The CD40-CD154 dyad has a central role in the development of immune-inflammatory processes. Therefore, disruption of CD40 signaling has the potential to be therapeutically useful in a number of disease indications, including autoimmune syndromes, atherosclerosis, and allograft rejection. Blocking antibodies to CD154 have been successfully employed in experimental animal models, and recently in clinical trials, to prevent or treat these immunologically induced diseases. However, the thrombotic events observed in some of these studies raise important issues regarding future use of anti-CD154 antibodies in humans. In this study, we demonstrate that a small interfering RNA (siRNA) can effectively reduce the surface expression of the human CD40 costimulatory receptor. Moreover, by rendering endothelial cells unresponsive to CD154+ Jurkat cell–mediated activation through RNA interference, induction of endothelial cell-adhesion molecule expression and leukocyte adhesion is prevented in vitro. Thus, anti-CD40 siRNA may become a safe and effective therapeutic option for interfering with CD40-CD154–mediated acute or chronic immune-inflammatory conditions.
Introduction
The 48 kD type I transmembrane glycoprotein CD40 (tumor necrosis factor receptor 5 [TNFR5]) holds a spectrum of signaling activities. Activation of endothelial cells via CD40-CD154 interaction triggers proinflammatory cytokine and chemokine production; matrix metalloproteinase and tissue factor expression; an increased density of leukocyte adhesion molecules CD62E, CD106, and CD54; and thrombomodulin down-regulation.1-7 These events play an important role in the promotion of extravasation and accumulation of activated T cells at the sites of inflammation. Interruption of CD40 signaling is, thus, being considered as a potential pharmacologic target for therapy. Indeed, preclinical studies in several animal models have shown that blockade of the CD40-CD154 pathway by antibodies to CD154 can prevent or treat a variety of autoimmune diseases, vascular disease, and allograft rejection without inducing generalized immune suppression.8 Remarkably, a recent clinical trial has demonstrated that passive administration of a humanized monoclonal antibody (mAb) specific for CD154 ameliorates disease in systemic lupus erythematosus (SLE) patients.9,10 However, the association of anti-CD154 mAb therapy with thromboembolic complications in human and nonhuman primates has provoked concern, leading to a halt in all clinical studies using anti-CD154 antibodies.10-12 Thus, the development of alternative approaches to abrogate CD40-CD154 signaling seems mandatory for an effective and safer prevention or treatment of human immune-inflammatory disorders.
RNA interference (RNAi) mediated by small interfering RNAs (siRNAs), a type of posttranscriptional gene silencing, is revolutionizing the field of functional genomics owing to its powerful, sequence-specific capability to knock down gene expression and shows great promise for therapeutic applications.13 In this study, we demonstrate that efficient blockade of the CD40-CD154 signaling by RNAi-mediated silencing of human CD40 expression on vascular endothelial cells leads to inhibition of CD62E, CD106, and CD54 expression and to a concomitant reduction of leukocyte adherence on these cells. This suggests an effective and potentially therapeutic anti-inflammatory effect mediated by anti-CD40 siRNA in the human endothelium.
Study design
The siRNA duplexes targeting the human CD40 mRNA (GenBank accession no. X60592.1) were designed according to previously described guidelines.14 For the initial screening of the most effective siRNA duplexes, the 21-nucleotide (21-nt) RNAs were synthesized by in vitro transcription (Silencer siRNA Construction Kit; Ambion, Austin, TX). Further experiments were performed with the use of the most efficient siRNA-2–duplex chemically synthesized (Qiagen, Hilden, Germany), with 2′-deoxythymidines instead of uridine residues in the 3′ overhangs to enhance nuclease resistance.
ECV-304 cells (American Type Culture Collection [ATCC], Manassas, VA) (CRL 1998) were seeded in 6-well culture plates at a density of 1 × 105 cells per well 16 to 18 hours prior to RNAi treatment. Transfection of siRNAs was performed by means of the cationic lipid OligofectAMINE in OptiMEM medium (Invitrogen, San Diego, CA), according to the manufacturer's instructions. Exponentially growing human umbilical vein endothelial cells (HUVECs) (Advancell, Barcelona, Spain) (4 × 105 cells per well in 6-well culture plates) underwent lipid-mediated transfection with the use of Targefect (Targeting Systems, Santee, CA), as recommended by the manufacturer. Two consecutive rounds of transfection with siRNA were performed to ensure maximum CD40 silencing on these primary cells.
RNA from siRNA-transfected cells was isolated and reverse transcribed (3 μg DNAse I–treated RNA per sample) by means of the RNeasy RNA Isolation and Omniscript RT kits (Qiagen). Quantification of CD40 mRNA levels was performed by real-time polymerase chain reaction (PCR) with the use of the LightCycler technology (Roche Molecular Biochemicals, Indianapolis, IN) and the human CD40-specific primers: CD40-forward (CD40-f) (5′-CAGCCAGGACAGAAACTGGTGAGT-3′) and CD40-reverse (CD40-r) (5′-CTTCTTCACAGGTGCAGATGGTGTC-3′). All samples were normalized with the use of the following primer set for the constitutively expressed human cyclophilin gene: Cyph-f (5′-CTCCTTTGAGCTGTTTGCAG-3′) and Cyph-r (5′-CACCACATGCTTGCCATCC-3′). PCR amplifications were performed in a 20 μL vol containing 2 μL ready-to-use reaction mix, 10 × DNA Master SYBR Green I (Roche Molecular Biochemicals); MgCl2 (3 mM for CD40 amplification; 4 mM for cyclophilin amplification); 0.15 μM each primer; 5% dimethyl sulfoxide (DMSO); and 75 ng cDNA as template. The amplification program used an initial denaturation at 95°C for 10 minutes, followed by 45 cycles: 95°C for 1 second; 64°C for 5 seconds; 72°C for 10 seconds.
Immunoblotting for human CD40 detection was performed with a monoclonal antibody (H-10; 1:1000 dilution) (Santa Cruz Biotechnology, Santa Cruz, CA). Membranes were reprobed with rabbit polyclonal antiactin antibody (1:2000 dilution) (Sigma-Aldrich, St Louis, MO).
Indirect immunofluorescence staining for cell adhesion molecule (CAM) expression included anti–intercellular adhesion molecule 1 (anti–ICAM-1), CD54 (HA58); anti–vascular cell adhesion molecule 1 (anti–VCAM-1), CD106 (51-10C9); and anti–E-selectin, CD62E (68-5H11) (all from BD Pharmingen, San Diego, CA).
Adhesion of calcein acetoxymethyl ester (calcein AM)–loaded HL-60 leukemia cells (ATCC CCL 240) to endothelial cells was performed essentially as previously described.4
Results and discussion
Selection and characterization of effective anti-CD40 siRNAs
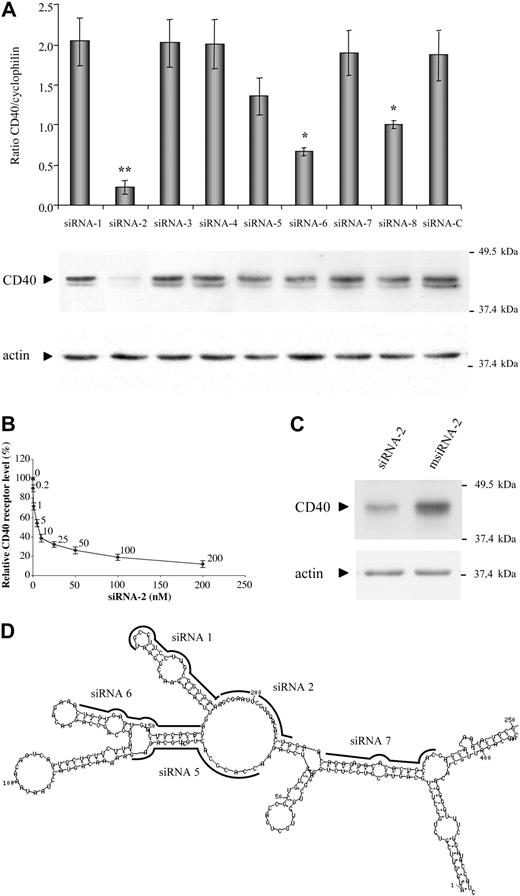
Eight siRNAs were designed and generated by in vitro transcription to target-defined positions within the coding region of the human CD40 mRNA (Table 1). The potencies of the 8 siRNAs relative to a randomized control siRNA were compared both at the posttranscriptional level by real-time RT-PCR and at the translational level by Western immunoblotting (Figure 1A). The fluorescence ratios of target CD40 mRNA level to endogenous cyclophilin mRNA level in the presence of 100 nM siRNA duplexes revealed that 3 of the 8 siRNAs tested (siRNA-2, siRNA-6, and siRNA-8) were capable of specifically and significantly reducing the basal expression of CD40 in the ECV-304 cells. However, siRNA-2 was the most active inhibitor, showing a nearly 9-fold reduction of the CD40-cyclophilin mRNA ratio with respect to that shown by treatment with both the control siRNA (siRNA-C) and the inactive siRNAs. To better discriminate the effect of the different siRNAs on the CD40 protein level, we performed the screening over ECV-304 cells activated through TNF-α and IFN-γ, which up-regulated CD40 expression. It is noteworthy that siRNA-2 was confirmed as the most potent CD40 silencer capable of reducing CD40 receptor expression below its basal level on cytokine-activated ECV-304 cells, giving an overall inhibition of 85% to 90% at 48 hours, which correlates with that observed at the posttranscriptional level. Certainly, a dose-response assay corroborated the potency of siRNA-2 (50% inhibitory concentration [IC50], approximately 5 nM) in ECV-304 cells (Figure 1B). In terms of local target accessibility, the superior efficiency of the siRNA-2 over alternative siRNA designs may reflect its targeting toward a stable internal loop within the secondary structure of the CD40 mRNA, determined by suboptimal folding (Figure 1D). It is being progressively recognized that the activity of siRNAs, similar to that of antisense oligonucleotides, is affected by the secondary structure of the target mRNA.17,18 Interestingly, recent comparisons of the effectiveness of antisense oligonucleotides and siRNAs, both in cell culture and in vivo, suggest a superior efficiency and inhibitory persistence of the siRNAs.19,20 Moreover, siRNAs are highly effective without any chemical modifications, resulting in a dramatically lower toxicity profile compared with antisense oligomers.
Sequence composition and target localization within the human CD40 mRNA of the siRNAs (siRNA-1 through siRNA-8) designed to screen for effective CD40 gene silencing
siRNA anti-hCD40 . | hDC40 mRNA targeted region* . | Sequence . |
|---|---|---|
| siRNA-1 | 173-193 | 5′-UGCCUUCCUUGCGGUGAAAUU-3′ |
| 3′-UUACGGAAGGAACGCCACUUU-5′ | ||
| siRNA-2 | 192-212 | 5′-GCGAAUUCCUAGACACCUGUU-3′ |
| 3′-UUCGCUUAAGGAUCUGUGGAC-5′ | ||
| siRNA-3 | 479-499 | 5′-UGUCACCCUUGGACAAGCUUU-3′ |
| 3′-UUACAGUGGGAACCUGUUCGU-5′ | ||
| siRNA-4 | 709-729 | 5′-UUUUCCCGACGAUCUUCCUUU-3′ |
| 3′-UUAAAAGGGCUGCUAGAAGGA-5′ | ||
| siRNA-5 | 62-82 | 5′-CCACCCACUGCAUGCAGAGUU-3′ |
| 3′-UUGGUGGGUGACGUACGUCUC-3′ | ||
| siRNA-6 | 137-157 | 5′-CUGGUGAGUGACUGCACAGUU-3′ |
| 3′-UUGACCACUCACUGACGUGUC-5′ | ||
| siRNA-7 | 214-234 | 5′-CAGAGAGACACACUGCCACUU-3′ |
| 3′-UUGUCUCUCUGUGUGACGGUG-5′ | ||
| siRNA-8 | 242-262 | 5′-UACUGCGACCCCAACCUAGUU-3′ |
| 3′-UUAUGACGCUGGGGUUGGAUC-5′ | ||
| msiRNA-2 | 192-212 | 5′-GCGAAUUCGAUCUCACCUGUU-3′ |
| 3′-UUCGCUUAAGCUAGAGUGGAC-5′ | ||
| siRNA-C | - | 5′-ACUACAAGACUCGUGACCAUU-3′ |
| 3′-UUUGAUGUUCUGAGCACUGGU-5′ |
siRNA anti-hCD40 . | hDC40 mRNA targeted region* . | Sequence . |
|---|---|---|
| siRNA-1 | 173-193 | 5′-UGCCUUCCUUGCGGUGAAAUU-3′ |
| 3′-UUACGGAAGGAACGCCACUUU-5′ | ||
| siRNA-2 | 192-212 | 5′-GCGAAUUCCUAGACACCUGUU-3′ |
| 3′-UUCGCUUAAGGAUCUGUGGAC-5′ | ||
| siRNA-3 | 479-499 | 5′-UGUCACCCUUGGACAAGCUUU-3′ |
| 3′-UUACAGUGGGAACCUGUUCGU-5′ | ||
| siRNA-4 | 709-729 | 5′-UUUUCCCGACGAUCUUCCUUU-3′ |
| 3′-UUAAAAGGGCUGCUAGAAGGA-5′ | ||
| siRNA-5 | 62-82 | 5′-CCACCCACUGCAUGCAGAGUU-3′ |
| 3′-UUGGUGGGUGACGUACGUCUC-3′ | ||
| siRNA-6 | 137-157 | 5′-CUGGUGAGUGACUGCACAGUU-3′ |
| 3′-UUGACCACUCACUGACGUGUC-5′ | ||
| siRNA-7 | 214-234 | 5′-CAGAGAGACACACUGCCACUU-3′ |
| 3′-UUGUCUCUCUGUGUGACGGUG-5′ | ||
| siRNA-8 | 242-262 | 5′-UACUGCGACCCCAACCUAGUU-3′ |
| 3′-UUAUGACGCUGGGGUUGGAUC-5′ | ||
| msiRNA-2 | 192-212 | 5′-GCGAAUUCGAUCUCACCUGUU-3′ |
| 3′-UUCGCUUAAGCUAGAGUGGAC-5′ | ||
| siRNA-C | - | 5′-ACUACAAGACUCGUGACCAUU-3′ |
| 3′-UUUGAUGUUCUGAGCACUGGU-5′ |
Selected sequences were of the type AA(N19)UU (N, any nucleotide) and were submitted to a basic local alignment search tool (BLAST)15 search against the human genome sequence to ensure gene specificity. The siRNA-C, with a scrambled sequence, was used as control for the initial screening on ECV-304 cells. The mismatched siRNA-2 (msiRNA-2), whose sequence was identical to that siRNA-2 except for the underlined 5-nt mismatch mutated by transversion, was used as control in the endothelial cell assays. The 21 nucleotide RNA sequences displayed in each row are arranged by complementary homology yielding the corresponding siRNA duplexes, with 2 unpaired uridine nucleotides at the 3′ end of each strand.
indicates absence of homology with respect to the hCD40 mRNA sequence
Numbers indicate the position of the targeted region within the hCD40 mRNA sequence from ATG initiation codon
RNAi-mediated silencing of endogenous human CD40 expression. (A) Screening for effective siRNA inhibitors of endogenous CD40 expression. ECV-304 cells were transfected for 4 hours with 100 nM of the siRNAs as indicated in “Study Design.” Total cellular RNA was extracted 48 hours after transfection and analyzed by real-time semiquantitative reverse-transcription PCR (RT-PCR). Human CD40 mRNA expression levels were assessed relative to the constitutively expressed cyclophilin gene (ratio of CD40 to cyclophilin mRNA). Data are presented as means and standard error of the mean (SEM) for 3 independent experiments (**P < .01, *P < .05, versus siRNA-C). The lower parts of panel A depict the evaluation of CD40 silencing at the protein level. The siRNA-transfected ECV-304 cells were stimulated with proinflammatory cytokines TNF-α (100 U/mL) and interferon-γ (IFN-γ) (1000 U/mL) 24 hours prior to protein extraction and Western immunoblotting, performed 48 hours after transfection. (B) Potency of siRNA-2. ECV-304 cells were stimulated as previously described and transfected with the indicated concentration of siRNA-2, and the CD40 receptor levels relative to the β-actin protein levels were determined by Western immunoblotting and densitometry by means of Phoretix 10 software (Nonlinear Dynamics, Newcastle upon Tyne, United Kingdom). Data are shown as means and SEM from 2 independent assays. (C) Specific inhibition by siRNA-2 of CD40 expression on human endothelial cells. HUVECs were lipofected for 2 hours with 100 nM siRNA-2 or its corresponding siRNA control, msiRNA-2, complexed with Targefect as described in “Study Design,” and activated with TNF-α (100 U/mL) and IFN-γ (1000 U/mL) 16 hours prior to protein extraction. Representative Western immunoblotting was performed 48 hours after transfection. The housekeeping β-actin protein was included in the Western analyses to normalize for equal loading of the gel lanes. Relative mobility of molecular weight markers is shown in kilodaltons. (D) Predicted secondary structure of human CD40 mRNA. The predicted secondary structure of CD40 mRNA (residues 1-250 from the coding region are displayed) was determined by means of the suboptimal folding program of Michael Zucker, Mfold,16 based on the energy minimization method. The targeting locations from 5 of the 8 designed siRNAs are indicated over the optimal lowest free energy structure.
RNAi-mediated silencing of endogenous human CD40 expression. (A) Screening for effective siRNA inhibitors of endogenous CD40 expression. ECV-304 cells were transfected for 4 hours with 100 nM of the siRNAs as indicated in “Study Design.” Total cellular RNA was extracted 48 hours after transfection and analyzed by real-time semiquantitative reverse-transcription PCR (RT-PCR). Human CD40 mRNA expression levels were assessed relative to the constitutively expressed cyclophilin gene (ratio of CD40 to cyclophilin mRNA). Data are presented as means and standard error of the mean (SEM) for 3 independent experiments (**P < .01, *P < .05, versus siRNA-C). The lower parts of panel A depict the evaluation of CD40 silencing at the protein level. The siRNA-transfected ECV-304 cells were stimulated with proinflammatory cytokines TNF-α (100 U/mL) and interferon-γ (IFN-γ) (1000 U/mL) 24 hours prior to protein extraction and Western immunoblotting, performed 48 hours after transfection. (B) Potency of siRNA-2. ECV-304 cells were stimulated as previously described and transfected with the indicated concentration of siRNA-2, and the CD40 receptor levels relative to the β-actin protein levels were determined by Western immunoblotting and densitometry by means of Phoretix 10 software (Nonlinear Dynamics, Newcastle upon Tyne, United Kingdom). Data are shown as means and SEM from 2 independent assays. (C) Specific inhibition by siRNA-2 of CD40 expression on human endothelial cells. HUVECs were lipofected for 2 hours with 100 nM siRNA-2 or its corresponding siRNA control, msiRNA-2, complexed with Targefect as described in “Study Design,” and activated with TNF-α (100 U/mL) and IFN-γ (1000 U/mL) 16 hours prior to protein extraction. Representative Western immunoblotting was performed 48 hours after transfection. The housekeeping β-actin protein was included in the Western analyses to normalize for equal loading of the gel lanes. Relative mobility of molecular weight markers is shown in kilodaltons. (D) Predicted secondary structure of human CD40 mRNA. The predicted secondary structure of CD40 mRNA (residues 1-250 from the coding region are displayed) was determined by means of the suboptimal folding program of Michael Zucker, Mfold,16 based on the energy minimization method. The targeting locations from 5 of the 8 designed siRNAs are indicated over the optimal lowest free energy structure.
Efficient blockade of CD40-CD154 signaling by anti-CD40 siRNA-2 mitigates proinflammatory events on human endothelial cells
CD40 receptor silencing by siRNA-2 in HUVECs was further recognized by immunoblotting (Figure 1C). Careful optimization of cationic lipid–mediated transfection of cyanine 3 (Cy3)–labeled siRNAs on these cells yielded efficiencies in the range of 50% determined by fluorescence microscopy, with an acceptable toxicity (data not shown). This correlates with the moderate effect of siRNA-2, a 45% inhibition of endogenous CD40 receptor expression, detected in TNF-α/IFN-γ–activated HUVECs.
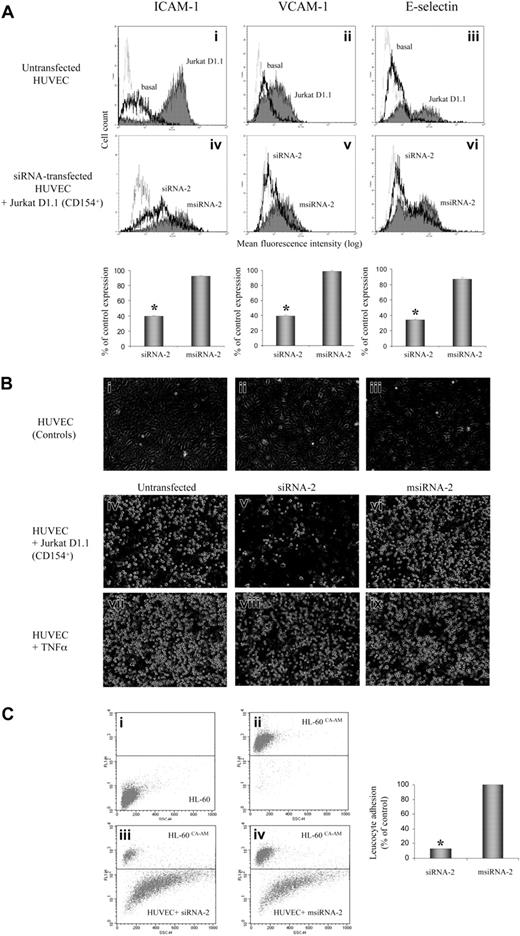
A major step of the immune-mediated inflammatory process, endothelial cell activation via CD40-CD154 interaction, leads to induction of cell surface adhesion molecule expression, which orchestrates the egress of inflammatory cells from the vessel lumen.4-6 Thus, we investigated whether siRNA-2 could interfere with the CD40-CD154 activation pathway in HUVECs. We assessed the specific silencing effect of siRNA-2, compared with its corresponding mismatched control msiRNA-2, analyzing by flow cytometry both CAM expression and leukocyte adhesion in CD154-activated endothelial cells (Figure 2). The siRNA-2–treated, but not control msiRNA-2–treated, HUVECs showed a strong reduction of CAM expression when activated via CD154 (Figure 2A). Moreover, direct evaluation of leukocyte adhesion on CD154-activated HUVECs using HL-60 cells, which express the ligands for all 3 adhesion molecules ICAM-1, VCAM-1, and E-selectin, demonstrated that pretreatment with siRNA-2, but not with msiRNA-2, reduced leukocyte adherence by 87% (Figure 2B-C). Furthermore, the anti-CD40 siRNA-2 had no effect on either CAM expression (data not shown) or leukocyte adhesion induced by TNF-α–mediated HUVEC activation (Figure 2B), demonstrating its inability to interfere with the family member CD120 (TNF-α receptor), and hence its pathway specificity.
Effect of CD40 gene silencing by siRNA-2 on HUVEC leukocyte adhesion. CD40 gene silencing by siRNA-2 prevents CD154-mediated induction of leukocyte adhesion in human endothelial cells. (A) RNAi-mediated inhibition of CAM expression on CD154-activated endothelial cells. HUVECs, either untransfected or transfected for 2 hours with 100 nM siRNA-2 or msiRNA-2 as indicated in “Study Design,” were activated by coincubation with Jurkat D1.1 (CD154+) cells (ATCC CRL 1095; a Jurkat T cell subclone that constitutively expresses CD154) at a T/EC ratio of 5:1 at 6 hours (E-selectin) or 16 hours (ICAM-1 and VCAM-1) prior to analysis. At 48 hours after transfection, the cocultures were washed extensively with warm phosphate-buffered saline (PBS) to release the attached Jurkat D1.1 cells from the HUVEC monolayer. The endothelial cells were analyzed for CAM expression in the MoFlo flow cytometer (DakoCytomation, Fort Collins, CO) and were readily distinguished from residual T cells by light scatter. In i-iii, solid-line histograms represent basal CAM expression from untreated HUVECs, and filled histograms represent CAM expression from D1.1 (CD154+)–stimulated, untreated HUVECs. In iv-vi, solid-line histograms represent CAM expression from D1.1 (CD154+)–stimulated, siRNA-2–transfected HUVECs, and filled histograms represent CAM expression from D1.1 (CD154+)–stimulated, msiRNA-2–transfected HUVECs. Dotted-line histograms display the corresponding immunoglobulin G (IgG) isotype-matching controls in each panel. Bar diagrams quantify the percentage of CAM expression after siRNA treatment, calculated as follows: ((CAM expression for siRNA–treated, D1.1 (CD154+)–induced cells) - (basal CAM expression)) / ((D1.1 (CD154+)–induced CAM expression) - (basal CAM expression)) × 100. CAM expression data are the mean fluorescence intensity (MFI) values of the corresponding histograms from 3 independent experiments (*P < .05, versus msiRNA-2). (B) RNAi-mediated inhibition of leukocyte adhesion on CD154-activated, but not in TNF-α–activated, endothelial cells. HUVECs, either untransfected or transfected for 2 hours with 100 nM siRNA-2 or msiRNA-2 as indicated in “Study Design,” were activated either by coculture with Jurkat D1.1 (CD154+) cells at a T/EC ratio of 5:1 (iv-vi), or by incubation with TNF-α (300 U/mL) (vii-ix) at 16 hours prior to the assay. At 48 hours after transfection, the cocultures were washed extensively with warm PBS to release the attached Jurkat D1.1 cells from the HUVEC monolayer, which was further coincubated with HL-60 cells for 30 minutes (2 × 106 cells per well from 6-well plate, in 2 mL endothelial cell culture medium). Supernatant aspiration plus 2 rounds of brief washing with PBS removed nonadherent cells. Shown are phase-contrast microscopic images of PBS-washed, adherent cells taken at × 40 magnification. The images were acquired as monochromatic JPEG files using a SPOT camera and SPOT 3.2.4 software (Diagnostic Instruments, Sterling Heights, MI) mounted on an Olympus IX-70 inverted microscope (Olympus America, Melville, NY) and linked to an iMac G4 Apple computer (Apple Computers, Cupertino, CA). Controls refer to untreated HUVEC monolayer (i); untreated HUVECs coincubated with HL-60 cells (ii); untreated Jurkat D1.1 (CD154+)–activated HUVECs (iii). (C) To quantify the percentage of leukocyte adhesion to siRNA-2–transfected HUVECs, the same assay as in panel B was performed with the use of calcein AM–loaded HL-60 cells (5 μM calcein AM for 60 minutes at 37°C). Finally, attached cells were harvested by mild trypsinization and the ratio of adherent, fluorescence-labeled HL-60 cells to unlabeled endothelial cells was determined by flow cytometry after counting 10 000 events per sample. Shown are density plots corresponding to the following: unlabeled HL-60 cells (i); calcein AM–loaded HL-60 cells (ii); calcein AM–loaded HL-60 cells plus siRNA-2–transfected HUVEC mix (iii); calcein AM–loaded HL-60 cells plus msiRNA-2–transfected HUVEC mix (iv), all from one representative assay. The mean ratio obtained from msiRNA-2–treated HUVECs was assigned a 100% adhesion. Data are from 5 independent experiments (*P < .05, versus msiRNA-2).
Effect of CD40 gene silencing by siRNA-2 on HUVEC leukocyte adhesion. CD40 gene silencing by siRNA-2 prevents CD154-mediated induction of leukocyte adhesion in human endothelial cells. (A) RNAi-mediated inhibition of CAM expression on CD154-activated endothelial cells. HUVECs, either untransfected or transfected for 2 hours with 100 nM siRNA-2 or msiRNA-2 as indicated in “Study Design,” were activated by coincubation with Jurkat D1.1 (CD154+) cells (ATCC CRL 1095; a Jurkat T cell subclone that constitutively expresses CD154) at a T/EC ratio of 5:1 at 6 hours (E-selectin) or 16 hours (ICAM-1 and VCAM-1) prior to analysis. At 48 hours after transfection, the cocultures were washed extensively with warm phosphate-buffered saline (PBS) to release the attached Jurkat D1.1 cells from the HUVEC monolayer. The endothelial cells were analyzed for CAM expression in the MoFlo flow cytometer (DakoCytomation, Fort Collins, CO) and were readily distinguished from residual T cells by light scatter. In i-iii, solid-line histograms represent basal CAM expression from untreated HUVECs, and filled histograms represent CAM expression from D1.1 (CD154+)–stimulated, untreated HUVECs. In iv-vi, solid-line histograms represent CAM expression from D1.1 (CD154+)–stimulated, siRNA-2–transfected HUVECs, and filled histograms represent CAM expression from D1.1 (CD154+)–stimulated, msiRNA-2–transfected HUVECs. Dotted-line histograms display the corresponding immunoglobulin G (IgG) isotype-matching controls in each panel. Bar diagrams quantify the percentage of CAM expression after siRNA treatment, calculated as follows: ((CAM expression for siRNA–treated, D1.1 (CD154+)–induced cells) - (basal CAM expression)) / ((D1.1 (CD154+)–induced CAM expression) - (basal CAM expression)) × 100. CAM expression data are the mean fluorescence intensity (MFI) values of the corresponding histograms from 3 independent experiments (*P < .05, versus msiRNA-2). (B) RNAi-mediated inhibition of leukocyte adhesion on CD154-activated, but not in TNF-α–activated, endothelial cells. HUVECs, either untransfected or transfected for 2 hours with 100 nM siRNA-2 or msiRNA-2 as indicated in “Study Design,” were activated either by coculture with Jurkat D1.1 (CD154+) cells at a T/EC ratio of 5:1 (iv-vi), or by incubation with TNF-α (300 U/mL) (vii-ix) at 16 hours prior to the assay. At 48 hours after transfection, the cocultures were washed extensively with warm PBS to release the attached Jurkat D1.1 cells from the HUVEC monolayer, which was further coincubated with HL-60 cells for 30 minutes (2 × 106 cells per well from 6-well plate, in 2 mL endothelial cell culture medium). Supernatant aspiration plus 2 rounds of brief washing with PBS removed nonadherent cells. Shown are phase-contrast microscopic images of PBS-washed, adherent cells taken at × 40 magnification. The images were acquired as monochromatic JPEG files using a SPOT camera and SPOT 3.2.4 software (Diagnostic Instruments, Sterling Heights, MI) mounted on an Olympus IX-70 inverted microscope (Olympus America, Melville, NY) and linked to an iMac G4 Apple computer (Apple Computers, Cupertino, CA). Controls refer to untreated HUVEC monolayer (i); untreated HUVECs coincubated with HL-60 cells (ii); untreated Jurkat D1.1 (CD154+)–activated HUVECs (iii). (C) To quantify the percentage of leukocyte adhesion to siRNA-2–transfected HUVECs, the same assay as in panel B was performed with the use of calcein AM–loaded HL-60 cells (5 μM calcein AM for 60 minutes at 37°C). Finally, attached cells were harvested by mild trypsinization and the ratio of adherent, fluorescence-labeled HL-60 cells to unlabeled endothelial cells was determined by flow cytometry after counting 10 000 events per sample. Shown are density plots corresponding to the following: unlabeled HL-60 cells (i); calcein AM–loaded HL-60 cells (ii); calcein AM–loaded HL-60 cells plus siRNA-2–transfected HUVEC mix (iii); calcein AM–loaded HL-60 cells plus msiRNA-2–transfected HUVEC mix (iv), all from one representative assay. The mean ratio obtained from msiRNA-2–treated HUVECs was assigned a 100% adhesion. Data are from 5 independent experiments (*P < .05, versus msiRNA-2).
A recently completed preclinical trial has demonstrated the safety and efficacy of an siRNA therapeutic agent against vascular endothelial growth factor in a validated primate model of age-related macular degeneration.21 The anti-CD40 siRNA is expected to have significant advantages over alternative approaches for CD40-CD154 costimulatory pathway blockade based on ligand/receptor binding, such as anti-CD40 or anti-CD154 antibodies, or the soluble CD40 form CD40Ig.22 By activating the enzymatic RNAi mechanism, one anti-CD40 siRNA can destroy hundreds of targeted CD40 mRNAs, which will result in the silencing of potentially thousands of CD40 protein receptors. Conversely, systemic administration of therapeutic antibodies or recombinant protein fusions may induce undesired immune responses against the foreign protein by the host or result in serious side effects such as thromboembolism.10,23 The precise mechanism leading to the prothrombotic effects of anti-CD154 antibodies is not known, although it has been related either to inhibition of CD154-β3 platelet integrin interaction24 or to their direct interaction with platelet Fc receptors. The anti-CD40 siRNA may become instrumental for discarding the involvement of CD40 signaling in anti-CD154 antibody-mediated thromboembolism. In all, targeted delivery of anti-CD40 siRNA to specific CD40-expressing cells should avert these unwanted effects while exerting a powerful upstream down-regulation of the CD40 pathway.
Prepublished online as Blood First Edition Paper, August 17, 2004; DOI 10.1182/blood- 2004-03-0817.
Supported by grants 01/0097 and 03/0516 from the Fondo de Investigaciones Sanitarias (FIS); grant 00/4031 from Fundació La Marató de TV3; and grants from the Red de Centros del Instituto de Salud Carlos III (refs C03/03 and C03/07).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal