Abstract
Postnatal thymic involution occurs progressively throughout the first 3 decades of life. It predominantly affects T-cell receptor (TCR) αβ-lineage precursors, with a consequent proportional increase in multipotent thymic precursors. We show that T-acute lymphoblastic leukemias (T-ALLs) demonstrate a similar shift with age from predominantly TCR expressing to an immature (IM0/δ/γ) stage of maturation arrest. Half demonstrate HOX11, HOX11L2, SIL-TAL1, or CALM-AF10 deregulation, with each being associated with a specific, age-independent stage of maturation arrest. HOX11 and SIL-TAL represent αβ-lineage oncogenes, whereas HOX11L2 expression identifies an intermediate αβ/γδ-lineage stage of maturation arrest. In keeping with preferential αβ-lineage involution, the incidence of SIL-TAL1 and HOX11L2 deregulation decreased with age. In contrast, HOX11 deregulation became more frequent, suggesting longer latency. TAL1/LMO1 deregulation is more frequent in αβ-lineage T-ALL, when it is predominantly due to SIL-TAL1 rearrangements in children but to currently unknown mechanisms in adolescents and adults. LMO2 was more frequently coexpressed with LYL1, predominantly in IM0/δ/γ adult cases, than with TAL1. These age-related changes in phenotype and oncogenic pathways probably reflect progressive changes in the thymic population at risk of malignant transformation.
Introduction
Human T lymphocytes are derived from pluripotent hemopoietic progenitors that migrate throughout life from the bone marrow to the thymus, where the majority of T-cell development occurs. Progressive thymic atrophy during the first 2 to 3 decades of life leads to a reduction of thymic mass and, to a lesser extent, function.1,2 The earliest thymic progenitor corresponds to a minor (< 1% of thymocytes) sCD3-, CD4/CD8 double-negative (DN) population that does not alter with age.3-6 In the neonatal murine thymus, the majority of lymphocytes belong to the αβ lineage; successful T-cell receptor β (TCRβ) rearrangement in a DN precursor in the presence of pTα allows expression of the pre-TCR in association with CD3, progression to the CD4/8 double-positive (DP) stage, and massive thymocyte expansion, a process known as β selection7 ; this population diminishes markedly with age, with a consequent increase in the proportion of DN cells.
T-acute lymphoblastic leukemias (T-ALLs) correspond to a heterogeneous group of acute leukemias arrested at various stages of lymphoid development. They account for 10% to 15% of pediatric and 25% of adult ALLs, with a relatively constant incidence up to the third decade.8-10 Recognized T-ALL oncogenic pathways include transcriptional deregulation by juxtapositioning to one of the TCR loci, resulting in transcriptional deregulation of genes such as HOX11/TLX1, LMO2, LMO1, LYL1, and TAL1/SCL, each of which is present in less than 10% of cases.11-22 More common genetic defects in T-ALL are submicroscopic deletions of p16/INK4 in 40% to 80%23,24 or SIL-TAL1 in 10% to 25%.25-27 Although the latter leads to a fusion transcript, there is no SIL-TAL1 chimeric protein, only deregulated TAL1 expression.28 Cryptic translocations are also recognized, with the most common being the t(5;14)(q35;q32), leading to overexpression of HOX11L2/TLX3, an orphan homeobox factor very similar to HOX11, in 25% to 30% of pediatric and 13% of adult T-ALLs.22,29-31 HOX11 deregulation in the t(10;14)(q24;q11) or t(7;10)(q35;q24) has been described in 4% to 10% of children with T-ALLs and up to 14% of adults, with a predominantly cortical phenotype and a favorable outcome.12,13,32-35 Although most HOX11 and HOX11L2+ T-ALLs express CD1a, the immunophenotype of the latter is heterogeneous and the precise stage of maturation arrest within early cortical thymocytes is not known.22,30,36
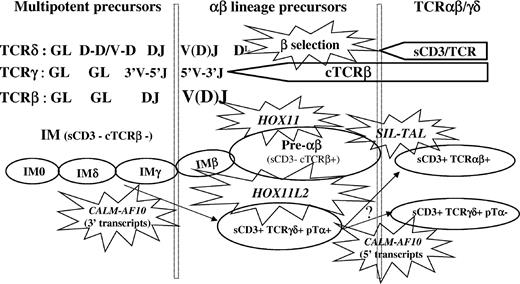
T-ALLs are usually classified immunophenotypically on the basis of CD1a expression.37 We have recently described a TCR-based classification of T-ALLs, which demonstrates that T-ALLs largely reproduce normal thymic development, and allows separation of cases into αβ-lineage, TCRγδ-expressing, and immature/uncommitted (IM), TCR, and cytoplasmic TCRβ (cTCRβ)-negative cases.38 The latter were further classified on the basis of their TCRδ, γ, and β gene configurations: IM0 T-ALLs demonstrate a germline configuration at all 3 loci; IMδ T-ALLs demonstrate TCRδ rearrangement; IMγ T-ALLs show TCRδ and TCRγ rearrangements but, at most, incomplete TCRβ DJ and IMβ T-ALLs have undergone TCRβ VDJ. The αβ-lineage T-ALLs include surface TCRαβ+ cases, pre-αβ T-ALLs, which express cTCRβ and pTα in the absence of sCD3 and, as such, are likely to express a pre-TCR, and their IMβ immediate precursors, which only differ by the absence of detectable cTCRβ. The γδ-lineage T-ALLs include mature surface TCRγδ-expressing cases, 40% of which express pTα, RAG-1, and V(D)J TCRβ rearrangement, with or without cTCRβ and, as such, appear to represent precursors arrested at an intermediate stage between the TCRαβ and TCRγδ lineages. Precursor TCRγδ T-ALLs are found within the IMδ and IMγ subsets.38,39
We then undertook to determine whether T-ALL oncogenic pathways are associated with specific subsets, initially demonstrating that PICALM-MLLT10/AF10 (referred to as CALM-AF10 here) is restricted to T-ALLs of the TCRγδ lineage.39 In the present study we have analyzed the effect of age and thymic involution on the incidence and TCR-based stage of maturation arrest of T-ALLs demonstrating HOX11, HOX11L2, TAL1, LMO1, LMO2, and LYL1 expression. Both TAL1 and LYL1 are tissue-specific bHLH transcription factors that bind to DNA as a heterodimer with HEB or one of the E2A proteins and that form a multiprotein complex also involving LMO1/2.21 TAL1, LYL1, and LMO2 are transcribed in immature hematopoietic cells, when they are essential to normal development, but not in the T-lymphoid lineage.21,40-42 TAL1 overexpression rarely induces oncogenic transformation, but accelerates LMO1-induced oncogenesis. It is not yet clear whether the predominant oncogenic role of TAL1 in T-ALL is via a complex with LMO1/2 or by titrating out partner bHLH proteins, and both are possible.43,44
We show that genetic subtype correlates closely with stage of maturation arrest, regardless of age, whereas the incidence of the different subtypes differs markedly, in keeping with age-related changes in the thymocytes at risk of oncogenic transformation or variable latency.
Patients, materials, and methods
Diagnostic peripheral blood or bone marrow samples from 173 T-ALLs, defined by expression of c/sCD3 and surface CD7 and negativity of CD19, were analyzed. Approval was obtained from the participating institutional review boards for these studies. Informed consent was provided according to the Declaration of Helsinki. The majority had more than 90% blasts. A total of 165 unselected patients, for whom availability of material was the only potential bias, came from 25 clinical centers. The majority of adults were treated on the LALA94 (Leucémies Aigues Lymphoblastiques de l'Adult) multicenter protocol (coordinator Denis Fière) and the majority of children on the FRALLE93/2000 (French ALL de l'Enfant study) protocols (coordinator André Baruchel). Eight TCRγδ T-ALLs (3 adults, 2 adolescents, and 3 children) from a single European Organisation for the Research and Treatment of Cancer (EORTC) center were included for certain analyses. Normal and cell line controls are detailed in Figure 2. All cell lines were maintained in RPMI 1640 supplemented with 10% fetal calf serum.
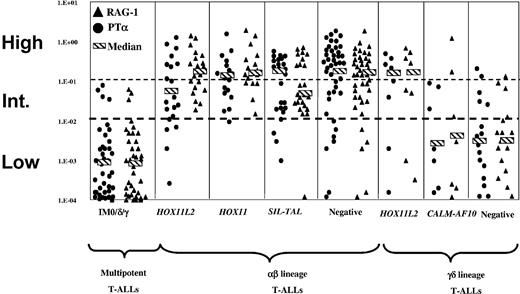
RAG-1. RAG-1 (▴) and pTα (•) expression in different T-ALL genetic subgroups. The HPB-ALL cell line expressed high-level pTα and RAG-1 and was arbitrarily attributed a value of 100%; all results were expressed relative to this value. Cases are divided into low-, intermediate-, and high-level expression by the dotted lines.
RAG-1. RAG-1 (▴) and pTα (•) expression in different T-ALL genetic subgroups. The HPB-ALL cell line expressed high-level pTα and RAG-1 and was arbitrarily attributed a value of 100%; all results were expressed relative to this value. Cases are divided into low-, intermediate-, and high-level expression by the dotted lines.
Immunophenotyping was performed in the diagnostic center on fresh material and completed at the Necker Hospital from cryopreserved material, as previously described.38 Samples with more than 20% labeled cells after correction for the proportion of blasts and normal T lymphocytes were considered positive. DNA and RNA were extracted from fresh or cryopreserved cells.45 TCR loci were analyzed as previously described.38,46
cDNA synthesis was performed at the Necker facility,38 and RNA quality assessed and normalized by quantification of ABL on an ABI PRISM 7700 (Perkin-Elmer Applied Biosystems, Branchburg, NJ), using guidelines of the Europe Against Cancer program.47 Samples with cycle threshold (Ct) above 32 (threshold 0.1) were excluded. Amplification efficiency was assessed by the slope obtained from logarithmic dilutions of the HPB-ALL cell line (pTα, RAG-1)38 or plasmids (HOX11, HOX11L2, LYL1, TAL1/SCL, LMO1/2 with 102-105 copies). Polyacrylamide gel electrophoresis (PAGE) and sequence analysis to evaluate product size and specificity were performed from a positive sample for each real-time quantitative polymerase chain reaction (RQ-PCR). Each experiment included 2 nontemplate controls for contamination and all RQ-PCRs were performed in duplicate. All primers spanned an intron and absence of genomic amplification was confirmed by RQ-PCR from peripheral blood lymphocyte (PBL) DNA. Transcript quantification was performed after normalization by the ABL housekeeping gene from the standard curves using the delta of delta Ct method.47 Primers and RQ-PCR probes are detailed in Table 1.
RQ-PCR primer and probe sequences
Transcript . | Sense . | Antisense . | Probe (FAM-TAMRA) . |
|---|---|---|---|
| ABL | TGGAGATAACACTCTAAGCATAACTAAAGGT | GATGTAGTTGCTTGGGACCCA | CCATTTTTGGTTTGGGCTTCACACCATT |
| RAG-1 | AGCCTGCTGAGCAAGGTACC | GAACTGAGTCCCAAGGTGGG | AGCCAGCATGGCAGCCTCTTTCC |
| pTα | TTGGGTGTCCAGCCCTACC | GCCATAGGTGAAGGCATCCA | CAGCCGGCAATGGCAGTGCA |
| HOX11 | CGCAGATACACAAAGGACAGGTT | CGTGCGCGGCTTCTTCT | AGAACCGGACGCCCCCCAAG |
| TAL1 | CTCTACAGCCTCAGCCAGCC | CAAAGGCCCCGTTCACATT | CGGTAGCCTTCCCTATGTTCACCACC |
| LYL1 | TCACCCCTTCCTCAACAGTGT | CGGGCCACCTTCTGGG | ACATTGGGCCAGCAGGACCTTTTAGC |
| LMO2 | GCCATCGAAAGGAAGAGCCT | AAGTAGCGGTCCCCGATGTT | CCTGCTGACATGCGGCT |
| LMO1 | CCAAGGCCAACCTCATCCT | AGTTCCCTGTGGTGCCAAAG | TGCCGACGCGACTACCTGAGGC |
Transcript . | Sense . | Antisense . | Probe (FAM-TAMRA) . |
|---|---|---|---|
| ABL | TGGAGATAACACTCTAAGCATAACTAAAGGT | GATGTAGTTGCTTGGGACCCA | CCATTTTTGGTTTGGGCTTCACACCATT |
| RAG-1 | AGCCTGCTGAGCAAGGTACC | GAACTGAGTCCCAAGGTGGG | AGCCAGCATGGCAGCCTCTTTCC |
| pTα | TTGGGTGTCCAGCCCTACC | GCCATAGGTGAAGGCATCCA | CAGCCGGCAATGGCAGTGCA |
| HOX11 | CGCAGATACACAAAGGACAGGTT | CGTGCGCGGCTTCTTCT | AGAACCGGACGCCCCCCAAG |
| TAL1 | CTCTACAGCCTCAGCCAGCC | CAAAGGCCCCGTTCACATT | CGGTAGCCTTCCCTATGTTCACCACC |
| LYL1 | TCACCCCTTCCTCAACAGTGT | CGGGCCACCTTCTGGG | ACATTGGGCCAGCAGGACCTTTTAGC |
| LMO2 | GCCATCGAAAGGAAGAGCCT | AAGTAGCGGTCCCCGATGTT | CCTGCTGACATGCGGCT |
| LMO1 | CCAAGGCCAACCTCATCCT | AGTTCCCTGTGGTGCCAAAG | TGCCGACGCGACTACCTGAGGC |
Results
Adult T-ALLs are arrested at an earlier stage than pediatric cases
T-ALLs from 165 unselected adults (n = 80; aged > 20 years), adolescents (n = 45; aged 11-20 years), and children (n = 40; aged ≤ 10 years) were divided into 2 major categories: mature TCR-expressing, and TCR-negative T-ALLs. The latter were divided into αβ precursors (pre-αβ and IMβ) and immature (IM0/δ/γ) T-ALLs (Table 2). Half of the childhood T-ALLs expressed a TCR compared to only 21% of adult cases (P = .0005). Conversely, IM0/δ/γ represented 38% of adult but only 8% of childhood T-ALLs (P = .0005). IM0/δ/γ was not seen in children under 9 years of age. The frequency of TCR+ and IM T-ALLs was intermediate in adolescents, in keeping with a progressive transition. The incidence of pre-αβ and IMβ T-ALLs did not change with age.
Incidence of HOX11, HOX11L2, SIL-TAL1, and CALM-AF10 within pediatric, adolescent, and adult T-ALLs, classified by immunophenotype
. | . | TCR- T-ALLs . | . | TCR+ T-ALLs . | . | ||
|---|---|---|---|---|---|---|---|
. | Total . | IM0/δ/γ . | IMβ and pre-αβ . | TCRαβ . | TCRγδ . | ||
| Children | 40 | 3 (8) | 16 (40) | 12 (30) | 9 (22) | ||
| Mean age, y (range)* | 6.3 (0.3-10) | 9.7 (9-10) | 5.7 (2-9) | 6.2 (0.3-10) | 6.2 (1-10) | ||
| SIL-TAL1, no (%) | 9 (23) | (0) | 4 (25) | 5 (42) | (0) | ||
| HOX11, no (%) | 1 (3) | (0) | 1 (6) | (0) | (0) | ||
| HOX11L2, no (%) | 9 (23) | (0) | 8 (50) | (0) | 1 (11) | ||
| CALM-AF10, no (%) | 2 (5) | (0) | (0) | (0) | 2 (22) | ||
| MG-, no (%) | 19 (48) | 3 (100) | 3 (19) | 7 (58) | 6 (67) | ||
| Adolescents | 45 | 10 (22) | 21 (46) | 9 (20) | 5 (11) | ||
| Mean age, y (range)† | 15.5 (11-20) | 15.0 (12-20) | 16.1 (12-20) | 15.2 (11-20) | 14.4 (11-20) | ||
| SIL-TAL1, no (%) | 7 (16) | (0) | 4 (19) | 3 (33) | (0) | ||
| HOX11, no (%) | 4 (9) | (0) | 4 (19) | (0) | (0) | ||
| HOX11L2, no (%) | 7 (16) | (0) | 4 (19) | (0) | 3 (60) | ||
| CALM-AF10, no (%) | 6 (13) | 4 (40) | (0) | (0) | 2 (40) | ||
| MG-, no (%) | 21 (47) | 6 (60) | 9 (43) | 6 (67) | (0) | ||
| Adults | 80 | 30 (38) | 33 (41) | 7 (9) | 10 (12) | ||
| Mean age, y (range)‡ | 33.9 (21-78) | 32.9 (21-78) | 34.0 (22-66) | 37.3 (24-53) | 31.2 (26-40) | ||
| SIL-TAL1, no (%) | 4 (5) | (0) | 2 (6) | 2 (29) | (0) | ||
| HOX11, no (%) | 14 (18) | 1 (3) | 12 (36) | (0) | 1 (10) | ||
| HOX11L2, no (%) | 9 (11) | (0) | 6 (18) | 2 (29) | 1 (10) | ||
| CALM-AF10, no (%) | 10 (13) | 8 (27) | (0) | (0) | 2 (20) | ||
| MG-, no (%) | 43 (54) | 21 (70) | 13 (39) | 3 (42) | 6 (60) | ||
. | . | TCR- T-ALLs . | . | TCR+ T-ALLs . | . | ||
|---|---|---|---|---|---|---|---|
. | Total . | IM0/δ/γ . | IMβ and pre-αβ . | TCRαβ . | TCRγδ . | ||
| Children | 40 | 3 (8) | 16 (40) | 12 (30) | 9 (22) | ||
| Mean age, y (range)* | 6.3 (0.3-10) | 9.7 (9-10) | 5.7 (2-9) | 6.2 (0.3-10) | 6.2 (1-10) | ||
| SIL-TAL1, no (%) | 9 (23) | (0) | 4 (25) | 5 (42) | (0) | ||
| HOX11, no (%) | 1 (3) | (0) | 1 (6) | (0) | (0) | ||
| HOX11L2, no (%) | 9 (23) | (0) | 8 (50) | (0) | 1 (11) | ||
| CALM-AF10, no (%) | 2 (5) | (0) | (0) | (0) | 2 (22) | ||
| MG-, no (%) | 19 (48) | 3 (100) | 3 (19) | 7 (58) | 6 (67) | ||
| Adolescents | 45 | 10 (22) | 21 (46) | 9 (20) | 5 (11) | ||
| Mean age, y (range)† | 15.5 (11-20) | 15.0 (12-20) | 16.1 (12-20) | 15.2 (11-20) | 14.4 (11-20) | ||
| SIL-TAL1, no (%) | 7 (16) | (0) | 4 (19) | 3 (33) | (0) | ||
| HOX11, no (%) | 4 (9) | (0) | 4 (19) | (0) | (0) | ||
| HOX11L2, no (%) | 7 (16) | (0) | 4 (19) | (0) | 3 (60) | ||
| CALM-AF10, no (%) | 6 (13) | 4 (40) | (0) | (0) | 2 (40) | ||
| MG-, no (%) | 21 (47) | 6 (60) | 9 (43) | 6 (67) | (0) | ||
| Adults | 80 | 30 (38) | 33 (41) | 7 (9) | 10 (12) | ||
| Mean age, y (range)‡ | 33.9 (21-78) | 32.9 (21-78) | 34.0 (22-66) | 37.3 (24-53) | 31.2 (26-40) | ||
| SIL-TAL1, no (%) | 4 (5) | (0) | 2 (6) | 2 (29) | (0) | ||
| HOX11, no (%) | 14 (18) | 1 (3) | 12 (36) | (0) | 1 (10) | ||
| HOX11L2, no (%) | 9 (11) | (0) | 6 (18) | 2 (29) | 1 (10) | ||
| CALM-AF10, no (%) | 10 (13) | 8 (27) | (0) | (0) | 2 (20) | ||
| MG-, no (%) | 43 (54) | 21 (70) | 13 (39) | 3 (42) | 6 (60) | ||
MG- T-ALLs are negative by molecular genetics (MG) for HOX11, HOX11L2, SIL-TAL1, and CALM-AF10.
Mean ages for children were as follows: SIL-TAL1, 5.1 years (3-9 years); HOX11, 4.0 years; HOX11L2, 6.3 years (5-8 years); CALM-AF10, 4.5 years (3-6 years); and MG-, 6.8 years (0.3-10 years)
Mean ages for adolescents were as follows: SIL-TAL1, 15.9 years (11-20 years); HOX11, 15.5 years (12-20 years); HOX11L2, 13.9 years (11-17 years); CALM-AF10, 13.8 years (11-20 years); and MG-, 16.3 years (12-20 years)
Mean ages for adolescents were as follows: SIL-TAL1, 29.5 years (22-40 years); HOX11, 40.4 years (24-66 years); HOX11L2, 28.1 years (22-37 years); CALM-AF10, 28.1 years (23-43 years); and MG-, 36.4 years (21-78 years)
The incidence of HOX11, HOX11L2, SIL-TAL1, and CALM-AF10 deregulation is age-dependent
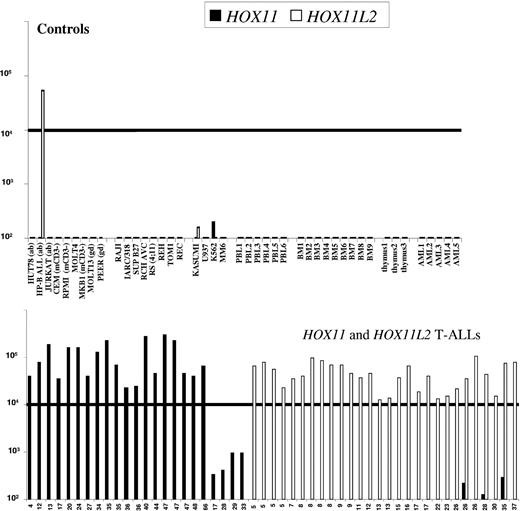
Normal peripheral blood mononuclear cells (PBMCs), bone marrow, and thymic controls showed absent or low-level expression of both HOX11 and HOX11L2 (< 100 copies), as did acute myeloid leukemias (AMLs; Figure 1) and 10 B-lineage ALLs (data not shown). T-ALLs expressing greater than 104 copies of HOX11 or HOX11L2 were classified as positive. Seven T-ALLs expressed 1 to 2 log lower levels of HOX11. Sequence analysis confirmed that these corresponded to true HOX11 transcripts (data not shown) but 0 of 5 analyzed showed 10q24 abnormalities; they have been classified as HOX11-. No sample showed both HOX11 and HOX11L2 positivity.
HOX11 and HOX11L2 RT-PCR quantification. RQ-PCR quantification of HOX11 (▪) and HOX11L2 (□) transcripts from controls (upper panel) and T-ALLs (lower panel). Controls include 6 normal peripheral mononuclear cell (PBL), 9 bone marrow (BM) and 3 thymi, 5 acute myeloid leukemias (AMLs), and 7-T and 8-B lymphoid and 4 myeloid (My) cell lines, as indicated. Amplified transcripts were normalized for RNA quality using the ABL housekeeping gene and were expressed as equivalent plasmid copy numbers on a logarithmic scale. Samples expressing more than 104 copies of HOX11 or HOX11L2 were classified as positive (bold line). Six adult and 1 adolescent T-ALLs expressed 1 to 2 log lower levels of HOX11, 3 in conjunction with HOX11L2 expression. T-ALLs demonstrating fewer than 100 copies of HOX11 and HOX11L2 are not shown.
HOX11 and HOX11L2 RT-PCR quantification. RQ-PCR quantification of HOX11 (▪) and HOX11L2 (□) transcripts from controls (upper panel) and T-ALLs (lower panel). Controls include 6 normal peripheral mononuclear cell (PBL), 9 bone marrow (BM) and 3 thymi, 5 acute myeloid leukemias (AMLs), and 7-T and 8-B lymphoid and 4 myeloid (My) cell lines, as indicated. Amplified transcripts were normalized for RNA quality using the ABL housekeeping gene and were expressed as equivalent plasmid copy numbers on a logarithmic scale. Samples expressing more than 104 copies of HOX11 or HOX11L2 were classified as positive (bold line). Six adult and 1 adolescent T-ALLs expressed 1 to 2 log lower levels of HOX11, 3 in conjunction with HOX11L2 expression. T-ALLs demonstrating fewer than 100 copies of HOX11 and HOX11L2 are not shown.
The incidence of HOX11L2 and SIL-TAL1 decreased with age, whereas that of HOX11 increased (Table 2). IM CALM-AF10+ cases were exclusively adolescents and young adults (median age, 25.5 years), whereas the rare pediatric CALM-AF10 cases expressed TCRγδ.39 Among the 43 adults older than 30 years the incidences of HOX11L2, SIL-TAL1, and CALM-AF10 were all 5%, whereas HOX11 was found in 28%.
HOX11 and SIL-TAL1 deregulation are virtually restricted to αβ-lineage T-ALLs
SIL-TAL1 and HOX11 deregulation was virtually absent from IM0/δ/γ T-ALLs (Table 2). SIL-TAL1 was more common in TCRαβ-expressing (36%; 10 of 28) than αβ-precursor T-ALLs (14%; 10 of 70; P = .02), irrespective of age group (Table 2). In keeping with a late cortical stage of αβ-lineage arrest, SIL-TAL1+ cases showed relatively low levels of RAG-1 expression, particularly in cases positive for TCRαβ (Figure 2). Despite the fact that enforced TAL1/SCL expression inhibits pTα expression,44 pTα levels in SIL-TAL1 T-ALLs were similar to other αβ-lineage cases (Table 3; Figure 2). TAL1 inhibits the CD4 proximal enhancer by titrating E2A/HEB.43 SIL-TAL1 clearly does not prevent expression of CD4 on CD4/8 DP cells but CD4 single-positive (SP) SIL-TAL1+ T-ALLs were not seen and CD8 SP cells were relatively frequent, suggesting that TAL1 prevents the DP to CD4 SP transition rather than the DN to CD4 immature single-positive (ISP)/DP transition.
Immunophenotype, TCRβ gene configuration, and RAG-1 and pTα expression of T-ALLs
. | . | αβ lineage: IMβ, pre-αβ, and TCRαβ . | . | . | . | TCRγδ . | . | . | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
. | IM 0/δ/γ . | HOX11L2 . | HOX11 . | SIL-TAL1 . | MG- . | HOX11L2 . | CALM-AF10 . | MG- . | |||||
| T-ALLs, no. | 42 | 20 | 17 | 20 | 41 | 8 | 7 | 16 | |||||
| Phenotype | |||||||||||||
| CD34, % | 75 | 70 | 0 | 20 | 27 | 50 | 17 | 31 | |||||
| CD13/33, % | 61 | 40 | 0 | 5 | 8 | 38 | 14 | 31 | |||||
| CD4/8 DN, % | 86 | 35 | 0 | 15 | 12 | 14 | 14 | 56 | |||||
| CD8 SP, % | 5 | 10 | 0 | 30 | 10 | 0 | 28 | 13 | |||||
| CD4 SP, % | 9 | 15 | 12 | 0 | 15 | 29 | 28 | 25 | |||||
| CD4/8 DP, % | 0 | 40 | 88 | 55 | 63 | 57 | 28 | 6 | |||||
| CD1a, % | 17 | 60 | 100 | 55 | 58 | 38 | 14 | 25 | |||||
| CD10, % | 15 | 55 | 73 | 25 | 35 | 63 | 28 | 44 | |||||
| cTCRβ, % | 0 | 55 | 76 | 95 | 95 | 50 | 10 | 8 | |||||
| TCRβ | |||||||||||||
| At least 1 VDJ, % | 0 | 100 | 100 | 100 | 100 | 100 | 28 | 38 | |||||
| RAG1/pTα | |||||||||||||
| RAG1 median | 0.1 | 18 | 15 | 6 | 15 | 17.5 | 0.5 | 0.5 | |||||
| pTα median | 0.1 | 7 | 11 | 21 | 29 | 16 | 0.1 | 0.4 | |||||
. | . | αβ lineage: IMβ, pre-αβ, and TCRαβ . | . | . | . | TCRγδ . | . | . | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
. | IM 0/δ/γ . | HOX11L2 . | HOX11 . | SIL-TAL1 . | MG- . | HOX11L2 . | CALM-AF10 . | MG- . | |||||
| T-ALLs, no. | 42 | 20 | 17 | 20 | 41 | 8 | 7 | 16 | |||||
| Phenotype | |||||||||||||
| CD34, % | 75 | 70 | 0 | 20 | 27 | 50 | 17 | 31 | |||||
| CD13/33, % | 61 | 40 | 0 | 5 | 8 | 38 | 14 | 31 | |||||
| CD4/8 DN, % | 86 | 35 | 0 | 15 | 12 | 14 | 14 | 56 | |||||
| CD8 SP, % | 5 | 10 | 0 | 30 | 10 | 0 | 28 | 13 | |||||
| CD4 SP, % | 9 | 15 | 12 | 0 | 15 | 29 | 28 | 25 | |||||
| CD4/8 DP, % | 0 | 40 | 88 | 55 | 63 | 57 | 28 | 6 | |||||
| CD1a, % | 17 | 60 | 100 | 55 | 58 | 38 | 14 | 25 | |||||
| CD10, % | 15 | 55 | 73 | 25 | 35 | 63 | 28 | 44 | |||||
| cTCRβ, % | 0 | 55 | 76 | 95 | 95 | 50 | 10 | 8 | |||||
| TCRβ | |||||||||||||
| At least 1 VDJ, % | 0 | 100 | 100 | 100 | 100 | 100 | 28 | 38 | |||||
| RAG1/pTα | |||||||||||||
| RAG1 median | 0.1 | 18 | 15 | 6 | 15 | 17.5 | 0.5 | 0.5 | |||||
| pTα median | 0.1 | 7 | 11 | 21 | 29 | 16 | 0.1 | 0.4 | |||||
MG- T-ALLs are negative by molecular genetics for HOX11, HOX11L2, SIL-TAL1, and CALM-AF10, pTα and RAG1 transcripts were evaluated relative to the levels observed in the HPB-ALL cell line.
DN indicates double negative; SP, single positive; DP, double positive.
HOX11 T-ALLs were virtually all arrested at the IMβ or pre-αβ stage, with a strikingly uniform immunophenotype: CD1a+, CD10+, CD4/8 DP (or rarely CD4 SP) with high levels of pTα and RAG-1 and frequent CD10 positivity but CD34, CD13, and CD33 negativity. HOX11 and SIL-TAL1 are therefore virtually restricted to the αβ lineage, with an earlier stage of maturation arrest in HOX11 cases compared to SIL-TAL1 T-ALLs (Figure 3).
Schematic representation of a TCR-based classification of T-ALL. This diagram shows the stage of maturation arrest of cases expressing HOX11, HOX11L2, SIL-TAL1, and CALM-AF10, relative to TCRβ selection.
Schematic representation of a TCR-based classification of T-ALL. This diagram shows the stage of maturation arrest of cases expressing HOX11, HOX11L2, SIL-TAL1, and CALM-AF10, relative to TCRβ selection.
HOX11L2 deregulation is associated with a maturation arrest intermediate between the αβ and γδ lineages
HOX11L2 expression was most common in IMβ (10 of 17; 59%), pre-αβ (8 of 47; 17%), and TCRγδ (7 of 33; 21%) T-ALLs (Table 2). HOX11L2+ αβ-lineage T-ALLs were more likely to be CD34+, CD13/33+, and CD4/8 DN, suggestive of relative immaturity. In keeping with this, expression of cTCRβ was less common (45% arrested at the IMβ stage), despite universal TCRβ VDJ rearrangement. Despite their relative immaturity, HOX11L2 expression was completely absent from IM0/δ/γ cases. It was also rare in TCRαβ cases, occurring in only 2 of 28, which, interestingly, corresponded to the oldest HOX11L2+ cases. To further analyze HOX11L2+ T-ALLs, we included 8 additional TCRγδ T-ALLs (Table 3). Within TCRγδ T-ALLs, HOX11L2-expressing cases were more likely to express CD34, CD4/8 DP, CD1a, CD10, and strikingly, RAG-1, pTα, and cTCRβ, than CALM-AF10 or negative cases. cTCRβ was found in 50%, compared to only 6% of HOX11L2- cases (P = .01). All HOX11L2+ TCRγδ+ T-ALLs had undergone complete TCRβ V(D)J rearrangement, compared to only 35% (8 of 23) of HOX11L2- TCRγδ cases (P = .0005). These are all features of αβ-lineage cortical thymocytes. HOX11L2+ T-ALLs therefore differentiate toward the αβ lineage, with an earlier stage of maturation arrest than HOX11 cases, associated with differentiation to unusual TCRγδ-expressing cells that are intermediate between the αβ and γδ lineages.
The immunophenotypic features of IM CALM-AF10 T-ALLs have been fully described.39 CALM-AF10 was found in 29% (7 of 24) of TCRγδ T-ALLs. It was not seen in HOX11L2-expressing cases and was found in pTα and RAG-1-negative and -positive cases (Table 3; Figure 2). CALM-AF10 TCRγδ T-ALLs differed from HOX11L2- TCRγδ cases by less frequent CD34, CD13/33, CD1a, and CD10 positivity and more frequent expression of CD8 or CD4, in keeping with a relatively mature TCRγδ-lineage stage of maturation arrest.
Taken together, approximately 50% of T-ALLs demonstrate deregulation of HOX11, HOX11L2, SIL-TAL1, or CALM-AF10, with no overlap. These cases are collectively referred to as “molecular genetic” (MG) positive. We demonstrate for the first time that the incidence of these genetic abnormalities differs with age, but that the stage of maturation arrest is determined by the genotype, independent of age.
Quantitative analysis of LYL1, LMO2, TAL1, and LMO1 transcripts
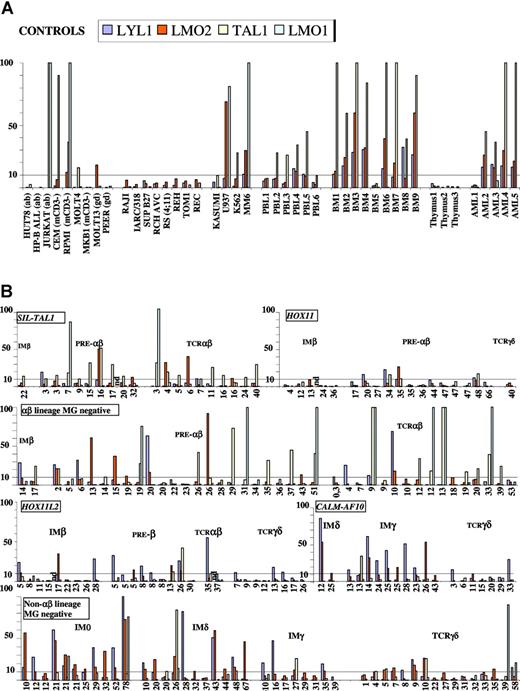
Expression levels for each T-ALL, classified by genotype and immunophenotype and sorted for age are shown in Figure 4. Overall, LYL1, LMO2, TAL1, and LMO1 were expressed by 35%, 31%, 30%, and 8% of T-ALLs, respectively (Table 4).
LYL1, LMO1/2, and TAL1 transcript quantification. Results of RQ-PCR quantification are shown for LYL1 (dark blue), LMO2 (red), TAL1 (cream), and LMO1 (light blue) transcripts from controls (A) and T-ALLs (B). (A) Controls include normal peripheral mononuclear cells (PBL), bone marrow (BM), and thymus, acute myeloid leukemias (AML), and several T- and B-lymphoid and myeloid (My) cell lines, as indicated. LYL1, LMO2, TAL1, and LMO1 quantification in normal, neonatal thymi showed 4000, 500, 60, and 500 mean copy numbers, respectively, whereas higher levels were observed in normal bone marrow (mean: 8.103, 1.104, 4.104, and 4.101, respectively). Because many T-ALL samples were of bone marrow origin and virtually all demonstrated at least 90% blasts, positive cut-off values were set at 10% of the mean for bone marrow samples for TAL1 and 10-fold of mean thymic values for LMO1/2 and LYL1. To compare levels of expression between transcripts, copy numbers were adjusted to give uniform positivity at 10 (4.104 copies for LYL1, 5.103 for LMO1/2, and 4.103 for TAL1). All B-cell lines were negative; 4 T-cell lines expressed TAL1, including 2 with SIL-TAL1 (CEM and RPMI.8402); 2 expressed LMO2 (Molt13 and RPMI.8402), and 2 expressed LMO1 (RPMI.8402 and Jurkat), but none expressed LYL1. (B) Linear, standardized, normalized copy numbers for each T-ALL, classified by genotype and immunophenotype and sorted for age, as shown in years below each histogram, are indicated. ND indicates LMO1 not quantified.
LYL1, LMO1/2, and TAL1 transcript quantification. Results of RQ-PCR quantification are shown for LYL1 (dark blue), LMO2 (red), TAL1 (cream), and LMO1 (light blue) transcripts from controls (A) and T-ALLs (B). (A) Controls include normal peripheral mononuclear cells (PBL), bone marrow (BM), and thymus, acute myeloid leukemias (AML), and several T- and B-lymphoid and myeloid (My) cell lines, as indicated. LYL1, LMO2, TAL1, and LMO1 quantification in normal, neonatal thymi showed 4000, 500, 60, and 500 mean copy numbers, respectively, whereas higher levels were observed in normal bone marrow (mean: 8.103, 1.104, 4.104, and 4.101, respectively). Because many T-ALL samples were of bone marrow origin and virtually all demonstrated at least 90% blasts, positive cut-off values were set at 10% of the mean for bone marrow samples for TAL1 and 10-fold of mean thymic values for LMO1/2 and LYL1. To compare levels of expression between transcripts, copy numbers were adjusted to give uniform positivity at 10 (4.104 copies for LYL1, 5.103 for LMO1/2, and 4.103 for TAL1). All B-cell lines were negative; 4 T-cell lines expressed TAL1, including 2 with SIL-TAL1 (CEM and RPMI.8402); 2 expressed LMO2 (Molt13 and RPMI.8402), and 2 expressed LMO1 (RPMI.8402 and Jurkat), but none expressed LYL1. (B) Linear, standardized, normalized copy numbers for each T-ALL, classified by genotype and immunophenotype and sorted for age, as shown in years below each histogram, are indicated. ND indicates LMO1 not quantified.
Incidence of LYL1, LMO2 and TAL1 and LMO1 transcript positivity in T-ALL by stage of maturation arrest
. | No. . | LYL1, % . | LMO2, % . | TAL1, % . | LMO1*, % . | None, % . |
|---|---|---|---|---|---|---|
| αβ lineage MG- | 41 | 20 | 17 | 44 | 17 | 20 |
| SIL-TAL | 20 | 15 | 25 | 85 | 10 | 0 |
| HOX11 | 19 | 28 | 6 | 17 | 0 | 74 |
| HOX11L2 | 28 | 43 | 18 | 11 | 0 | 50 |
| CALM-AF10 | 19 | 53 | 37 | 5 | 0 | 26 |
| TCRγδ MG- | 16 | 25 | 38 | 31 | 6 | 50 |
| IMγ MG- | 8 | 38 | 25 | 13 | 0 | 50 |
| IMδ MG- | 11 | 55 | 73 | 27 | 9 | 9 |
| IM0 MG- | 11 | 82 | 91 | 9 | 18 | 9 |
| Total | 173 | 35 | 31 | 30 | 8 | 32 |
. | No. . | LYL1, % . | LMO2, % . | TAL1, % . | LMO1*, % . | None, % . |
|---|---|---|---|---|---|---|
| αβ lineage MG- | 41 | 20 | 17 | 44 | 17 | 20 |
| SIL-TAL | 20 | 15 | 25 | 85 | 10 | 0 |
| HOX11 | 19 | 28 | 6 | 17 | 0 | 74 |
| HOX11L2 | 28 | 43 | 18 | 11 | 0 | 50 |
| CALM-AF10 | 19 | 53 | 37 | 5 | 0 | 26 |
| TCRγδ MG- | 16 | 25 | 38 | 31 | 6 | 50 |
| IMγ MG- | 8 | 38 | 25 | 13 | 0 | 50 |
| IMδ MG- | 11 | 55 | 73 | 27 | 9 | 9 |
| IM0 MG- | 11 | 82 | 91 | 9 | 18 | 9 |
| Total | 173 | 35 | 31 | 30 | 8 | 32 |
Transcript positivity was defined as being superior to the higher of 10% of mean normal bone marrow or thymic samples. Among TAL1- expressing cases, 8 of 10 children, 6 of 15 adolescents, and 3 of 16 adults demonstrated a SIL-TAL1 deletion.
Four patients (identified by ND in Figure 4B) were not analyzed for LMO1
Deregulation of TAL1, as defined by increased expression or SIL-TAL1 rearrangement, was more frequent within αβ-lineage T-ALLs (42%; 41 of 98) than all other categories (13%, 10 of 75, P = .00005). Within the αβ lineage, the mechanism of TAL1 deregulation was strikingly different in children and adults; it was due to SIL-TAL1 in 8 of 10 TAL1-deregulated pediatric cases, compared to only 9 of 31 adolescents and adults (P = .004), when “idiopathic” TAL1 expression predominated. It is noteworthy that IM T-ALLs, particularly IM0, which might represent expansions of cells close to multipotential/myeloid precursors were TAL1-, as were the majority of HOX11 and HOX11L2 T-ALLs. Using the criteria defined here, only 85% of SIL-TAL1+ T-ALLs were TAL1+ (Figure 4). The 3 TAL1- cases demonstrated the lowest levels of SIL-TAL1 transcripts by RQ-PCR (data not shown) and were all LMO2+, compared to only 2 of 17 TAL1+SIL-TAL1+ cases (P = .01). LMO1 expression was found in 2 SIL-TAL1+ cases.
LMO2 expression was most common in IM T-ALLs (Table 4), demonstrating an abrupt diminution at the IMγ-to-TCRγδ transition in CALM-AF10 T-ALLs and at the IMδ to IMγ transition in MG- cases (Figure 4). LMO1 expression was less frequent and predominated within αβ-lineage cases. Among SIL-TAL1+ and MG- αβ-lineage T-ALLs, LMO2 and LMO1 expression were mutually exclusive. Eight of 9 LMO1+ cases were also TAL1+, whereas only 3 of 13 LMO2+ T-ALLs were (P = .005). Conversely, of the 30 TAL1+ αβ-lineage T-ALLs analyzed for all 3 transcripts, 19 expressed only TAL1,8 TAL1, and LMO1 and only 3 TAL1 and LMO2.
LYL1 expression profiles were similar to those of LMO2, with which it was frequently coexpressed. LYL1 expression with relatively low LMO2 levels was characteristic of HOX11L2+ T-ALLs.
Taken together, expression of the bHLH transcripts occurs in different T-ALL populations, with TAL1 predominating in those of the αβ lineage and LYL1 in IM0/δ/γ T-ALLs. TAL1 is either isolated or preferentially coexpressed with LMO1, particularly in SIL-TAL1- cases, whereas LYL1 is commonly coexpressed with LMO2. The only clear influence of age, independent of genotype, was the deregulation of TAL1 by SIL-TAL1 in children, but by unknown mechanisms in older T-ALLs.
Discussion
We have recently shown that T-ALLs accurately reproduce thymic maturation and can be separated into expansions of the TCR αβ and γδ lineages, with the CALM-AF10 fusion transcript being specific for the latter.38,39 In the present study we demonstrate that TAL1 and HOX11 are specific to the αβ lineage, whereas HOX11L2 identifies an intermediate αβ/γδ population. The stage of maturation arrest of each of these abnormalities is determined by genotype and is independent of age, whereas their incidence and their mechanism of deregulation varies with age, probably reflecting evolution in the population at risk or variable latency.
The vast majority of neonatal murine thymocytes belong to the αβ lineage, reflecting the massive cellular expansion induced by β selection following pre-TCR engagement. This process diminishes with age, probably secondary to the effect of interleukin 7 (IL-7).48 Because the incidence of DN thymocytes does not diminish, their proportion increases with age.3,6 This process may also be operational in human thymic atrophy because we show that leukemic transformation in pediatric T-ALL predominantly results in a mature, TCR+ stage of maturation arrest compared to a much more immature arrest in adult T-ALL, prior to completion of TCRβ rearrangement, which roughly corresponds to a DN1/DN2 murine thymocyte stage.48,49 The proportion of pre-TCR-expressing, pre-αβ T-ALLs, did not vary with age, although their genetic subtypes evolved markedly.
Ferrando et al22 recently used gene expression profiles in pediatric T-ALL to demonstrate a closely related early cortical thymocyte stage of maturation arrest in HOX11 and HOX11L2 cases, earlier than the late cortical stage found in SIL-TAL1 cases. We have used complementary immunophenotypic and TCR genotypic analyses to confirm and extend these data. HOX11 deregulation leads to a uniform, cortical thymocyte stage of maturation arrest (RAG-1+, pTα+, cTCRβ+/V(D)J rearranged, sCD3-,CD1a+, CD4/8 DP, TdT+, CD34-), in keeping with a population undergoing, or just about to undergo, β selection. HOX11L2+ T-ALLs differ from HOX11+ T-ALLs by a more immature phenotype, more frequent LYL1 expression, and a potential for expression of both pTα and TCRγδ, in keeping with a population intermediate between the αβ and γδ TCR lineages. Despite expression of a surface TCRγδ, these cells rearrange TCRβ VDJ, retain a CD1a+, CD4/8 DP cortical phenotype, and fail to down-regulate RAG-1 and pTα. Dissociation of LMO2 and LYL1 expression argues against simple immaturity as the explanation for LYL1 expression in HOX11L2+ T-ALLs.
Maturation of murine DN precursors to the DP stage requires expression of a pre-TCR. Because normal TCRγδ lymphocytes are not thought to express a pre-TCR or to undergo β selection, expression of pTα is not necessary for their development.50 In the absence of either pTα or TCRβ, some, albeit inefficient, maturation to the DP stage is possible by replacement of the pre-TCR by TCRγδ.51 Gounari et al52 interpreted expression of a pTα-driven reporter in TCRγδ thymocytes as evidence of early TCRγδ cells that have recently derived from a pTα+ common αβ/γδ precursor. It is possible that the pTα+ TCRγδ HOX11L2 T-ALLs described here correspond to these cells. The frequency of this category suggests either that common αβ/γδ precursors are particularly susceptible to leukemic transformation by HOX11L2 or, more likely, that HOX11L2 expression leads to a specific maturation arrest at this stage. Only half of the pTα+ TCRγδ T-ALLs expressed HOX11L2, demonstrating that HOX11L2 expression is not the only mechanism leading to coexpression of TCRγδ and pTα.
The mechanisms leading to HOX11L2 deregulation are still unclear. Almost all are due to a cryptic translocation involving chromosome 5q35 and the CTIP2/BCL11B locus at chromosome14q32,29 but 2 t(5;14)(q32;q11) involving HOX11L2 and TCRα/δ have been described.53 Loss of BCL11B expression by homologous recombination leads to a DN block in murine αβ thymic development, prior to complete TCRβ rearrangement and pTα expression,54 with no evidence of leukemic transformation. Our data demonstrating universal TCRβ rearrangement and pTα expression are in keeping with BCL11B involvement in T-cell oncogenesis as a transcriptional activator of HOX11L2, rather than as a tumor suppressor gene.
bHLH and their partner LMO1/2 proteins showed distinct profiles, with TAL1 and LMO1 predominating in αβ-lineage T-ALLs and LYL1 and LMO2 in immature T-ALLs, as described for pediatric cases.22 TAL1 deregulation in αβ-lineage T-ALLs (approximately 40% of cases) was predominantly due to recombinase-mediated SIL-TAL1 deletions in children but to currently unknown mechanisms in adults, shown to be due to deregulation in cis in the Jurkat cell line.55 They are unlikely to be due to alternative SIL-TAL1 breakpoints, because the reverse transcription-PCR (RT-PCR) system used allows detection of the vast majority of recognized breakpoints.56 Although the αβ-lineage nature of SIL-TAL1 T-ALLs has long been recognized,25,27 this is the first demonstration of a clear αβ-lineage restriction for “idiopathic” TAL1 expression and suggests either that the mechanisms inducing both categories of TAL1 expression are specific to the αβ lineage or that TAL1 is preferentially oncogenic in this lineage. Using our criteria, 15% of SIL-TAL1+ T-ALLs were TAL1-. These cases demonstrated relatively high levels of LMO2 compared to their TAL1+ counterparts, as if a “critical mass” of rate-limiting proteins necessary for formation of a E2A/TAL1/LMO/GATA/LDB complex was necessary for oncogenic conversion, in keeping with murine models of TAL1/LMO oncogenic synergy.57,58 TAL1 and LMO1/2 expression are maximal in DN1/2 murine thymocytes and are down-regulated as E2A/HEB increase. Enforced TAL1/LMO1/2 expression prevents the DN-to-DP transition and pTα expression by titration of HEB, although the DP thymocytes that develop in these mice express higher pTα levels than controls.44 Our data would suggest that it is this latter population that is at risk of leukemic transformation because the majority of TAL1-expressing T-ALLs were DP/CD8 SP and expressed pTα. They suggest that TAL1-deregulated T-ALLs are arrested either during β selection or at the DP-to-CD4 SP transition. These are precisely the checkpoints at which the levels of functional E2A proteins decrease, due either to an increase of Id proteins or diminution of E2A/HEB.59 Our data support an “Id-like” role for TAL1, with its capacity to titrate out E2A/HEB being critical at these 2 checkpoints. Such a titration effect may not require an LMO protein, which would fit with the fact that we often found TAL1 expression in the absence of LMO1 and LMO2.
We have previously shown that IMδ/γ T-ALLs include expansions of TCRγδ precursors.38,39 Coexpression of LMO2 and LYL1 was reported to reflect immaturity in pediatric T-ALL22 ; our data are in keeping with maintenance of these transcripts in γδ precursors, which by extrapolation suggest that γδ precursors are closer to a population with multilineage potential. Both transcripts were down-regulated with TCRγδ expression, particularly in adults. IM0/δ/γ T-ALLs were not seen before 10 years of age. Their phenotype, their low level RAG-1 expression, and the absence of recombinase-mediated oncogenic rearrangements are all suggestive of an extrathymic origin. The absence of TAL1 expression is against transformation of a bone marrow multilineage precursor. Extrathymic γδ-lineage maturation is well recognized, predominantly in the gut or the fetal liver,60,61 although the relatively late age of onset of these T-ALLs argues against a fetal onset.
We found LYL1 expression in 35% of cases, compared to 38% and 22% of pediatric T-ALL series.22,30 Because LYL1 expression is most common in IM, CALM-AF10, and HOX11L2 T-ALLs, its incidence is dependent on the age range studied. TAL1/LYL1 amino acid homology is restricted to the bHLH domain,40 so it is likely that the consequences of LYL1 and TAL1 expression in T-ALLs is different, particularly if their action is not merely to titrate out E2A/HEB. The distinctive profiles of expression shown here are in support of this. Our data are in keeping with oncogenic synergy for LYL1 and LMO2 in IM0/δ/γ T-ALLs, although we cannot exclude that these signatures are merely evidence of physiologic expression of these transcripts by cCD3+, CD7+ precursors arrested prior to, or at an early stage of, γδ-lineage development. It is probable that these T-ALLs are similar to immature AML and undifferentiated leukemia and that their clinical behavior differs markedly from αβ lineage-restricted T-ALLs, particularly regarding cortico-sensitivity.
We show that the predominant pediatric oncogenic pathways are SIL-TAL1 and HOX11L2, which are progressively replaced by idiopathic TAL1 deregulation, CALM-AF10, and, lastly, HOX11. The diminution in the incidence of SIL-TAL1 and HOX11L2 with age is in keeping with progressive diminution in the thymic αβ lineage, but it does not explain the late onset of HOX11 T-ALL (Table 2). HOX11 is, at least partly, deregulated by the TCRα/δ or TCRβ locus,11,12,32,62 whose activity are also likely to diminish with age. An increasing number of “idiopathic” HOX11-expressing T-ALLs are recognized.33,36 We consider that the age difference is more likely to reflect different oncogenic potential, in keeping with their different prognoses.22,30,34,63 HOX11L2 expression may provide a sufficiently strong oncogenic signal for development of leukemias after a relatively short latency, whereas HOX11 may require a further event. Newborn HOX11-/- mice exhibit asplenia and HOX11 is essential for the survival of splenic precursors during organogenesis but not for the initiation of splenic development.64 Its enforced expression immortalizes embryonic yolk sac erythroid precursors,65 blocks maturation of the J2E erythroid cell line,66 and transforms immature bone marrow precursors into IL-3-dependent myeloid lines. It induces lymphoid tumors at low frequency with long latency.67,68 Taken together, HOX11 appears to immortalize, rather than transform, a variety of hematopoietic lineages. The second event could either correspond to the proliferative signal of β-selection, but this is common to HOX11- and HOX11L2-expressing T-ALL, or to an additional oncogenic abnormality. Thymocytes undergoing β selection demonstrate considerable apoptosis. It is possible that HOX11 overexpression is initially compensated for by this apoptotic potential. Leukemic development would occur when this equilibrium is lost.
In conclusion, we demonstrate that 50% of T-ALLs demonstrate HOX11, HOX11L2, SIL-TAL1, or CALM-AF10 deregulation, with each being associated with a specific stage of maturation arrest: HOX11, SIL-TAL1, and HOX11L2 and represent αβ-lineage oncogenes, despite TCRγδ expression in the latter, and CALM-AF10 a γδ-lineage oncogene (Figure 3). The frequency of each genetic subgroup varies with age, suggesting variable latency or that thymic atrophy has an impact on the population at risk of leukemic transformation and the mechanisms involved.
Prepublished online as Blood First Edition Paper, March 30, 2004; DOI 10.1182/blood-2003-11-3944.
Supported by the Fondation contre la leucémie de la Fondation de France, la Ligue Nationale Contre le Cancer, l'Association de la Recherche sur le Cancer (ARC), and the Biomed-2 BMH4-CT98-3936 concerted action.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank all the biologists and clinicians of the adult and pediatric French FRALLE and LALA groups. We particularly thank C. Bayle and Anne-Lyse Bennaceur (Institut Gustave-Roussy, Villejuif), F-X. Mahon and C. Bilhou-Nabera (Bordeaux), X. Troussard (Caen), M. Dupont (Montpellier), R. Garand (Nantes), J. M. Cayela (Saint-Louis, Paris), B. Lenormand (Rouen), P. Cornillet-Lefevre (Reims), F. Davi (La Pitie-Salpêtrière), M. H Estienne (Tours), S. Hayette (Lyon), and C. Schmitt (Nancy) for providing T-ALL samples and results.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal