Abstract
The xeroderma pigmentosum group D (XPD) gene encodes a DNA helicase that functions in nucleotide excision repair of chemotherapy-induced DNA damage, the efficiency of which is predicted to be affected by a lysine to glutamine variant at codon 751. We hypothesized that this constitutive genetic variant may modify clinical response to chemotherapy, and we have examined its association with outcome following chemotherapy for acute myeloid leukemia (AML) in 341 elderly patients entered into the United Kingdom Medical Research Council AML 11 trial, and with the risk of developing chemotherapy-related AML. Among subjects treated for AML, disease-free survival at one year was 44% for lysine homozygotes, compared with 36% for heterozygotes and 16% for glutamine homozygotes (hazard ratio [HR], 1.30; 95% confidence interval [CI], 1.01-1.70; P = .04). Similarly, overall survival at one year was 38% for lysine homozygotes, 35% for heterozygotes, and 23% for glutamine homozygotes (HR, 1.18; 95% CI, 0.99-1.41; P = .07). Furthermore, homozygosity for the XPD codon 751 glutamine variant was associated with a significantly increased risk of developing AML after chemotherapy (odds ratio, 2.22 for Gln/Gln vs Lys/Lys; 95% CI, 1.04-4.74). These data suggest that the XPD codon 751 glutamine variant protects against myeloid cell death after chemotherapy. (Blood. 2004;104:3872-3877)
Introduction
The affect of somatic alterations in genes such as RAS, FLT3, and P53 on the prognosis of acute myeloid leukemia (AML) has been well documented.1-4 In contrast, relatively little is known about how constitutive genetic variation may influence the response of either leukemic or normal myeloid cells to chemotherapy. Recent efforts have identified polymorphisms in chemotherapy-metabolizing genes, including the glutathione S-transferase genes, which modulate AML prognosis after chemotherapy and also the etiology of chemotherapy-related AML.5,6 Genetic variation in other pathways that mediate cellular response to chemotherapy, such as DNA repair, may also modulate prognosis and etiology of AML after chemotherapy.
Nucleotide excision DNA repair (NER) protects against mutagenicity and toxicity by removing deleterious DNA lesions from the genome, including those induced by nitrogen mustards and several other classes of chemotherapy.7-10 The chemotherapeutic nitrogen mustards, including melphalan and cyclophosphamide, are not only used in the treatment of AML,11,12 but are also suspected myeloid leukemogens.13 As such, constitutive variation in NER activity is predicted to affect how leukemic and normal myeloid cells respond to mustard-based therapies.
The xeroderma pigmentosum group B (XPB) and group D (XPD) genes encode DNA helicases that mediate DNA unwinding required for initiation of both NER and basal transcription.14 Such is the complexity of their roles in cellular function that rare constitutive mutations in XPB and XPD can give rise to 3 distinct human disorders: xeroderma pigmentosum, Cockayne syndrome, and trichothiodystrophy,14,15 all of which are characterized by deficiencies in NER and/or transcription.14,16 There is evidence to suggest that DNA helicases and NER may be involved in the pathogenesis and prognosis of some myeloid leukemias. First, XPB is bound and modified by P210 BCR-ABL,17 the oncoprotein responsible for pathogenesis of Philadelphia chromosome-positive myeloid leukemias, suggesting that deregulated DNA helicase activity may contribute to the genetic instability observed during blast crisis of chronic myeloid leukemia and in Philadelphia chromosome-positive AML.18,19 Second, expression of P210 BCR-ABL in human myeloid cells and primary human bone marrow up-regulates NER activity and renders cells highly resistant to the killing effects of genotoxins,20 suggesting a plausible mechanism for the antiapoptotic phenotype that is characteristic of Philadelphia chromosome-positive myeloid leukemia.21,22
Common polymorphisms in NER components, including the DNA helicases, may give rise to subtle alterations in the DNA repair proficiency of otherwise healthy individuals. Of particular interest is the XPD lysine to glutamine variant at codon 751, which is predicted to affect protein function and has been shown to modulate cellular response to genotoxins,23-26 and also outcome in patients treated with chemotherapy.27 However, conflicting results have led to suggestions that functionality of the codon 751 polymorphism may be exposure- and pathway-specific, affecting both DNA repair and cell death.28 Consistent with a role for XPD in cell death, P53-mediated apoptosis is attenuated in XPD mutated fibroblasts.29,30 Furthermore, P53 interacts directly with the carboxy terminus of XPD, which includes the polymorphic codon 751 residue.28
We hypothesized that any functional effect of the XPD codon 751 variant in myeloid cells, both leukemic and normal, may be amplified following exposure to chemotherapy, giving rise to an altered clinical response. In order to test this hypothesis, we determined association of the XPD codon 751 polymorphism with outcome of 341 AML patients treated with chemotherapy in the United Kingdom Medical Research Council AML 11 trial, and also with susceptibility to developing AML after chemotherapy.
Patients, materials, and methods
Subjects
We examined XPD codon 751 polymorphism status in 4 groups of individuals (Table 1): 341 AML cases enrolled in the United Kingdom Medical Research Council AML 11 trial, 420 de novo AML cases and 729 noncancer controls recruited as part of a United Kingdom Leukaemia Research Fund case-control study, and 91 therapy-related AML (t-AML) cases.
Description of study subjects
. | . | . | . | t-AML study . | . | . | ||
|---|---|---|---|---|---|---|---|---|
| . | . | Leukaemia Research Fund study . | . | . | Previous therapy . | . | ||
| Variable . | AML 11 . | Controls . | De novo AML . | All cases . | Radiotherapy only . | Chemotherapy* . | ||
| No. | 341 | 729 | 420 | 91 | 40 | 51 | ||
| Sex | ||||||||
| Male, no. (%) | 143 (42) | 393 (54) | 227 (54) | 36 (40) | 11 (28) | 25 (49) | ||
| Female, no. (%) | 198 (58) | 336 (46) | 193 (46) | 55 (60) | 29 (73) | 26 (51) | ||
| Age, y | ||||||||
| Mean (standard deviation) | 66.9 (5.5) | 48.1 (14.3) | 47.5 (14.5) | 55.3 (13.6) | 61.0 (8.5) | 50.8 (14.8) | ||
| Median | 66.6 | 50.3 | 49.9 | 58.7 | 62.1 | 52.7 | ||
| Age groups | ||||||||
| 16 to 39 y, no. (%) | 0 | 218 (30) | 132 (31) | 16 (18) | 0 | 16 (31) | ||
| 40 to 59 y, no. (%) | 38 (11) | 322 (44) | 180 (43) | 34 (37) | 17 (43) | 17 (33) | ||
| 60 to 69 y, no. (%) | 206 (60) | 189 (26) | 108 (26) | 41 (45) | 23 (58) | 18 (35) | ||
| 70 to 74 y, no. (%) | 72 (21) | 0 | 0 | 0 | 0 | 0 | ||
| 75 y or more, no. (%) | 25 (7) | 0 | 0 | 0 | 0 | 0 | ||
. | . | . | . | t-AML study . | . | . | ||
|---|---|---|---|---|---|---|---|---|
| . | . | Leukaemia Research Fund study . | . | . | Previous therapy . | . | ||
| Variable . | AML 11 . | Controls . | De novo AML . | All cases . | Radiotherapy only . | Chemotherapy* . | ||
| No. | 341 | 729 | 420 | 91 | 40 | 51 | ||
| Sex | ||||||||
| Male, no. (%) | 143 (42) | 393 (54) | 227 (54) | 36 (40) | 11 (28) | 25 (49) | ||
| Female, no. (%) | 198 (58) | 336 (46) | 193 (46) | 55 (60) | 29 (73) | 26 (51) | ||
| Age, y | ||||||||
| Mean (standard deviation) | 66.9 (5.5) | 48.1 (14.3) | 47.5 (14.5) | 55.3 (13.6) | 61.0 (8.5) | 50.8 (14.8) | ||
| Median | 66.6 | 50.3 | 49.9 | 58.7 | 62.1 | 52.7 | ||
| Age groups | ||||||||
| 16 to 39 y, no. (%) | 0 | 218 (30) | 132 (31) | 16 (18) | 0 | 16 (31) | ||
| 40 to 59 y, no. (%) | 38 (11) | 322 (44) | 180 (43) | 34 (37) | 17 (43) | 17 (33) | ||
| 60 to 69 y, no. (%) | 206 (60) | 189 (26) | 108 (26) | 41 (45) | 23 (58) | 18 (35) | ||
| 70 to 74 y, no. (%) | 72 (21) | 0 | 0 | 0 | 0 | 0 | ||
| 75 y or more, no. (%) | 25 (7) | 0 | 0 | 0 | 0 | 0 | ||
Nineteen patients also received radiotherapy.
AML 11 cases
DNA was available from 341 patients enrolled into the United Kingdom Medical Research Council AML 11 trial between November 1990 and June 1998 (Table 1). All patients signed an informed consent before entering the study. The demographic and clinical characteristics of the 341 patients who took part in this study were not significantly different from the 1314 patients enrolled in the AML11 study as a whole. Full details of this trial have been published elsewhere.31,32 Briefly, elderly patients (> 60 years of age) diagnosed with any form of de novo or secondary AML (secondary disease is defined as AML after a prior diagnosis of myelodysplastic syndrome or after previous therapy) were enrolled in the AML 11 trial, which incorporated 3 randomizations. Patients were initially randomized to receive induction therapy with either DAT (daunorubicin, cytarabine, 6-thioguanine), ADE (cytarabine, daunorubicin, etoposide), or MAC (mitoxantrone, cytarabine) for the first 2 courses. Patients who were in remission after 2 courses of induction therapy were given DAT consolidation therapy. The second randomization was between short consolidation (no further treatment) and long consolidation (3 additional courses: COAP [cyclophosphamide, vincristine, cytosine arabinoside, prednisolone], DAT, and COAP). The third randomization was between interferon-α maintenance for one year versus no maintenance.
A bone marrow aspirate containing less than 5% leukemic blast cells and showing normal maturation of other cell lineages was the criterion for the achievement of complete remission. Remission failure was classified by the investigating clinician as either induction death (related to treatment and/or hypoplasia) or resistant disease. Overall survival was defined as the time from entry into the trial to death, and disease-free survival was defined as the time from complete remission to either disease relapse or death in remission. The date of the last follow-up was June 1, 2003. Patients lost to follow-up (n = 3) were censored at the date they were last known to be alive.
The results of the AML11 trial have been published elsewhere.31 Briefly, the complete remission rate of patients allocated to DAT (62%) was significantly higher than that of patients allocated to either ADE (50%) or MAC (55%). However, there were no differences between the induction schedules with respect to disease-free survival or overall survival. There were also no differences between short consolidation versus long consolidation or interferon maintenance versus no maintenance with respect to disease-free survival or overall survival. As such, for the purposes of the present study we have collapsed all induction and consolidation groups.
Overall survival was affected by cytogenetics (karyotype), presenting white blood cell count, age, performance status, and type of AML (de novo disease [n = 241] vs secondary disease [n = 97]; 3 cases were unclassified).32 As such, we tested for association between these prognostic markers and XPD codon 751 polymorphism status.
Leukaemia Research Fund cases and unaffected controls
De novo AML (n = 420) and control group participants (n = 729) donated blood as part as part of a large population-based case-control study of acute leukemia, which has been fully described elsewhere.33 All patients signed an informed consent before entering the study. All patients were between 16 and 69 years of age, and were diagnosed with AML between April 1991 and December 1996, while resident in parts of the north and southwest of England. All diagnoses were pathologically confirmed. Patients were considered ineligible if, prior to a diagnosis of acute leukemia, they had been diagnosed with chronic myeloid leukemia or myelodysplastic syndrome within the previous 6 months, or any malignancy within the previous 2 years. From the general practice where the patient was registered, 2 controls per patient, individually matched on sex, age, and ethnic origin, were randomly selected. All patients and controls signed an informed consent before entering the study.
t-AML case series
For this study, t-AML is defined as AML diagnosed at least 2 months after the start of initial chemotherapy and/or radiotherapy. All 91 t-AML patients have been described elsewhere.5,34 Of the t-AML patients, 24 were identified in the Leukaemia Research Fund acute leukemia case-control study.33 DNA samples from an additional 67 individuals with t-AML were obtained from subjects enrolled in the United Kingdom Medical Research Council AML trials 10 (n = 6), 11 (n = 33), and 12 (n = 28).31,35 There were 40 t-AML patients who had previous exposure to radiotherapy only. There were 51 t-AML cases that occurred after treatment with chemotherapy, with 19 also receiving radiotherapy. Among the 35 cases for which specific chemotherapeutic agents were documented, the majority (n = 30) had prior exposure to combination chemotherapy, of which alkylating agents were the most common component.5
DNA extraction
DNA was extracted from whole frozen blood (AML incidence case-control study and AML 11 prognostic study) using proteinase K digestion and phenol/chloroform extraction, or from archived bone marrow smears (for some t-AML cases) using the QIAamp mini DNA extraction kit according to the manufacturer's recommendations for archived bone marrow (Qiagen, Hilden, Germany).
XPD codon 751 genotyping
XPD genotyping was performed using a polymerase chain reaction restriction fragment length polymorphism (RFLP). Briefly, exon 23 of XPD was amplified using Amplitaq Gold (Applied Biosystems, Foster City, CA), forward primer (5′ atcctgtccctactggccattc 3′) and reverse primer (5′ tgtggacgtgacagtgagaaat 3′), with the following amplification conditions: 1 cycle of 95°C for 15 minutes; 36 cycles of 95°C for 1 minute, 56°C for 1 minute, and 72°C for 1 minute; followed by 1 cycle of 72°C for 10 minutes. Digestion of the resulting 324-bp amplicon with PstI that resulted in generation of 100-bp and 224-bp fragments corresponded to individuals homozygous for the lysine-encoding allele. Generation of 66-bp, 100-bp, and 158-bp fragments after digestion corresponded to individuals homozygous for the glutamine-encoding allele. Generation of all 4 fragments after digestion corresponded to individuals heterozygous for the codon 751 polymorphism. Approximately 4% of all cases and controls were genotyped on 2 independent occasions; concordance between repeats was 100%. Furthermore, 18 randomly selected DNA samples were also genotyped using direct sequencing; concordance with RFLP genotyping was 100%.
Statistical analysis
In the AML 11 prognostic study, χ2 tests for heterogeneity and for trend were used to test for association between XPD genotype and prognostic markers (age, sex, presenting white blood cell count, performance status, French-American-British (FAB) type, cytogenetics, and diagnosis [de novo AML vs secondary AML]) and dichotomous outcomes (remission, induction death, resistant disease). For time to event data, Kaplan-Meier survival curves were constructed for each XPD codon 751 genotype group and compared using the log-rank test. Tests for trend were used for comparisons of outcome by the 3 XPD codon 751 genotype groups. Disease-free survival and overall survival analyses were adjusted for cytogenetic status, age, performance status, and white blood cell count. All P values are 2-tailed. All analyses were conducted using SAS (Cary, NC).
For the AML and t-AML incidence case-control analysis, odds ratios and 95% confidence intervals, adjusted for age and sex, were estimated using unconditional logistic regression.36 XPD codon 751 was analyzed as a trichotamous variable (Lys/Lys, Lys/Gln, Gln/Gln) using the lysine homozygotes as the reference group. Due to a considerably stronger association with leukemogenesis,13 chemotherapy was given precedence over radiotherapy; t-AML cases who received both modalities (n = 19) were grouped with those who received chemotherapy alone, as described previously.5 All analyses were conducted using Stata, 1999 (Stata, College Station, TX).
Results
XPD polymorphism and outcome after chemotherapy
We first determined association of the XPD codon 751 variant with outcome after chemotherapy in 341 elderly patients with AML. Of the patients, 134 (39%) were homozygous for the lysine-encoding allele, 163 (48%) were heterozygous, and 44 (13%) were homozygous for the glutamine-encoding allele. There were no significant differences between the polymorphism subgroups according to age, sex, presenting white blood cell count, secondary status, FAB type, or performance status (data not shown). However, there was an association between XPD genotype and cytogenetic status. Of 341 patients, 308 were successfully tested for cytogenetic status, and individuals who were carriers of at least one glutamine-encoding allele were more likely to have a karyotype associated with adverse prognosis (test for trend, P = .008; Table 2).32 The association between XPD genotype and prognostic cytogenetic status was not seen in the t-AML case series as a whole (test for trend, P = .62; Table 2), but was evident in those cases whose leukemia developed after any form of chemotherapy (test for trend, P = .02; Table 2).
XPD codon 751 polymorphism status and association with prognostic cytogenetic status
. | . | Prognosis . | . | . | . | ||
|---|---|---|---|---|---|---|---|
| XPD codon 751 . | All cases, no. . | Favorable, no. (%) . | Intermediate, no. (%) . | Adverse, no. (%) . | Cytogenetics not known, no. . | ||
| AML11 | 308 | 19 (100) | 235 (100) | 54 (100) | 33 | ||
| Lys/Lys | 122 | 13 (68) | 93 (40) | 16 (30) | 12 | ||
| Lys/Gln | 144 | 6 (32) | 109 (46) | 29 (54) | 19 | ||
| Gln/Gln | 42 | 0 | 33 (14) | 9 (17) | 2 | ||
| t-AML (all cases)* | 71 | 9 | 40 | 22 | 20 | ||
| Lys/Lys | 29 | 4 (44) | 17 (43) | 8 (36) | 7 | ||
| Lys/Gln | 28 | 5 (56) | 16 (40) | 7 (32) | 12 | ||
| Gln/Gln | 14 | 0 | 7 (18) | 7 (32) | 1 | ||
| t-AML after chemotherapy | 38 | 3 | 20 | 15 | 13 | ||
| Lys/Lys | 13 | 2 (67) | 9 (45) | 2 (13) | 4 | ||
| Lys/Gln | 13 | 1 (33) | 6 (30) | 6 (40) | 8 | ||
| Gln/Gln | 12 | 0 | 5 (25) | 7 (47) | 1 | ||
. | . | Prognosis . | . | . | . | ||
|---|---|---|---|---|---|---|---|
| XPD codon 751 . | All cases, no. . | Favorable, no. (%) . | Intermediate, no. (%) . | Adverse, no. (%) . | Cytogenetics not known, no. . | ||
| AML11 | 308 | 19 (100) | 235 (100) | 54 (100) | 33 | ||
| Lys/Lys | 122 | 13 (68) | 93 (40) | 16 (30) | 12 | ||
| Lys/Gln | 144 | 6 (32) | 109 (46) | 29 (54) | 19 | ||
| Gln/Gln | 42 | 0 | 33 (14) | 9 (17) | 2 | ||
| t-AML (all cases)* | 71 | 9 | 40 | 22 | 20 | ||
| Lys/Lys | 29 | 4 (44) | 17 (43) | 8 (36) | 7 | ||
| Lys/Gln | 28 | 5 (56) | 16 (40) | 7 (32) | 12 | ||
| Gln/Gln | 14 | 0 | 7 (18) | 7 (32) | 1 | ||
| t-AML after chemotherapy | 38 | 3 | 20 | 15 | 13 | ||
| Lys/Lys | 13 | 2 (67) | 9 (45) | 2 (13) | 4 | ||
| Lys/Gln | 13 | 1 (33) | 6 (30) | 6 (40) | 8 | ||
| Gln/Gln | 12 | 0 | 5 (25) | 7 (47) | 1 | ||
Favorable defined as cases with t(8;21), t(15;17), or inv(16), irrespective of the presence of additional cytogenetic abnormalities; adverse defined as cases lacking a favorable abnormality but with 5 or more unrelated abnormalities, −7, −5, del(5q), or any 3q abnormality; all remaining cases composed the intermediate risk group, and included cases with normal karyotype, + 8 or 11q23 abnormalities. In the test for trend between XPD genotype and prognostic cytogenetic status, for AML 11, P = .008; for t-AML, P = .62; for t-AML after chemotherapy, P = .02.
Includes 30 cases common with the AML11 case series.
The complete remission rate of all cases was 55%. There was no significant difference in complete remission rates between individuals homozygous for the glutamine-encoding allele (45%), lysine homozygotes (56%), and heterozygotes (58%) (test for trend, P = .8; Table 3). There were 152 (45%) patients who failed to achieve complete remission (Table 3), with 83 of these due to resistant disease and 69 due to induction death. XPD genotype was not significantly associated with either resistant disease (test for trend, P = .8) or induction death (test for trend, P = 1.0; Table 3).
XPD codon 751 polymorphism status and clinical outcome after chemotherapy of 341 patients enrolled in the United Kingdom Medical Research Council AML 11
XPD codon 751 . | All cases, no. . | Complete remission, no. (%) . | Resistant disease, no. (%) . | Induction death, no. (%) . | Disease-free survival at 12 mo, % . | Overall survival at 12 mo, % . |
|---|---|---|---|---|---|---|
| All patients | 341 | 189 (55) | 83 (24) | 69 (20) | 37 | 35 |
| Lys/Lys | 134 | 75 (56) | 29 (22) | 30 (22) | 44 | 38 |
| Lys/Gln | 163 | 94 (58) | 42 (26) | 27 (17) | 36 | 35 |
| Gln/Gln | 44 | 20 (45) | 12 (27) | 12 (27) | 16 | 23 |
| P | NA | .8 | .8 | 1.0 | .04* | .07* |
XPD codon 751 . | All cases, no. . | Complete remission, no. (%) . | Resistant disease, no. (%) . | Induction death, no. (%) . | Disease-free survival at 12 mo, % . | Overall survival at 12 mo, % . |
|---|---|---|---|---|---|---|
| All patients | 341 | 189 (55) | 83 (24) | 69 (20) | 37 | 35 |
| Lys/Lys | 134 | 75 (56) | 29 (22) | 30 (22) | 44 | 38 |
| Lys/Gln | 163 | 94 (58) | 42 (26) | 27 (17) | 36 | 35 |
| Gln/Gln | 44 | 20 (45) | 12 (27) | 12 (27) | 16 | 23 |
| P | NA | .8 | .8 | 1.0 | .04* | .07* |
All P values are for trend; NA indicates not applicable.
Adjusted for cytogenetic status, age, performance status, and white blood cell count.
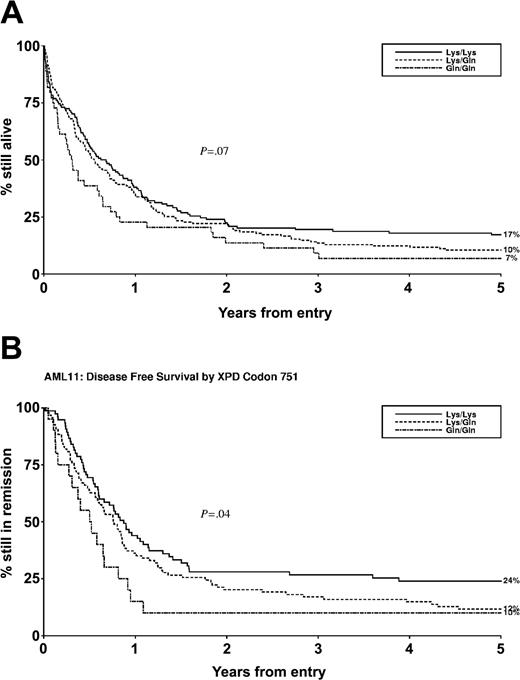
Disease-free survival and overall survival of all patients at one year was 37% and 35%, respectively (Table 3). The XPD codon 751 variant was associated with both disease-free survival and overall survival. Specifically, disease-free survival at one year for lysine homozygotes, heterozygotes, and glutamine homozygotes was 44%, 36%, and 16%, respectively (adjusted hazard ratio [HR], 1.30; 95% confidence interval [CI], 1.01-1.70; test for trend, P = .04) (Table 3; Figure 1). Consistent with this, overall survival at one year for lysine homozygotes, heterozygotes, and glutamine homozygotes was 38%, 35%, and 23%, respectively (adjusted HR, 1.19; 95% CI, 0.99-1.41; P = .07) (Table 3; Figure 1).
XPD codon 751 polymorphism status and outcome after chemotherapy for AML. (A) Overall survival (n = 341). (B) Disease-free survival (n = 189).
XPD codon 751 polymorphism status and outcome after chemotherapy for AML. (A) Overall survival (n = 341). (B) Disease-free survival (n = 189).
These data suggest a role for the XPD codon 751 polymorphism in modulating clinical outcome after chemotherapy for AML, and specifically that the glutamine variant confers greater protection against chemotherapy-induced leukemic blast cell death relative to the lysine variant, giving rise to earlier disease relapse and ultimately to shorter overall survival.
XPD polymorphism and risk of AML
Given that the codon 751 polymorphism may modulate myeloid leukemic blast cell death after chemotherapy, we next wished to address association of this variant with risk of developing myeloid leukemia, particularly t-AML arising after chemotherapy. In order to do this, we determined the codon 751 polymorphism status of 420 cases of de novo AML, 91 cases of t-AML, and 729 noncancer controls. XPD codon 751 polymorphism was not significantly associated with risk of de novo AML (Table 4). However, it is possible that cell death does not significantly contribute to determining de novo AML risk, or that NER is not involved in repairing DNA damage induced by endogenous or environmental leukemogens. In order to further examine these 2 possibilities, we determined the XPD codon 751 genotype distribution in a case series of 51 individuals who developed t-AML as a consequence of chemotherapy for a prior condition. The majority of these cases had prior exposure to alkylating agents, including melphalan, chlorambucil, mitomycin C, and cisplatin derivatives,5 many of which are human leukemogens13 and all of which induce DNA damage that is repaired by NER.7-10 Furthermore, therapeutic doses of many of these chemotherapy agents cause acute and severe bone marrow toxicity.37 Thus, the ability to appropriately engage apoptosis after chemotherapy, eliminating mutated myeloid precursor cells, is likely to play a critical role in protecting against the development of t-AML. Consistent with a role for the XPD glutamine variant in protecting against chemotherapy-induced myeloid cell death, and potentiating the survival of mutated clones, individuals homozygous for the glutamine-encoding allele were significantly overrepresented in t-AML cases after chemotherapy, compared with controls (odds ratio [OR], 2.22 [Gln/Gln vs Lys/Lys]; 95% CI, 1.04-4.74; Table 4).
XPD codon 751 polymorphism and risk of de novo and therapy-related AML
. | . | De novo AML cases . | . | t-AML cases, previous therapy . | . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| . | . | . | . | Radiotherapy only . | . | Chemotherapy* . | . | ||||
. | Controls, no. (%) . | No. (%) . | OR (95% CI) . | No. (%) . | OR no. (95% CI) . | No. (%) . | OR (95% CI) . | ||||
| No. | 729 (100) | 420 (100) | NA | 40 (100) | NA | 51 (100) | NA | ||||
| XPD codon 751 | |||||||||||
| Lys/Lys | 293 (42) | 144 (38) | 1 | 19 (48) | 1 | 17 (33) | 1 | ||||
| Lys/Gln | 299 (43) | 176 (46) | 1.20 (0.91-1.57) | 19 (48) | 1.06 (0.54-2.12) | 21 (41) | 1.22 (0.63-2.36) | ||||
| Gln/Gln | 104 (15) | 63 (16) | 1.22 (0.84-1.78) | 2 (5) | 0.34 (0.07-1.51) | 13 (25) | 2.22 (1.04-4.74) | ||||
| ND | 33 | 37 | NA | 0 | NA | 0 | NA | ||||
. | . | De novo AML cases . | . | t-AML cases, previous therapy . | . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| . | . | . | . | Radiotherapy only . | . | Chemotherapy* . | . | ||||
. | Controls, no. (%) . | No. (%) . | OR (95% CI) . | No. (%) . | OR no. (95% CI) . | No. (%) . | OR (95% CI) . | ||||
| No. | 729 (100) | 420 (100) | NA | 40 (100) | NA | 51 (100) | NA | ||||
| XPD codon 751 | |||||||||||
| Lys/Lys | 293 (42) | 144 (38) | 1 | 19 (48) | 1 | 17 (33) | 1 | ||||
| Lys/Gln | 299 (43) | 176 (46) | 1.20 (0.91-1.57) | 19 (48) | 1.06 (0.54-2.12) | 21 (41) | 1.22 (0.63-2.36) | ||||
| Gln/Gln | 104 (15) | 63 (16) | 1.22 (0.84-1.78) | 2 (5) | 0.34 (0.07-1.51) | 13 (25) | 2.22 (1.04-4.74) | ||||
| ND | 33 | 37 | NA | 0 | NA | 0 | NA | ||||
Odds ratios (OR) and 95% confidence intervals (CI) were estimated using logistic regression adjusting for sex and age at diagnosis in comparison with controls.
NA indicates not applicable; ND, not determined.
Chemotherapy alone, or in combination with radiotherapy.
We also determined the XPD codon 751 genotype distribution in a series of 40 t-AML cases whose disease developed subsequent to radiotherapy only. The toxicity and mutagenicity of radiotherapy is mediated predominantly via the direct induction of DNA single- and double-strand breaks, which are not substrates for NER. Consistent with this repair specificity, the XPD genotype distribution between the postradiotherapy t-AML case series and controls was not significantly different (Table 4).
Discussion
We have shown that the XPD codon 751 polymorphism is an independent prognostic marker for disease-free survival and overall survival in elderly AML patients treated with chemotherapy, and specifically that the glutamine variant was associated with a poorer prognosis relative to the lysine variant. It is likely, therefore, that at least one chemotherapy agent used in the United Kingdom Medical Research Council AML 11 trial induces DNA damage that is repaired by NER. Recognition and repair of DNA damage induced by other nitrogen mustards7,10 suggest that NER may also repair cyclophosphamide-induced DNA damage. If this is the case, the use of cyclophosphamide as consolidation therapy in the AML 11 trial, but not as induction therapy, may explain why the XPD codon 751 variant was not associated with remission, resistant disease, or induction death (Table 3). Further work to identify those chemotherapy agents that interact with NER may allow for the development of individualized treatment regimes for AML in the elderly.
These results are consistent with data reported in a small study of patients treated with platinum-based chemotherapy for colorectal cancer,27 where 10 subjects homozygous for the glutamine-encoding allele had significantly shorter overall survival (median survival, 3.3 months) than 22 subjects homozygous for the lysine-encoding allele (median survival, 17.4 months). As in our study, heterozygotes had an intermediate overall survival (median survival, 12.8 months), suggesting an allele dosage effect with respect to outcome. It is possible that the XPD codon 751 polymorphism may be an independent prognostic marker for outcome after chemotherapy for other forms of cancer, including other hematologic malignancies treated using nitrogen mustards, such as multiple myeloma.
We have also shown that homozygosity for the codon 751 glutamine variant is associated with a significantly increased risk of developing t-AML after chemotherapy, but is not associated with risk of t-AML after radiotherapy or with risk of de novo AML. However, we recognize that this association is modest and suggest that it be viewed in the context of other risk factors, including other genetic polymorphisms.
The findings reported in this study are consistent with a model whereby the XPD codon 751 variant modulates cellular ability to engage cell death in response to chemotherapy, and specifically that the XPD codon 751 glutamine variant protects against cell death relative to the lysine variant. We can postulate 2 general mechanisms by which the XPD codon 751 variant may modulate myeloid cell death in response to chemotherapy: either via a direct role for XPD in signaling cell death, or indirectly via XPD repair of protoxic DNA lesions.
The codon 751 polymorphism may exert a direct effect on the myeloid cell death machinery via interaction between XPD, through its carboxy terminus domain, and P53.28 Indeed, evidence exists to support this model, in that P53-dependent apoptosis is compromised in fibroblasts from XPD patients, a phenotype that can be restored by expression of wild-type XPD.29 Thus, we can hypothesize that the codon 751 polymorphism modulates the ability of XPD to signal P53-dependent cell death, possibly following stalled transcription or repair. If the codon 751 polymorphism modulates myeloid cell death only, and does not affect the efficiency of other cellular functions such as DNA repair, then its contribution to t-AML risk is readily predicted. The inability to appropriately engage cell death is predicted to result in the persistence of genetically unstable and potentially mutated preleukemic cells, a model suggested by others.28
The codon 751 variant may also modulate myeloid cell death indirectly by affecting the efficiency of NER to repair chemotherapy-induced protoxic DNA lesions. Like P53, P44 also interacts with the carboxy terminus of XPD, stimulating its helicase activity.38 Mutations in the carboxy terminus of XPD, including R722W and delta716-730, can prevent P44 binding, weakening the helicase activity of XPD.38 Indeed, attenuation of this interaction is responsible for the NER-deficient phenotype of xeroderma pigmentosum patients with these constitutive XPD carboxy terminus mutations. Thus, it might be alternatively hypothesized that the codon 751 polymorphism weakens but does not abolish P44 binding, which in turn compromises NER by attenuating the helicase activity of XPD. Because XPD is a component of both transcription-coupled and global genomic repair, a generic effect on NER capacity is likely to affect the repair of both protoxic and promutagenic chemotherapy-induced DNA lesions. In this model, predicting the overall effect on t-AML risk is made difficult because repair of protoxic and promutagenic DNA lesions contribute in opposing directions to cancer risk. This model predicts that any modulatory effect of the codon 751 polymorphism on the direction of t-AML risk will be determined not only by whether cell death, and the elimination of mutated clones, plays an important role in protecting against therapy-related leukemogenesis, but also by the extent of cell death, which will be a function of chemotherapy dose to the bone marrow.
Overrepresentation of individuals homozygous for the glutamine-encoding allele in the postchemotherapy t-AML case series (Table 4) is consistent with both of these models. As such, further work is required to determine which, if either, of these models may be operating in myeloid cells. Nevertheless, our results identify the XPD codon 751 polymorphism as an independent prognostic marker in elderly AML patients treated with chemotherapy. Furthermore, the observed association with t-AML risk provides additional evidence for functionality of the XPD codon 751 polymorphism in the myeloid lineage, and also highlights cell death as a potentially important mechanism protecting against therapy-related leukemogenesis.
Prepublished online as Blood First Edition Paper, August 31, 2004; DOI 10.1182/blood-2004-06-2161.
Supported by grants from the Kay Kendall Leukaemia Fund (J.M.A.) and the Leukaemia Research Fund (G.J.M.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We are grateful to Rachel Clack (Clinical Trial Service Unit, University of Oxford), the staff at the Leukaemia Research Fund Epidemiology and Genetics Unit (York University), and the Medical Research Council Adult Leukemia Working Party for their expert assistance. We acknowledge the support of Stephen Langabeer at the Medical Research Council DNA/RNA bank, University College London Hospital, a facility funded by the Kay Kendall Leukaemia Fund of the United Kingdom. The Epidemiology and Genetics Unit and the Haematological Malignancy Diagnostic Service are members of the Haematology Research Network for Yorkshire and Humber.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal