We previously identified a guanosine triphosphatase (GTPase)–activating protein (GAP) male germ cell Rac GAP (MgcRacGAP) that enhanced interleukin-6 (IL-6)–induced macrophage differentiation of murine M1 leukemia cells. Later, MgcRacGAP was found to play crucial roles in cell division. However, how MgcRacGAP enhanced IL-6–induced differentiation remained elusive. Here we show that MgcRacGAP enhances IL-6–induced differentiation through enhancement of signal transducer and activator of transcription–3 (STAT3) activation. MgcRacGAP, Rac, and STAT3 formed a complex in IL-6–stimulated M1 cells, where MgcRacGAP interacted with Rac1 and STAT3 through its cysteine-rich domain and GAP domain. In reporter assays, the wild-type MgcRacGAP enhanced transcriptional activation of STAT3 while a GAP-domain deletion mutant (ΔGAP) did not significantly enhance it, suggesting that the GAP domain was required for enhancement of STAT3-dependent transcription. Intriguingly, M1 cells expressing ΔGAP had no effect on the differentiation signal of IL-6, while forced expression of MgcRacGAP rendered M1 cells hyperresponsive to the IL-6–induced differentiation. Moreover, knockdown of MgcRacGAP by RNA interference profoundly suppressed STAT3 activation, implicating MgcRacGAP in the STAT3-dependent transcription. All together, our data not only reveal an important role for MgcRacGAP in STAT3 activation, but also demonstrate that MgcRacGAP regulates IL-6–induced cellular differentiation in which STAT3 plays a pivotal role.
Introduction
The signal transducer and activator of transcription (STAT) family members STAT1-4, STAT5A, STAT5B, and STAT6 are activated through phosphorylation by the Janus kinase (JAK) family upon cytokine stimulation. The phosphorylated STATs form homodimers or heterodimers and translocate into the nucleus, where they regulate expression of their target genes.1-4 Among them, STAT3 is activated mainly by the interleukin-6 (IL-6) family cytokines including IL-6, oncostatin M, and leukemia inhibitory factor (LIF), and is implicated in a wide range of biologic processes, including nephrogenesis, gliogenesis, hepatogenesis, T-cell proliferation, inflammation, and oncogenesis.5-11 The critical role of STAT3 in myeloid differentiation was demonstrated by the use of dominant negative mutants.12-14 In contrast, in embryonic stem cells, STAT3 is required for self-renewal.15-19 In addition, STAT3 is activated in a broad spectrum of human hematologic malignancies.20 STAT3 can also be negatively regulated. Among the known inhibitors of STAT proteins are the suppressor of cytokine signaling (SOCS) proteins,21 also known as Janus kinase binding (JAB) proteins22 or STAT-induced STAT inhibitors (SSIs).23 While SOCS proteins interact with JAKs and reduced their tyrosine kinase activity,21-23 a STAT3 inhibitor protein inhibitor of activated STAT3 (PIAS3) directly binds to STAT3 and inhibits its activity.24 A zinc finger protein Gfi-1 enhances STAT3 signaling by preventing this binding of PIAS3 to STAT3.25 Several other molecules have been found to interact with activated STAT3. Among them, cellular Jun (c-Jun) forms a complex with STAT3 and activates the α2-macroglobulin promoter that contains both STAT3- and c-Jun–binding sites.26,27 Like many other transcription factors, STAT3 associates with a transcriptional cofactor, cAMP response element binding protein–binding protein/p300 (CBP/p300), to form a transcriptional complex.28 A protein called gene-associated with retinoid interferon-induced mortality (GRIM)–19 suppresses STAT3 activity through cytoplasmic retention of STAT3, whereas an endothelial cell–derived zinc finger protein (EZI) enhances STAT3 activity through nuclear retention of STAT3.29,30 There may be more such regulators in various steps of STAT3 action, such as translocation to the nucleus, induction of chromatin remodeling, and proteolysis.
In a search for key molecules that prevent IL-6–induced terminal differentiation of murine myeloid leukemia M1 cells, we identified an antisense DNA for the full-length form of male germ cell Rac guanosine triphosphatase–activating protein (MgcRacGAP) through functional cloning.31 An N-terminus-truncated form of MgcRacGAP had been isolated and named male germ cell Rac GAP, as its expression was highest in testis.32 Rac, Cdc42, and RhoA, the Rho family of small guanosine triphosphates (GTPases), play pleiotropic roles in a variety of cell functions such as transformation, migration, cytokinesis, and transcriptional activation.33-35 Recently, MgcRacGAP and Cyk4, a counterpart of Caenorhabditis elegans, have been proven to be essential for cell cycle progression, in particular completion of cytokinesis.36-40 MgcRacGAP localized to the mitotic spindles in metaphase and accumulates to the midbody in cytokinesis.36-38 MgcRacGAP associated with microtubules through its N-terminal myosinlike domain, and overexpression of the N-terminal domain–deletion mutant or a GAP activity–defective mutant of MgcRacGAP (R385A) halted cell division and led to the formation of multinucleated cells.37 Gene depletion of MgcRacGAP in mice lead to death during preimplantation development caused by impaired mitosis and cytokinesis with binucleated blastomeres in which the nuclei were partially interconnected.38 Thus, available data indicated that MgcRacGAP plays essential roles in the mitotic (M) phase, especially in cytokinesis, through association with the microtubules. We recently demonstrated that a serine/threonine kinase Aurora B phosphorylated MgcRacGAP at the midbody, thereby inducing its latent GAP activity toward RhoA during mitosis and that MgcRacGAP associated with RhoA on the contractile ring. We also demonstrated that the Aurora B–induced MgcRacGAP phosphorylation at Ser387 was essential for its RhoGAP activity and for the completion of cytokinesis.40
In interphase, MgcRacGAP localizes in both the nucleus and cytoplasm, and the biologic functions of MgcRacGAP in the interphase remained elusive. Although overexpression of the antisense cDNA for MgcRacGAP efficiently inhibited the IL-6–induced differentiation of M1 cells,31 the underlying molecular mechanism was not clear. In this study, we demonstrate that overexpression of MgcRacGAP rendered M1 cells hyperresponsive to IL-6–induced differentiation and that MgcRacGAP and STAT3 functionally associate with each other in vivo and in vitro. Moreover, MgcRacGAP was required for the transcriptional activation of STAT3. Thus, in addition to the critical role in completion of cytokinesis in the interphase, MgcRacGAP also plays a distinct role in transcriptional activation of STAT3 in IL-6–induced differentiation of M1 cells.
Materials and methods
Culture, cytokines, and antibodies
The M1, HeLa, and 293T cells were grown in Dulbecco modified Eagle medium (DMEM) (GIBCO, Grand Island, NY) containing 10% fetal calf serum. An affinity-purified anti-MgcRacGAP antibody (Ab) was produced, as described.37 The anti-Rac1 monoclonal Ab (mAb) was purchased from Transduction Laboratories (Newington, NH). The anti-Rac2 Ab, anti-STAT3 Ab (C-20), anti-STAT3 mAb (F-2), antiphospho-STAT3 Ab (B-7), anti–mitotic kinesin-like protein (anti-MKLP) Ab, and anti-hemagglutinin (anti-HA) Ab (Y-11) were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-Flag (M2) mAb was purchased from Sigma (St Louis, MO). Recombinant human IL-6 and soluble IL-6R (sIL-6R) were obtained from R&D Systems (Minneapolis, MN).
Retrovirus vectors
A bicistronic retrovirus vector pMX-IRES-EGFP (pMX-IG) was constructed to transduce a gene together with an enhanced green fluorescent protein (EGFP).41 A complementary DNA for the dominant negative STAT3-Y705F (STAT3F), the Flag-tagged full-length MgcRacGAP (FL), and the deletion mutant of MgcRacGAP lacking the GAP domain (ΔGAP) or Cys domain (ΔCys) were inserted into EcoRI and NotI sites of the pMX-IG to construct pMX-IG/STAT3F, pMX-IG/FL, pMX-IG/ΔGAP, or pMX-IG/ΔCys, respectively.
Production of retrovirus and virus infection
High-titer retroviruses carrying STAT3F, FL, ΔGAP, or ΔCys were produced in a transient retrovirus-packaging cell line PLAT-E.42 Briefly, PLAT-E cells were cotransfected with 3 μg of each retrovirus vector plasmid with the use of FuGene6 Transfection Reagent (Roche Molecular Diagnostics, Indianapolis, IN). At 48 hours after transfection, the supernatants were harvested as viral stock solutions. For infection, M1 cells (1 × 106) were incubated for 6 hours with 10 mL supernatants in the presence of 10 μg/mL hexadimethrine bromide (Sigma). First, 10 mL fresh growth medium was added to the culture, and incubation was continued for 18 hours. The cells were resuspended with growth medium and allowed to grow for another 24 hours before the cells were sorted.
Cell sorting and flow cytometry
Briefly, 2 days after virus infection, cells were washed twice with phosphate-buffered saline (PBS) and suspended in PBS containing 1% bovine serum albumin (BSA). The infected cells were sorted on the basis of GFP expression on FACS Vantage (Becton Dickinson, San Jose, CA). The sorted cells (1 × 104) were resuspended in growth medium, and cultured for 5 days (7 days after virus infection). Half of the sorted population was used to confirm GFP expression by means of FACS Calibur (Becton Dickinson), and the other half was expanded and used for further analysis.
Immunoprecipitation and Western blotting
Immunoprecipitation, gel electrophoresis, and immunoblotting were performed as described,41 but with minor modifications. Exponentially growing cells were lysed in a buffer (0.5% Triton X-100, 50 mM Tris-HCl [tris(hydroxymethyl)aminomethane-HCl] [pH 7.5], 0.1 mM EDTA [ethylenediaminetetraacetic acid], 150 mM NaCl, 200 μMNa3VO4,50mMNaF, 1 mM dithiothreitol [DTT], 0.4 mM phenylmethylsulfonyl fluoride [PMSF], 3 μg/mL aprotinin, 2 μg/mL pepstatin A, 1 μg/mL leupeptin) (1 × 107 per milliliter cells), and incubated on ice for 30 minutes. Cell lysates were clarified by centrifugation for 15 minutes at 12 000g prior to incubation at 4°C for 2 hours with the anti-Rac2 (or Rac1) Ab, anti-STAT3 Ab, or the control rabbit whole immunoglobulin G (IgG), and protein A–sepharose (Amersham, Arlington Heights, IL). The immunoprecipitates were subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and electrophoretically transferred onto Immobilon filters (Millipore, Billerica, MA). After blocking in a solution containing 5% BSA, the filter was probed with an anti-STAT3 Ab or anti-MgcRacGAP Ab.
Preparation of recombinant MBP-fusion proteins and MBP pull-down assays
A full-length cDNA for human MgcRacGAP (FL), either of myosin-like domain (Myo), internal domain (INT), cystine-rich domain (Cys), or GAP domain (GAP), was inserted 3′ of and in frame to the maltose binding protein (MBP) coding sequence in a bacterial expression vector pMal (New England Biolabs, Beverly, MA). Similarly, each of the murine STAT3 domains was inserted into pMal. The integrity of each coding sequence was confirmed by DNA sequencing. Escherichia coli BL21 (DE3) cells harboring the recombinant expression vectors were induced with 1 mM isopropyl β-D-thiogalactopyranoside (IPTG) (American Bioanalytical, Natick, MA) at 25°C for 4 hours. Cells suspended in 4 mL ice-cold suspension buffer (20 mM Tris-HCL, 200 mM NaCl, 1 mM EDTA, 1 mM DTT, and 2 mM PMSF). The cell suspension was sonicated, and insoluble debris was pelleted by centrifugation (12 000g for 15 minutes at 4°C). The supernatants were mixed with amylose resin beads (New England Biolabs) at 4°C for 30 minutes. The beads were washed 3 times and resuspended in 1 mL suspension buffer. The purity and quantity of bound MBP-fusion proteins were examined by means of SDS-PAGE followed by Coomassie blue staining. A similar amount of MBP fusion proteins bound to amylose resin beads was incubated for the time indicated with 1 mL cell lysates (1 × 107/mL) from IL-6–stimulated (50 ng/mL) or unstimulated M1 cells. The pull-down binding reaction was done for 30 minutes at 4°C in 500 μL binding buffer (0.5% Triton X-100, 50 mM Tris-HCl [pH 7.5], 0.1 mM EDTA, 150 mM NaCl, 5 mM MgCl2, 200 μMNa3VO4,50mMNaF,1mM DTT, 0.4 mM PMSF, 3 μg/mL aprotinin, 2 μg/mL pepstatin A, 1 μg/mL leupeptin). The samples were resolved with the use of SDS-PAGE, followed by Western blotting with an anti-STAT3 Ab or an anti-MgcRacGAP Ab. For some experiments, the blots were stripped of bound antibodies and reprobed with anti-Rac1 mAb or anti-MKLP Ab.
Immunostaining
M1 cells, with or without stimulation of IL-6 (50 ng/mL for 12 hours), were plated on glass coverslips and fixed with 4% paraformaldehyde/PBS for 20 minutes at room temperature. The cells were then washed 3 times with ice-cold PBS followed by a 10-minute incubation at room temperature in PBS containing 0.1% Nonidet P-40. The anti-MgcRacGAP and anti-STAT3 (F-2) mAb were diluted in PBS containing 3% bovine serum albumin, placed as a drop on the coverslips, and incubated for 1 hour. The coverslips were incubated with a solution containing fluorescein isothiocyanate (FITC)–conjugated goat anti-rabbit IgG (Wako Pure Chemical Industries, Osaka, Japan) and Rhodamine-conjugated goat anti-mouse IgG (Sigma) for 1 hour. The coverslips were mounted with glycerin containing paraphenylenediamine (PPD) at 10 mg/mL and 4′,6-diamino-2-phenylindole (DAPI) at 1μg/mL for 30 minutes, then viewed by means of a fluorescence microscope IX70 (Olympus, Tokyo, Japan) equipped with SenSys/OL cold charge-coupled device (CCD) camera (Olympus) and IP-Lab software (Signal Analytics, Vienna, VA). The objective lens used was an LCPlanFI × 40/0.60 (Olympus).
Generation, expression, and purification of MgcRacGAP recombinant protein in Sf-9 cells
The cDNA encoding MgcRacGAP with the C-terminal Flag epitope tag was subcloned into the baculovirus transfer vector pBacPAK (BD Biosciences, San Jose, CA). The resulting construct was used to acquire recombinant baculoviruses by cotransfection with Bsu36 I–digested BacPAK viral DNA (BD Biosciences) according to the manufacturer's protocol. For protein expression, Sf-9 cells were infected with high-titer viral stocks for 96 hours and lysed in lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1.0% Nonidet P-40, 1 mM EDTA, 0.2 mM Na3Vo4, 2 mM PMSF, 2 μg/mL leupeptin, 10 μg/mL aprotinin). The lysate was clarified by centrifugation, and the supernatant was immunoprecipitated with the anti-Flag M2-agarose-affinity gel (Sigma) for 2 hours at 4°C. The agarose beads were washed 3 times with the lysis buffer, and the recombinant MgcRacGAP was eluted with 3 × Flag fusion protein (Sigma). To confirm the purity, the eluted MgcRacGAP was subjected to SDS-PAGE, followed by Coomassie blue staining (data not shown).
Luciferase reporter assay
HeLa cells were transfected with 0.6 μg pME, pME/FL, pME/ΔGAP, or pME/ΔCys together with 0.6 μg reporter plasmid carrying a firefly luciferase gene driven by the glial fibrillary acidic protein (GFAP) promoter43 and 0.6 μg internal control reporter plasmid with the Rous sarcoma virus long-terminal repeat promoter by means of Lipofectamine Plus Reagents (Life Technologies, Bethesda, MD). At 24 hours after transfection, cells were stimulated with IL-6 (20 ng/mL) and sIL-6R (20 ng/mL) for 12 hours or left untreated before cell lysates were prepared. Cell lysates were then subjected to a dual luciferase reporter system (Promega, Madison, WI). Transfection efficiency was normalized with Renilla luciferase activity.
RNA interference for MgcRacGAP
Expression of MgcRacGAP was selectively suppressed by means of the RNA interference method, as described.39 We used CCUCUUCUGACCUUUCGCC as a target sequence of MgcRacGAP and GCCCUCUUGUACUUCCCCU as a scrambled sequence, and 293T cells were incubated with the MgcRacGAP siRNA (10 nM) with the use of Lipofectamine 2000 (Life Technologies). After 48 hours, cells were subjected to the reporter assay.
Results
Overexpression of the sense cDNA for MgcRacGAP renders M1 cells hyperresponsive to IL-6–induced macrophage differentiation
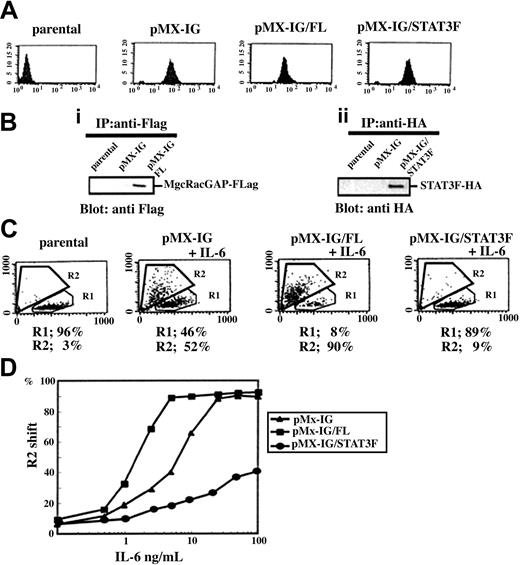
Expression of the antisense cDNA for MgcRacGAP significantly inhibited the IL-6–induced macrophage differentiation of murine myeloid leukemia M1 cells.31 However, how MgcRacGAP was involved in the IL-6–mediated cellular responses was not determined. To investigate the role of MgcRacGAP in IL-6–mediated cell differentiation, we overexpressed MgcRacGAP (FL) and, as a control, a dominant negative mutant STAT3 (STAT3F) in M1 cells using pMX-IRES-GFP (pMX-IG). After transduction of M1 cells with these vectors, GFP+ cells were collected by means of a cell sorter, and we confirmed that most of the sorted cells expressed GFP at high levels (Figure 1A). The expression of Flag-tagged FL and HA-tagged STAT3F was also confirmed in Western blot analysis (Figure 1B). Since overexpression of the FL alone did not induce detectable differentiation of M1 cells, we asked if it renders M1 cells more sensitive to IL-6–induced differentiation. The sorted cells were cultured for 4 days in the presence of 5 ng/mL IL-6. Flow cytometric analysis was done to quantify morphologic changes that occurred after the culture. Increase in cell size and granule content of the cytoplasm were evaluated on the basis of the increase in forward scatter (FSC) and side scatter (SSC), respectively (Figure 1C). After treatment with IL-6, while 52% of M1 cells transduced with the control vector pMX-IG showed a shift from region R1 to region R2, a hallmark of macrophage differentiation, 90% of the M1 cells transduced with pMX-IG/FL showed similar shifts. Conversely, only 9% of M1 cells transduced with pMX-IG/STAT3F shifted from region R1 to region R2. To further confirm the positive effect of MgcRacGAP overexpression in the induction of M1 differentiation, we also measured percentages of the cells that underwent differentiation in response to various concentrations of IL-6. Most of the M1 cells transduced with pMX-IG/FL underwent differentiation in response to 5 ng/mL IL-6, while higher concentrations of IL-6 (25 to 50 ng/mL) were required to achieve similar levels of differentiation in the control M1 cells. On the other hand, a dominant negative STAT3F potently inhibited the differentiation of M1 cells even at the higher concentrations of IL-6, up to 100 ng/mL (Figure 1D). These results indicated that the overexpression of MgcRacGAP rendered M1 cells hyperresponsive to the IL-6 and confirmed that STAT3F strongly suppressed the IL-6–induced macrophage differentiation of M1 cells, as reported.14
Effect of MgcRacGAP on M1 cell sensitivity to IL-6–induced differentiation. MgcRacGAP renders M1 cells more sensitive to the IL-6–induced differentiation signal. (A) Quantitation of GFP expression in the M1 cells at 4 days after retroviral gene transduction on flow cytometry in pMX-IG, pMX-IG/FL, or pMX-IG/STAT3F. The x-axis indicates fluorescence intensity as a log scale ranging from 100 to 104. The y-axis indicates the number of the cells. Parental M1 cells were used as a control. (B) Expression of the Flag-tagged FL and HA-tagged STAT3F in M1 transfectants. Cell lysates from parental M1, M1/pMX-IG, and M1/pMX-IG/FL cells (1 × 107 per lane) were immunoprecipitated and examined by means of immunoblotting and an anti-Flag M2 monoclonal antibody (i). Cell lysates from parental M1, M1/pMX-IG, and M1/pMX-IG/FL cells (1 × 107 per lane) were immunoprecipitated with the use of the anti-HA monoclonal antibody (12CA5) and immunoblotted with anti-HA rabbit polyclonal Ab (ii). (C) Quantitation of cell differentiation on flow cytometry in unstimulated parental M1 cells and M1 cells expressing pMX-IG, pMX-IG/FL, or pMX-IG/STAT3F at 4 days after IL-6 treatment (5 ng/mL). Differentiated M1 cells were detected in region R2. (D) M1 cells expressing pMX-IG, pMX-IG/FL, or pMX-IG/STAT3F were incubated for 4 days with various concentrations of IL-6. The cells were then analyzed for the differentiation on flow cytometry. The percentages of differentiated cells were evaluated by the percentages of the cells in the R2 region. The result shown is representative of 3 experiments.
Effect of MgcRacGAP on M1 cell sensitivity to IL-6–induced differentiation. MgcRacGAP renders M1 cells more sensitive to the IL-6–induced differentiation signal. (A) Quantitation of GFP expression in the M1 cells at 4 days after retroviral gene transduction on flow cytometry in pMX-IG, pMX-IG/FL, or pMX-IG/STAT3F. The x-axis indicates fluorescence intensity as a log scale ranging from 100 to 104. The y-axis indicates the number of the cells. Parental M1 cells were used as a control. (B) Expression of the Flag-tagged FL and HA-tagged STAT3F in M1 transfectants. Cell lysates from parental M1, M1/pMX-IG, and M1/pMX-IG/FL cells (1 × 107 per lane) were immunoprecipitated and examined by means of immunoblotting and an anti-Flag M2 monoclonal antibody (i). Cell lysates from parental M1, M1/pMX-IG, and M1/pMX-IG/FL cells (1 × 107 per lane) were immunoprecipitated with the use of the anti-HA monoclonal antibody (12CA5) and immunoblotted with anti-HA rabbit polyclonal Ab (ii). (C) Quantitation of cell differentiation on flow cytometry in unstimulated parental M1 cells and M1 cells expressing pMX-IG, pMX-IG/FL, or pMX-IG/STAT3F at 4 days after IL-6 treatment (5 ng/mL). Differentiated M1 cells were detected in region R2. (D) M1 cells expressing pMX-IG, pMX-IG/FL, or pMX-IG/STAT3F were incubated for 4 days with various concentrations of IL-6. The cells were then analyzed for the differentiation on flow cytometry. The percentages of differentiated cells were evaluated by the percentages of the cells in the R2 region. The result shown is representative of 3 experiments.
Rac, STAT3, and MgcRacGAP form a complex in hematopoietic M1 cells
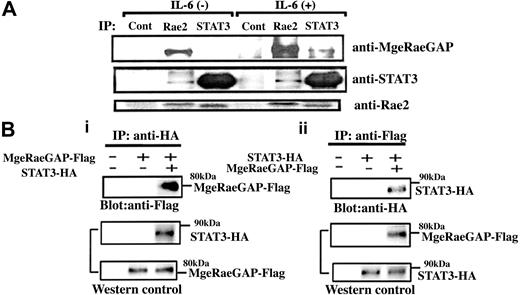
To understand how MgcRacGAP enhanced the IL-6–induced cellular differentiation, we next sought to identify molecules interacting with MgcRacGAP in the IL-6 signaling pathway. STAT3, which plays a central role in the IL-6–induced differentiation of M1 cells,17 directly binds to Rac1,44 and MgcRacGAP directly binds to, and serves as a GAP against Rac1, Rac2, and Cdc42 in vitro.31,32 Rac2 is 92% homologous to Rac1 and is highly expressed in hemopoietic cells, while Rac1 expression is ubiquitous.45,46 To determine if there are interactions among STAT3, Rac2, and MgcRacGAP, we performed the immunoprecipitation in M1 cells; we precipitated the endogenous proteins from M1 cells before and after IL-6 stimulation with anti-STAT3 Ab, anti-Rac2 Ab, or a control Ab. Both STAT3 and MgcRacGAP were coimmunoprecipitated with Rac2 in the cell lysate of M1 cells with or without IL-6 (Figure 2A). We also confirmed that STAT3 and MgcRacGAP coimmunoprecipitated with Rac1 (data not shown). In addition, MgcRacGAP coimmunoprecipitated with STAT3 in the cell lysate of M1 cells stimulated with IL-6. While the pre-existing association between MgcRacGAP and Rac2 was enhanced by IL-6, the association between MgcRacGAP and STAT3 seemed to be IL-6 dependent (Figure 2A). To confirm the binding using a different set of antibodies, 293T cells were transfected with Flag-tagged MgcRacGAP and either HA-tagged STAT3 or a control vector. Flag-tagged MgcRacGAP was found in anti-HA immunoprecipitates when HA-tagged STAT3 was coexpressed (Figure 2Bi, upper panel); conversely, HA-tagged STAT3 was detected in anti-Flag immunoprecipitates when Flag-tagged MgcRacGAP was coexpressed (Figure 2Bii, upper panel). All together, these results demonstrated that STAT3 and MgcRacGAP associated with each other in vivo.
In vivo interaction of MgcRacGAP with STAT3 and Rac GTPases. (A) Coprecipitation of STAT3, MgcRacGAP, and Rac2. M1 cells were incubated in the presence or absence of 50 ng/mL for 15 minutes, and the cell lysates were subjected to immunoprecipitation with anti-STAT3, anti-Rac2, and a control Ab, followed by the immunoblotting with anti-MgcRacGAP, anti-STAT3, or anti-Rac2 Ab. (B) (i) Coprecipitation of MgcRacGAP with STAT3 in 293T cells transfected with Flag-tagged MgcRacGAP and either the empty vector or HA-tagged STAT3. Cell lysates were immunoprecipitated with anti-HA, and immunoblotted with anti-Flag (top panel). Levels of transfected STAT3-HA and MgcRacGAP-Flag were assayed by blotting with anti-HA and anti-Flag (middle and bottom panels). Cells transfected with the empty vectors alone were used as a negative control. (ii) Coprecipitation of STAT3 with MgcRacGAP in 293T cells transfected with STAT3-HA and either the empty vector or MgcRacGAP-Flag. Cell lysates were immunoprecipitated with anti-Flag and immunoblotted with anti-HA (top panel). Levels of transfected MgcRacGAP-Flag and STAT3-HA were assayed by blotting with anti-Flag and anti-HA (middle and bottom panels). Cells transfected with the empty vector alone were also used as a negative control.
In vivo interaction of MgcRacGAP with STAT3 and Rac GTPases. (A) Coprecipitation of STAT3, MgcRacGAP, and Rac2. M1 cells were incubated in the presence or absence of 50 ng/mL for 15 minutes, and the cell lysates were subjected to immunoprecipitation with anti-STAT3, anti-Rac2, and a control Ab, followed by the immunoblotting with anti-MgcRacGAP, anti-STAT3, or anti-Rac2 Ab. (B) (i) Coprecipitation of MgcRacGAP with STAT3 in 293T cells transfected with Flag-tagged MgcRacGAP and either the empty vector or HA-tagged STAT3. Cell lysates were immunoprecipitated with anti-HA, and immunoblotted with anti-Flag (top panel). Levels of transfected STAT3-HA and MgcRacGAP-Flag were assayed by blotting with anti-HA and anti-Flag (middle and bottom panels). Cells transfected with the empty vectors alone were used as a negative control. (ii) Coprecipitation of STAT3 with MgcRacGAP in 293T cells transfected with STAT3-HA and either the empty vector or MgcRacGAP-Flag. Cell lysates were immunoprecipitated with anti-Flag and immunoblotted with anti-HA (top panel). Levels of transfected MgcRacGAP-Flag and STAT3-HA were assayed by blotting with anti-Flag and anti-HA (middle and bottom panels). Cells transfected with the empty vector alone were also used as a negative control.
Delineation of the regions of MgcRacGAP and STAT3 that mediate their interaction
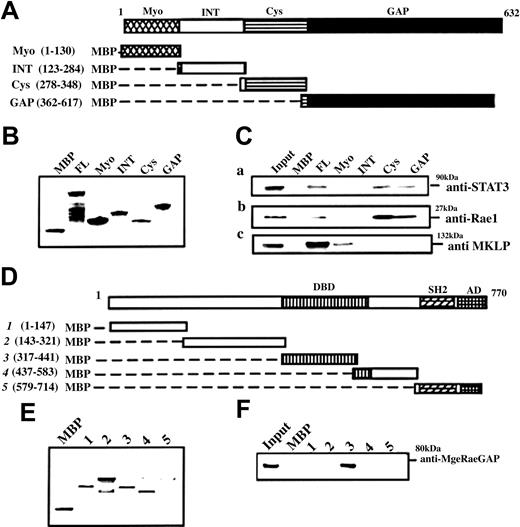
To map the interacting domains of MgcRacGAP with STAT3, we made a series of truncation constructs or FL of MgcRacGAP fused with MBP (Figure 3A), and performed the pull-down assay. Similar amounts of MBP-MgcRacGAP fusion proteins bound to amylose resin beads (Figure 3B) were incubated with cell lysates of the IL-6–stimulated M1 cells (50 ng/mL IL-6 for 30 minutes), and the retained proteins were analyzed by immunoblotting with anti-STAT3 Ab and anti-Rac1 Ab. Both STAT3 and Rac1 bound to the Cys and GAP domain as well as the FL of MgcRacGAP (Figure 3Ci-ii). MKLP, which interacts with the Myo domain (Myo) of MgcRacGAP,39 was used as a positive control (Figure 3Ciii). We next sought to define the binding domain of STAT3 to MgcRacGAP and prepared MBP-fused STAT3 truncations (Figure 3D). A similar amount of MBP-STAT3 truncations bound to beads (Figure 3E), and it was clear that DBD of STAT3 bound MgcRacGAP under a stringent condition using 0.5% Triton-X (Figure 3F). The activation domain of STAT3 could also weakly bind MgcRacGAP in a nonstringent condition using 0.1% Triton-X (data not shown). These results raise a possibility that both MgcRacGAP and STAT3 harbor 2 domains interacting with each other.
MgcRacGAP interactions at the Cys-rich and GAP domains with the DNA-binding domains of STAT3. STAT3 and Rac1 interact with MgcRacGAP at the Cys-rich domain and GAP domain, while MgcRacGAP interacts with the DNA-binding domain of STAT3. (A) Schematic diagram of various MBP-MgcRacGAP fusion proteins. (B) Purity and quantity of MBP and various MBP-MgcRacGAP fusion proteins were examined by means of SDS-PAGE followed by Coomassie blue staining. (C) Two regions required for MgcRacGAP-STAT3 and MgcRacGAP-Rac1 interactions. Lysates from IL-6–treated M1 (50 ng/mL for 30 minutes) were incubated with similar amounts of different MBP or MBP-MgcRacGAP fusion proteins bound to beads. Bound proteins were separated on SDS-PAGE and immunoblotted with anti-STAT3 Ab (i), anti-Rac1 Ab (ii), or anti-MKLP Ab (iii). (D) Schematic diagram of various MBP-STAT3 truncations. DBD and AD represent DNA-binding domain and activation domain, respectively. (E) The purity and quantity of MBP and various MBP-STAT3 truncations were examined by means of SDS-PAGE followed by Coomassie blue staining. (F) A region required for STAT3-MgcRacGAP interaction. Lysates from IL-6–treated M1 (50 ng/mL for 30 minutes) were incubated with a similar amount of different MBP or MBP-STAT3 truncations bound to beads. Bound proteins were separated on SDS-PAGE and immunoblotted with anti-MgcRacGAP Ab.
MgcRacGAP interactions at the Cys-rich and GAP domains with the DNA-binding domains of STAT3. STAT3 and Rac1 interact with MgcRacGAP at the Cys-rich domain and GAP domain, while MgcRacGAP interacts with the DNA-binding domain of STAT3. (A) Schematic diagram of various MBP-MgcRacGAP fusion proteins. (B) Purity and quantity of MBP and various MBP-MgcRacGAP fusion proteins were examined by means of SDS-PAGE followed by Coomassie blue staining. (C) Two regions required for MgcRacGAP-STAT3 and MgcRacGAP-Rac1 interactions. Lysates from IL-6–treated M1 (50 ng/mL for 30 minutes) were incubated with similar amounts of different MBP or MBP-MgcRacGAP fusion proteins bound to beads. Bound proteins were separated on SDS-PAGE and immunoblotted with anti-STAT3 Ab (i), anti-Rac1 Ab (ii), or anti-MKLP Ab (iii). (D) Schematic diagram of various MBP-STAT3 truncations. DBD and AD represent DNA-binding domain and activation domain, respectively. (E) The purity and quantity of MBP and various MBP-STAT3 truncations were examined by means of SDS-PAGE followed by Coomassie blue staining. (F) A region required for STAT3-MgcRacGAP interaction. Lysates from IL-6–treated M1 (50 ng/mL for 30 minutes) were incubated with a similar amount of different MBP or MBP-STAT3 truncations bound to beads. Bound proteins were separated on SDS-PAGE and immunoblotted with anti-MgcRacGAP Ab.
Augmentation of the association of MgcRacGAP with STAT3 upon IL-6 stimulation
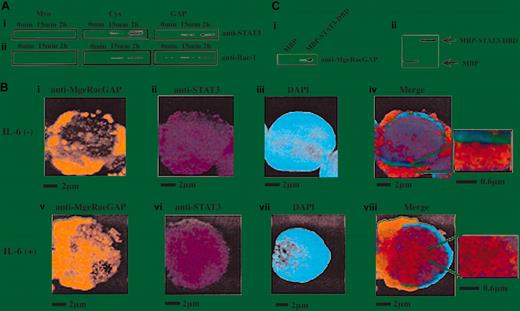
We next studied the kinetics of association between STAT3 and MgcRacGAP upon IL-6 stimulation using the MBP pull-down assay. The amount of STAT3 bound to the Cys and GAP domains of MgcRacGAP apparently increased upon IL-6 stimulation (Figure 4Ai). The amount of Rac1 bound to the same domains of MgcRacGAP also increased upon IL-6 stimulation (Figure 4Aii). These results suggested that MgcRacGAP bound STAT3 without IL-6 stimulation but that the binding was enhanced by IL-6 stimulation. To further confirm the cytokine-dependent augmentation of the interaction, we visualized MgcRacGAP and STAT3 in the M1 cells before and after the IL-6 stimulation using anti-STAT3 mAb and anti-MgcRacGAP Ab (Figure 4B). As shown in Figure 4Biv, MgcRacGAP and STAT3 partly colocalized in the cytoplasm of the unstimulated M1 cells. However, upon IL-6 stimulation, most STAT3 translocated to the nucleus, and a part of MgcRacGAP moved into the nucleus (Figure 4Bv-vi), resulting in colocalization of MgcRacGAP and STAT3 with a speckled pattern (Figure 4Bviii). Small insets to Figure 4Biv and 4Bviii showed the better details of colocalization of MgcRacGAP and STAT3 in the cytoplasm and nucleus. To confirm that the association between MgcRacGAP and STAT3 is direct, we produced and purified a Flag-tagged MgcRacGAP in Sf-9 cells and performed the pull-down assay. We found that the purified MgcRacGAP was pulled down by the MBP-STAT3-DBD but not by MBP alone, indicating that MgcRacGAP directly bound STAT3 (Figure 4C). Interaction of STAT3 with both Rac1 and MgcRacGAP was further confirmed with the use of a yeast 2-hybrid system (data not shown). Therefore, it is most likely that Rac, MgcRacGAP, and STAT3 interact directly with each other in a noninterdependent manner.
Enhancement of MgcRacGAP-STAT3 interaction by IL-6 stimulation. (A) Lysates from IL-6–stimulated M1 cells (50 ng/mL for the time indicated) were incubated with a similar amount of MBP-Myo, MBP-Cys, or MBP-GAP bound to beads. Bound proteins were separated on SDS-PAGE and immunoblotted with anti-STAT3 Ab or anti-Rac1 Ab. (B) MgcRacGAP partially colocalized with STAT3 at the cytoplasm and at the nucleus in M1 cells. With the use of unstimulated (i-iv) and IL-6–stimulated (v-viii) M1 cells, immunostaining for MgcRacGAP (i,v) and STAT3 (ii,vi) was done. For the merge figures (iv,viii), small insets on the right show the field at a high magnification to demonstrate the detail of the colocalization of MgcRacGAP and STAT3. The immunostained coverslips were viewed with a fluorescence microscope IX70 (Olympus). The scale bar indicates 2 μm or 0.6 μm. (C) Direct interaction of MgcRacGAP with STAT3-DNA–binding domain in vitro. Full-length MgcRacGAP was expressed in Sf9 cells with the use of the baculovirus vector and was purified from infected Sf9 cells. The recombinant MgcRacGAP was pulled down by MBP-STAT3-DBD– or MBP-bound beads, then subjected to Western blot analysis with anti-MgcRacGAP (i) or anti-MBP Ab for the loading control (ii).
Enhancement of MgcRacGAP-STAT3 interaction by IL-6 stimulation. (A) Lysates from IL-6–stimulated M1 cells (50 ng/mL for the time indicated) were incubated with a similar amount of MBP-Myo, MBP-Cys, or MBP-GAP bound to beads. Bound proteins were separated on SDS-PAGE and immunoblotted with anti-STAT3 Ab or anti-Rac1 Ab. (B) MgcRacGAP partially colocalized with STAT3 at the cytoplasm and at the nucleus in M1 cells. With the use of unstimulated (i-iv) and IL-6–stimulated (v-viii) M1 cells, immunostaining for MgcRacGAP (i,v) and STAT3 (ii,vi) was done. For the merge figures (iv,viii), small insets on the right show the field at a high magnification to demonstrate the detail of the colocalization of MgcRacGAP and STAT3. The immunostained coverslips were viewed with a fluorescence microscope IX70 (Olympus). The scale bar indicates 2 μm or 0.6 μm. (C) Direct interaction of MgcRacGAP with STAT3-DNA–binding domain in vitro. Full-length MgcRacGAP was expressed in Sf9 cells with the use of the baculovirus vector and was purified from infected Sf9 cells. The recombinant MgcRacGAP was pulled down by MBP-STAT3-DBD– or MBP-bound beads, then subjected to Western blot analysis with anti-MgcRacGAP (i) or anti-MBP Ab for the loading control (ii).
MgcRacGAP enhances the transactivation of STAT3, and the GAP domain is required for the enhancement
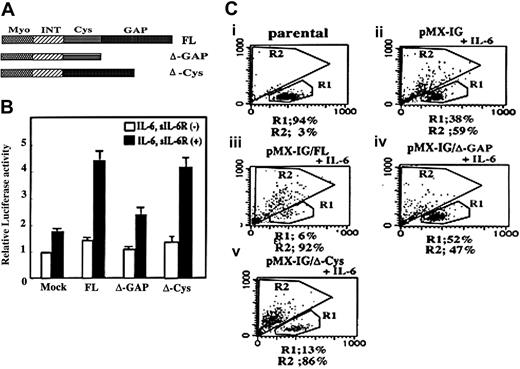
To determine if MgcRacGAP could alter the transcriptional activation of STAT3, we did the luciferase assay. HeLa cells were cotransfected with the luciferase reporter,43 the internal control, and a vector carrying the full-length wild-type MgcRacGAP with a Flag-tag (FL), the GAP domain deletion mutant (ΔGAP), the cystine-rich domain deletion mutant (ΔCys), or the vector alone (Mock) (Figure 5A). Flag tag did not affect the Rac-GAP activity of MgcRacGAP, and deletion of the GAP domain abolished the GAP activity (data not shown). As shown in Figure 5B, the IL-6–induced activation of STAT3 was enhanced by cotransfection with FL (approximately 4.5-fold) and ΔCys (approximately 4.0-fold) (Figure 5B). However, cotransfection of ΔGAP did not significantly enhance the IL-6–induced transactivation of STAT3. Thus, the GAP domain, but not the Cys domain, was required for the MgcRacGAP-mediated enhancement of-IL-6–induced transcriptional activation of STAT3.
Requirement of the GAP domain in the MgcRacGAP enhancement of IL-6–mediated STAT3 activation in HeLa cells and differentiation of M1 cells. The GAP domain is required for augmentation of IL-6–mediated STAT3 transactivation in HeLa cells and for IL-6–induced differentiation of M1 cells. (A) The structures of FL and deletion mutants of MgcRacGAP lacking the GAP domain (ΔGAP) or the cysteine-rich domain (ΔCys). (B) The ΔGAP did not augment the IL-6–mediated transactivation of STAT3. Luciferase activities were examined in lysates of unstimulated or IL-6– and sIL-6R–stimulated HeLa cells cotransfected with the internal control reporter plasmids and either the mock vector (pME) or the expression vector for the FL, ΔGAP, or ΔCys as described in “Materials and methods.” The results shown are the averages ± standard deviations of 3 independent experiments. (C) Quantitation of cell differentiation on flow cytometry in untreated parental M1 (i), and in M1 cells expressing pMX-IG (ii), pMX-IG-FL (iii), pMX-IG-ΔGAP (iv), or pMX-IG-ΔCys (v) at 4 days after IL-6 treatment (5 ng/mL). Differentiated M1 cells were detected in region R2.
Requirement of the GAP domain in the MgcRacGAP enhancement of IL-6–mediated STAT3 activation in HeLa cells and differentiation of M1 cells. The GAP domain is required for augmentation of IL-6–mediated STAT3 transactivation in HeLa cells and for IL-6–induced differentiation of M1 cells. (A) The structures of FL and deletion mutants of MgcRacGAP lacking the GAP domain (ΔGAP) or the cysteine-rich domain (ΔCys). (B) The ΔGAP did not augment the IL-6–mediated transactivation of STAT3. Luciferase activities were examined in lysates of unstimulated or IL-6– and sIL-6R–stimulated HeLa cells cotransfected with the internal control reporter plasmids and either the mock vector (pME) or the expression vector for the FL, ΔGAP, or ΔCys as described in “Materials and methods.” The results shown are the averages ± standard deviations of 3 independent experiments. (C) Quantitation of cell differentiation on flow cytometry in untreated parental M1 (i), and in M1 cells expressing pMX-IG (ii), pMX-IG-FL (iii), pMX-IG-ΔGAP (iv), or pMX-IG-ΔCys (v) at 4 days after IL-6 treatment (5 ng/mL). Differentiated M1 cells were detected in region R2.
The GAP domain of MgcRacGAP is required to render M1 cells hyperresponsive to IL-6–induced macrophage differentiation
Since overexpression of the MgcRacGAP rendered M1 cells hypersensitive to IL-6, we next investigated whether overexpression of ΔGAP or ΔCys could alter the IL-6 sensitivity of M1 cells. The ΔGAP and ΔCys mutants were expressed in M1 cells via retrovirus infection. As a positive and negative control, respectively, pMX-IG/FL and pMX-IG alone were also introduced into M1 cells. After infection of M1 cells with these vectors, the GFP+ cells were sorted on a fluorescence-activated cell sorter (FACS). We confirmed that the sorted GFP+ M1 cells expressed the Flag-tagged FL and mutants of MgcRacGAP at similar levels using Western blot analysis (data not shown). The sorted cells were cultured for 4 days in the presence of 5 ng/mL IL-6, and the cell differentiation was evaluated by flow cytometric analysis. Consistent with the results in Figure 1B, 59% of M1 cells transduced with the control vector pMX-IG showed a shift from region R1 to region R2. On the other hand, over 90% of the M1 cells transduced with pMX-IG/FL and 86% of those transduced with pMX-IG/ΔCys showed differentiation (shift from R1 to R2) (Figure 5Ciii,5Cv). Only 47% of M1 cells transduced with ΔGAP showed the shift (Figure 5Civ), indicating that ΔGAP did not significantly enhance the sensitivity of M1 cells to IL-6. These results paralleled the transcriptional activation of STAT3 when FL, ΔGAP, and ΔCys were overexpressed in the luciferase assay (Figure 5B).
MgcRacGAP is required for the transcriptional activation of STAT3
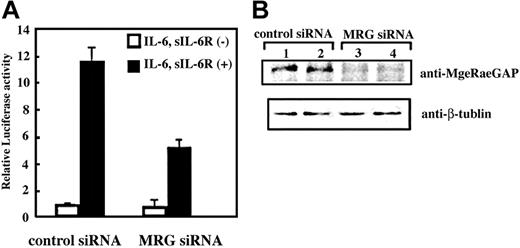
Finally, we asked if MgcRacGAP is essential for the transcriptional activation of STAT3. To this end, we used siRNA to knock down MgcRacGAP in 293T cells and performed the STAT3 reporter assay using the siRNA-treated cells. As shown in Figure 6A, the IL-6–induced transcriptional activation of STAT3 was profoundly suppressed by pretreatment of the cells with siRNA for MgcRacGAP, compared with those with the control siRNA (approximately 0.4-fold). We confirmed that the expression levels of MgcRacGAP protein were suppressed by siRNA for MgcRacGAP (Figure 6B, upper panel), but did not alter those of β-tubulin (Figure 6B, lower panel). The siRNA suppression of MgcRacGAP also attenuated IL-6–induced transcriptional activation of STAT3 in HeLa and Huh-7 cells (approximately 0.5-fold and approximately 0.6-fold, respectively), and expression of MgcRacGAP, but not STAT3, selectively decreased in these cells (data not shown). Thus, MgcRacGAP seems to play a critical role in IL-6–induced transcriptional activation of STAT3.
Suppression of STAT3-mediated transactivation by siRNA treatment for MgcRacGAP. (A) Luciferase activities were examined in the lysates of unstimulated or IL-6– and sIL-6R–stimulated 293T cells with pretreatment with siRNA for MgcRacGAP (right columns) or a control siRNA (left columns). At 24 hours after the siRNA treatment, the cells were transfected with the STAT3 reporter and internal control with Lipofectamine plus reagents. Another 24 hours after the transfection, cells were stimulated with IL-6 (20 ng/mL) and sIL-6R (20 ng/mL) for 12 hours before the cell lysates were prepared. The lysates were then subjected to a dual luciferase assay system, as described in “Materials and methods.” The error bars show the standard deviations of triplicates. The results shown are a representative result of 3 independent experiments. (B) Expression of MgcRacGAP (upper panel) and β-tubulin (lower panel) were examined in unstimulated (lanes 1 and 3) or IL-6/sIL-6R–stimulated (lanes 2 and 4) 293T cells pretreated with a control siRNA (lanes 1 and 2) or siRNA for MgcRacGAP (lanes 3 and 4) were examined.
Suppression of STAT3-mediated transactivation by siRNA treatment for MgcRacGAP. (A) Luciferase activities were examined in the lysates of unstimulated or IL-6– and sIL-6R–stimulated 293T cells with pretreatment with siRNA for MgcRacGAP (right columns) or a control siRNA (left columns). At 24 hours after the siRNA treatment, the cells were transfected with the STAT3 reporter and internal control with Lipofectamine plus reagents. Another 24 hours after the transfection, cells were stimulated with IL-6 (20 ng/mL) and sIL-6R (20 ng/mL) for 12 hours before the cell lysates were prepared. The lysates were then subjected to a dual luciferase assay system, as described in “Materials and methods.” The error bars show the standard deviations of triplicates. The results shown are a representative result of 3 independent experiments. (B) Expression of MgcRacGAP (upper panel) and β-tubulin (lower panel) were examined in unstimulated (lanes 1 and 3) or IL-6/sIL-6R–stimulated (lanes 2 and 4) 293T cells pretreated with a control siRNA (lanes 1 and 2) or siRNA for MgcRacGAP (lanes 3 and 4) were examined.
Discussion
As we earlier noted, the antisense MgcRacGAP profoundly inhibited the IL-6–induced macrophage differentiation of M1 cells.31 We and others later demonstrated that MgcRacGAP plays a critical role in cytokinesis.36,37 It was known that RhoA and serine/threonine kinase Aurora B play important roles in cytokinesis. We have recently clarified a molecular mechanism by which MgcRacGAP controls cytokinesis; MgcRacGAP is phosphorylated at Ser387 by Aurora B at the midbody and acquires RhoGAP activity for the completion of cytokinesis.40 However, the molecular mechanism by which MgcRacGAP affected IL-6–induced cell differentiation was still unclear. In the present work, we demonstrated that MgcRacGAP plays a role in STAT3 activation, thereby enhancing IL-6–induced differentiation of M1 cells, on the basis of the following: (1) MgcRacGAP directly bound STAT3 and Rac1/Rac2, and this interaction was enhanced by IL-6 stimulation. (2) Overexpression of MgcRacGAP rendered M1 cells hypersensitive to IL-6 stimulation, resulting in enhanced differentiation of M1 cells. (3) MgcRacGAP enhanced STAT3-induced transcriptional activation in a luciferase assay. (4) Knockdown of MgcRacGAP by siRNA profoundly inhibited STAT3-induced transcriptional activation in 293T, HeLa, and Huh-7 cells. Thus, while we and others reported that MgcRacGAP plays critical roles in cell division,36,37,40 the present results revealed an unexpected role for MgcRacGAP as a regulator of STAT3-mediated transcription in interphase.
GAPs for Rho GTPases constitute a class of regulatory proteins that can bind GTP-bound active forms of small G proteins and stimulate GTP hydrolysis.47,48 With this catalytic function, Rho-GAPs negatively regulate Rho-mediated signals. On the other hand, we demonstrated herein that the GAP domain was required for IL-6–induced STAT3 activation. It is possible that MgcRacGAP serves as an effecter molecule toward Rac-STAT3 complex in the IL-6 signaling pathway. For instance, certain GAPs, including p120RasGAP, n-chimaerin, and phospholipase C (a GAP for heterotrimeric G proteins), simultaneously function as effectors downstream of the GTPases.49-51
Concerning the cross-talk between STAT3 and a small GTPase Rac1, several groups suggested an indirect connection, and one indicated direct interaction.44,52,53 A constitutively active mutant RacV12 was reported to mediate STAT3 activation by autocrine IL-6 through the activation of nuclear factor–κB (NF-κB).52 It was also reported that in response to growth factor and cytokine stimulation, the activated Rac1 mediated generation of reactive oxygen species (ROS), which activated Src and JAKs, leading to STAT3 activation.48,53-55 In addition to the indirect connection between Rac and STAT3, it was reported that STAT3 bound directly to active but not inactive Rac1.44 In this case, expression of RacV12 induced both tyrosine and serine phosphorylation of STAT3, whereas overexpression of a dominant negative RacN17 inhibited either tyrosine or serine phosphorylation of STAT3 induced by epidermal growth factor. However, it was not determined whether the molecular mechanism by which Rac1 induced phosphorylation of STAT3 was dependent on Rac-STAT3 interaction; tyrosine phosphorylation of JAK2 was also induced by RacV12 in their system. Gu et al56 recently reported that Rac2 regulated transcriptional activation of c-Jun via activation of c-Jun N-terminal kinases (JNKs). Obviously, pleiotropic functions of Rac in the STAT3 activation pathway make it difficult to elucidate the biologic significance of STAT3-Rac interaction. Demonstration of the MgcRacGAP-Rac-STAT3 complex and identification of MgcRacGAP as an important regulator of STAT3 function provides new evidence for the cross-talk between STAT3 and Rac GTPases through direct interaction.
Although we observed that RacV12 enhanced the transcriptional activation of STAT3 induced by IL-6 and sIL-6R in HeLa cells but that RacN17 suppressed it (data not shown), we have been unable to identify the underlying mechanisms involved in the enhancement of STAT3 activation by MgcRacGAP. It is possible that MgcRacGAP plays some role in nuclear transport of STAT3. In fact, MgcRacGAP partly colocalized with STAT3 in the cytoplasm without cytokine stimulation, and in the nucleus upon cytokine stimulation, showing a speckled pattern (Figure 4Bviii). Alternatively, MgcRacGAP may somehow stabilize the STAT3-DNA complex, resulting in prolonged activation of STAT3, or may work as a scaffold protein that bridges between STAT3 and unidentified transcriptional coactivators. Interestingly, in a search for proteins that interact with MgcRacGAP using the yeast 2-hybrid screening system, we recently identified a transcriptional coactivator of activating protein-1 (AP-1) and estrogen receptors (CAPER), (Y. Minoshima, T. Kawashima, and T. Kitamura, unpublished results, 2004).
Although both GAP and Cys domains of MgcRacGAP can interact with STAT3, the deletion of the Cys domain did not significantly affect MgcRacGAP-mediated enhancement of IL-6–induced transcriptional activation of STAT3 (Figure 5B) and macrophage differentiation (Figure 5C). Our results suggested that Cys domain of MgcRacGAP is dispensable for IL-6–induced transcriptional activation of STAT3 and cell differentiation. The fact that the GAP domain of MgcRacGAP was required for the enhancement of STAT3-dependent transcription (Figure 5B) and IL-6–induced cell differentiation (Figure 5C) suggests that these functions of MgcRacGAP were mediated through Rho-family small GTPases. Coordinated activation and inactivation of small GTPases by GAPs and exchange factors (GEFs) are important in exerting their biologic functions. During M phase, MgcRacGAP and epithelial cell transforming sequence 2 (ECT2) oncogene proteins colocalize and orchestrate to control cell division.57 In addition, it has been reported that RacGAP50C, an ortholog of MgcRacGAP, binds pebble (PBL), an ortholog of ECT2, in drosophila cells58 although the direct interaction between MgcRacGAP and ECT2 was not detectable in mammalian cells (Y. Minoshima, T. Kawashima, and T. Kitamura, unpublished results, 2004). Therefore, it is possible that ECT2 also plays some role in transcriptional activation of STAT3. Although STAT3-dependent transcription was not affected by the overexpression of ECT2 (data not shown), this may be because ECT2 requires phosphorylation to be activated. The underlying mechanism of how MgcRacGAP regulates STAT3-induced gene expression awaits further investigation; however, the present data do reveal cross-talk between small GTPases and STAT3 downstream of IL-6 receptors in which MgcRacGAP plays pivotal roles. It is tempting to postulate that MgcRacGAP controls cell proliferation and differentiation in concert with the members of Rho-family GTPase by playing dual roles in M phase and interphase, completion of cytokinesis, and regulation of transcription.
Prepublished online as Blood First Edition Paper, July 29, 2004; DOI 10.1182/blood-2004-03-1066.
Supported by the Ministry of Education, Science, Technology, Sports and Culture and the Ministry of Health and Welfare, Japan; the Division of Hematopoietic Factors is supported by the Chugai Pharmaceutical Co.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank M. Itoh for excellent sorting on FACS; M. Ohara for language assistance; and T. Satoh for helpful discussions.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal