the majority of FLT3 mutations are internal tandem duplications (ITDs) located in the juxtamembrane domain of the receptor, some leukemias instead harbor point mutations in the activation loop of the kinase domain. By contrast, FLT3 is rarely mutated in leukemic lymphoblasts,6,8-10 with the exception of the FLT3 point mutations that are found in B-lineage ALLs containing mixed-lineage leukemia (MLL) gene rearrangements.11
In this study, we asked whether the subset of early T-lineage ALL cases that express CD117/KIT also express FLT3 and harbor activating mutations of this gene. By analyzing leukemic cells from 55 adult patients with T-cell ALL (T-ALL; Eastern Cooperative Oncology Group [ECOG] trial no. E2993), we identified 3 cases with high CD117/KIT expression, and demonstrated activating FLT3 mutations in each of them—a finding with therapeutic significance in view of the availability of new inhibitors of FLT3 tyrosine kinase activity.
Study design
Patient samples
Bone marrow or peripheral blood from 449 patients in ECOG trial no. E2993 was centrally immunophenotyped using multiparameter flow cytometry, as reported.1,12 Consent for collection and testing of samples was obtained at study entry. B-lineage ALL was diagnosed in 380 cases; T-lineage ALL in 69 cases. Sufficient material for reverse transcriptase-polymerase chain reaction (RT-PCR) studies was available from 341 B-precursor ALLs and 55 T-lineage T-ALLs. Morphology and cytogenetics were reviewed centrally by ECOG core facilities. To investigate the association of CD117/KIT expression with T-lineage ALL, the Fisher exact test was run.
All patients reported in this paper were accrued to ECOG phase 3 trial E2993. Until February 1998, patients accrued to E2993 were simultaneously entered on laboratory protocol E1485, which included written consent for immunophenotyping, cytogenetics, and selected molecular studies. After closure of this laboratory protocol, written consent for the collection and testing of bone marrow and peripheral blood was sought from each patient at the time he or she consented to participate in the clincal trial. E1485 and E2993 were reviewed and approved by each of the participating institutions' review boards. A.T.L. obtained a separate IRB approval for his study on transcription factors in T-ALL.
Qualitative and quantitative RT-PCR
Total RNA was extracted13 from mononuclear cells and tested by RT-PCR for BCR-ABL, MLL-AF4, E2A-PBX1, and TEL-AML1 fusion transcripts.14 Real-time RT-PCR analysis of the expression levels of the T-cell oncogenes, TAL1, LYL1, HOX11, HOX11L2, LMO1, and LMO2, and the control (glyceraldehyde-3-phosphate dehydrogenase) was performed as described.15 Samples were considered positive for a given oncogene if they expressed more than 3 × 105(LYL1), more than 104 (LMO1, TAL1, HOX11, HOX11L2, BHLHB1), or more than 105 (LMO2) mRNA copies per 100 ng total RNA.15
The regions of the FLT3 mRNA sequence encoding the juxtamembrane or the activation loop domains were amplified by RT-PCR.16 For ITD analysis, PCR products larger than wild-type FLT3 were subcloned into the PUC18 plasmid vector and sequenced. To detect mutations in the FLT3 activation loop domain, PCR products corresponding to this region were digested with EcoRV; digestion is disrupted by the most frequent point mutations in this region. DNA resistant to digestion was subcloned into the PUC18 plasmid vector and sequenced.
Results and discussion
Among 449 adult patients with de novo ALL, we identified 3 men (ages 22, 36, and 55 years) with CD117/KIT expression on more than 90% of T lymphoblasts. This corresponded to 3 of 69 T-ALL or 4% (90% confidence interval: 1%-11%) and reflected a strong association of CD117/KIT expression with T-lineage compared with B-lineage disease (0% CD117/KIT+ cases; P = .005). Leukemic blasts accounted for approximately 95% of mononuclear cells in every patient. By flow cytometry, all blasts were classified as T lymphoblasts based on intracytoplasmic CD3 expression. The remaining antigen profile was identical among the 3 CD117/KIT+ cases and was not found in any of the other E2993 patients: surface CD3-, CD34+, CD62L+, CD56-, CD2+, CD7+, CD1a-, CD5-, CD4/CD8-, TdT+, expressing one myeloid antigen, CD13. This immunophenotype fits into the most immature category of TALL,17 resembling multipotent thymic precursors.
Morphology was exclusively lymphoid in case 2, whereas cases 1 and 3 were predominantly lymphoid by the French-American-British (FAB) classification, but contained 2% to 10% M1 blasts with occasional Auer rods. By flow cytometric analysis, myeloperoxidase and intracytoplasmic CD3 were coexpressed in 7%, 0%, and 6% of intracytoplasmic CD3+ blast cells in cases 1, 2, and 3, respectively. Myeloperoxidase and intracytoplasmic CD3 are expressed by cells of the myeloid and the T-lymphoid lineage, respectively.18 Because of the predominance of T-lymphoid characteristics, we classified the immunophenotype of these leukemias as T-lymphoid. According to the World Health Organization (WHO) classification, cases 1 and 3 would fit the category of biphenotypic acute leukemia.18 The presence of CD13 as the sole surface myeloid antigen in CD117/KIT+ T-ALL has been previously noted.19,20
We have shown that the oncogenic transcription factor genes, HOX11, HOX11L2, TAL1, LYL1, LMO2, and MLL-ENL identify discrete molecular groups of T-ALL.15,21 The 3 CD117/KIT+ ALL cases shared high expression levels of LYL1 and LMO2 oncogenes. This agreed with our observation in pediatric T-ALL that LYL1 and LMO2 together are associated with an early CD34+ thymocyte phenotype,15 comparable to that of our adult CD117/KIT+ patients. As expected, the CD117/KIT+ cases were negative for leukemia transcripts typical for B-lineage ALL (BCR-ABL, MLL-AF4, E2APBX1, TEL-AML1).
Cytogenetic analysis of the CD117/KIT+ ALL cases showed the presence of a der(16)t(1;16)(q12;q11.2) in case 1, a t(7;14)(q22; q32) in case 2, and a complex karyotype including t(7;11)(p22;q23) and an add(11)(q23) in case 3, 2 abnormalities potentially involving the MLL gene. B-lineage11 but not T-lineage ALL with MLL rearrangements21 has been associated with high levels of FLT3 expression and the presence of FLT3 mutations.
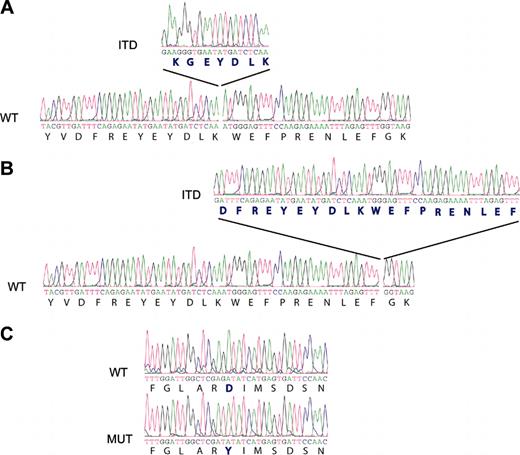
Antibody staining for FLT3 (CD135) indicated strong cell-surface expression of this protein in each of the 3 CD117/KIT+ ALLs. To determine whether FLT3 was mutated in CD117/KIT+ T-ALL, we performed RT-PCR analysis looking for abnormalities in the FLT3 juxtamembrane region. FLT3 transcripts were readily detected and showed the FLT3-ITD abnormality in cases 1 and 2. Sequence analysis of RT-PCR products confirmed the presence of an insertion of 21 base pairs encoding 7 amino acids in the juxtamembrane region of FLT3 in case 1 and an insertion of 60 base pairs resulting in 20 extra amino acids in case 2. Sequence analysis of the activation-loop coding region of FLT3 showed that case 3 harbored a point mutation in this region, resulting in a D835Y amino acid change, which is the most frequently reported activating point mutation of FLT322 (Figure 1).
FLT3 tyrosine kinase activating mutations in CD117/KIT+ T-ALL samples. RT-PCR analysis of the juxtamembrane region suggested length mutations (ITDs) in cases 1 (A) and 2 (B). Sequence analysis of PCR products demonstrated the insertion of 21 (case 1) and 60 (case 2) base pairs in positions 1863 and 1894 of the juxtamembrane region of the FLT3 kinase gene, respectively. In case 3, the D835Y mutation (MUT) in the activation loop of FLT3 was detected when compared with the wild-type (WT) allele. (C) Mutation position annotation is based in the FLT3 reference sequence NM_004119.
FLT3 tyrosine kinase activating mutations in CD117/KIT+ T-ALL samples. RT-PCR analysis of the juxtamembrane region suggested length mutations (ITDs) in cases 1 (A) and 2 (B). Sequence analysis of PCR products demonstrated the insertion of 21 (case 1) and 60 (case 2) base pairs in positions 1863 and 1894 of the juxtamembrane region of the FLT3 kinase gene, respectively. In case 3, the D835Y mutation (MUT) in the activation loop of FLT3 was detected when compared with the wild-type (WT) allele. (C) Mutation position annotation is based in the FLT3 reference sequence NM_004119.
The mutant-to-wild-type ratio was 1.0 in cases 1 and 3, whereas the mutant level approached 75% in case 2, suggesting that in every case the FLT3 mutation was present in all blast cells, and that in case 2 heterozygosity was partially lost.22 FLT3 gene mutations were not detected in any of the CD117/KIT- T-lineage ALLs analyzed. In the majority of these cases, CD135 expression was weak or undetectable. The relationship between FLT3 mutation status and CD135 expression has not yet been established.
Thus, our results suggest that CD117/KIT expression in T-ALL lymphoblasts identifies a subset of patients in whom FLT3 gene mutations play an essential part of the multistep mutational pathway to oncogenesis. To date, FLT3 inhibitors have been considered for clinical trials exclusively in AML, due to the high frequency of activating FLT3 mutations in this disease.6,22 Based on our findings, we suggest that the analysis of CD117/KIT expression should be included in the immunophenotypic workup of all patients with T-ALL who relapse on current therapy. Importantly, CD117/KIT+ patients should be tested for FLT3 activating mutations so that these patients can be enrolled in trials to determine the efficacy of FLT3 inhibitors as single agents in refractory patients with this disease.
Prepublished online as Blood First Edition Paper, March 25, 2004; DOI 10.1182/blood-2004-01-0168.
Supported by the National Cancer Institute (NCI); Department of Health and Human Services (DHHS) grant nos. CA21115, CA23318, CA11083, CA68484, CA56771; and by the A.L. Levine Family Foundation.
E.P. and A.A.F. contributed equally to this work.
An Inside Blood analysis of this article appears in the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The authors wish to thank John-Paul Hezel and the staff of ECOG's Leukemia Translational Studies Laboratory for technical and editorial assistance. The continued commitment of ECOG physicians to submitting specimens for laboratory studies is highly appreciated.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal