Abstract
Two closely related casein kinase I (CKI) isoforms, CKIδ and CKIϵ, are ubiquitously expressed in many human tissues, but their specific biologic function remains to be clarified. Here, we provide the first evidence that CKIϵ is involved in hematopoietic cell differentiation. CKIϵ, but not CKIδ, was down-regulated along with human granulocytic differentiation. The specific down-regulation was observed in granulocyte colony-stimulating factor (G-CSF)–induced cell differentiation of murine interleukin-3 (IL-3)–dependent myeloid progenitor 32D cells. Introduction of wild-type (WT)–CKIϵ into 32D cells inhibited the G-CSF–induced cell differentiation, whereas kinase-negative (KN)–CKIϵ promoted the differentiation. Neither WT- nor KN-CKIϵ affected IL-3–dependent cell growth. Moreover, introduction of WT- or KN-CKIδ did not affect the cytokine-induced cell growth and differentiation. While G-CSF–induced activation of signal transducers and activators of transcription 3 (STAT3) was sustained by KN-CKIϵ, STAT3 activation was attenuated by WT-CKIϵ. This may be explained by the fact that the suppressor of cytokine signaling 3 (SOCS3) was stabilized by its physical association with CKIϵ. Such stabilization by CKIϵ was also seen in IL-3–induced β-catenin. The stabilization of downstream components of cytokine and Wnt signaling by CKIϵ might be critical for integration of several intracellular signaling pathways to a cell-specific biologic response in hematopoietic cell self-renewal. (Blood. 2004;103: 2997-3004)
Introduction
Members of the casein kinase I (CKI) family of monomeric serine/threonine kinases are highly conserved from yeast to human and are ubiquitously expressed in different cell types.1,2 In mammals, 7 isoforms (α, β, γ1-3, δ, and ϵ) have been identified.3-6 These isoforms share a high degree of similarity within the NH2-terminal catalytic domains but show considerable variation in their carboxy-terminal (C-terminal) noncatalytic domains. Their variable C-terminal domains are responsible for substrate specificity and serve to promote differential subcellular localization of individual isoforms and to modulate kinase activity.4,7-10
Studies of CKI homologs in yeast have shown the biologic role of CKI in the regulation of DNA repair and normal cell cycle progression, vesicular trafficking, and cytokinesis.11-15 The identification of potential substrates for CKI in vitro also possibly inferred that CKI might be involved in a wide variety of cellular functions in mammals. For example, CKI is likely to regulate DNA and RNA metabolism, cellular morphology, vesicular trafficking, DNA damage response and repair, and the activity of various transmembrane receptors.16-24 From the diverse cellular functions of CKI isoforms arose a possibility that CKI is likely to regulate the stability of their substrates, protein turnover, and transport-dependent cellular processes.25-28 However, there have been only a few reports in which the phosphorylation by CKI is shown to be essential for the biologic function of the substrates. Unexpectedly, recent genetic analyses in diverse fields have demonstrated that CKIϵ plays an essential role in regulating several critical in vivo processes such as circadian rhythm, embryogenesis, and morphogenesis in various species.29-32
The homolog of CKIδ and CKIϵ was first cloned in a screen for a budding yeast mutant, hrr25.11 Human CKIδ and CKIϵ encoded on 2 independent genes localized at chromosome 17q25 and 22q12-13 are basic polypeptides of 49 kDa and 47 kDa, respectively.4,5 They are the closest isoforms in the CKI family, because their amino acid sequences are 98% identical over their kinase domains and 53% identical over their C-terminal domains. The autophosphorylation of the C-terminal domain has been shown to inhibit its kinase activity.33 Successful complementation of hrr25 mutants in budding yeast by human CKIδ and CKIϵ, but not CKIα, suggested that these 2 isoforms might have similar functions in mammals.5 Indeed, CKIδ as well as CKIϵ also have been reported to phosphorylate the tumor suppressor p5334 and to regulate circadian clock and Wnt signaling.29-31,35,36 Taken together, the presence of an isoform-specific biologic function of CKIδ and CKIϵ remains to be elucidated.
Here, we describe a novel biologic function of CKIϵ associated with hematopoietic cell differentiation. The expression of CKIϵ, but not CKIδ, is down-regulated specifically along granulocytic differentiation. Thus, we studied the molecular function of CKIϵ in granulocytic differentiation by introducing wild-type or kinase-dead human CKIϵ cDNA into interleukin-3 (IL-3)–dependent myeloid progenitor cells.
Materials and methods
Cells
Human peripheral polymorphonuclear (PMN) cells were separated by gradient sedimentation over a Mono-Poly Resolving Medium (ICN Biomedicals, Aurora, OH). Mononuclear cells (MNCs) were separated by gradient sedimentation over Ficoll-Hypaque (Amersham Pharmacia Biotech, Buckinghamshire, United Kingdom).37 The cells were incubated in petri dishes in RPMI 1640 for 30 minutes (Life Technologies, Rockville, MD) and then nonadherent MNCs were separated from adherent MNCs. The former MNCs included mainly lymphocytes, and the latter monocytes. CD34-positive (CD34+) hematopoietic cells were isolated from mobilized peripheral blood cells by Isolex50 (Baxter, Deerfield, IL).38
The murine IL-3–dependent myeloid progenitor cell line 32D was maintained in RPMI 1640 containing 10% fetal calf serum (FCS) (JRH Biosciences, Lenexa, KS) and 15% WEHI-conditioned medium (WEHI-CM). The cells were induced to differentiate by treatment with human granulocyte colony-stimulating factor (G-CSF) (Chugai Pharmaceutical, Tokyo, Japan) in a differentiation medium containing 10% FCS and 0.5% WEHI-CM.
To investigate cytokine-induced signal transduction pathways, cells were starved in only RPMI 1640 for 60 minutes and then stimulated with mouse recombinant IL-3 (Genzyme, Cambridge, MA) or G-CSF in a medium containing 10% FCS in the presence or absence of a proteasome inhibitor, MG132 (Calbiochem-Novabiochem, Darmstadt, Germany).
cDNA cloning and recombinant retrovirus vectors
The full-length human CKIδ and CKIϵ cDNAs were cloned from a CD34+ hematopoietic progenitor cell cDNA library.38 The human CKIδ and CKIϵ cDNA nucleotide sequences were essentially identical to that in a previous report,5 with several differences that did not affect the predicted amino acid sequence. The changes of CKIϵ include 211G>T, 250C>T, and 943G>A, where the numbers indicate nucleotide numbers of the formally deposited sequence (accession no. AB091043). The changes of CKIδ were 529C>T, 1034T>C, 1315C>G, and 1414G>A (accession no. AB091044).
To facilitate subsequent subcloning, an NdeI site was created at the initiating ATG of wild-type (WT)-CKIδ and -CKIϵ cDNAs, and a BamHI and a claI site was created at the stop codon TGA of WT-CKIδ and -CKIϵ, respectively. Then, hemagglutinin (HA) nucleotide sequence was introduced at the NdeI site. To generate human kinase–negative (KN)–CKIϵ (human CKIϵ K38R) cDNA, PCR-based site-directed mutagenesis was performed in pUC18 containing HA-tagged WT-CKIϵ open reading frame by using the oligonucleotides 5′-CACACACTCCAGCCTGATGGCGACTTCCTC-3′ and 5′-GAGGAAGTCGCCATCAGGCTGGAGTGTGTG-3′ and by subsequent DpnI digestion. KN-CKIϵ was reported to interfere with the biologic activity in a dominant-negative manner.5,31 Similarly, human KN-CKIδ cDNA was generated by using the oligonucleotides 5′-GACACATTCAAGCCTGATGGCAACCTCTTCTCC-3′ and 5′-GGAGAAGAGGTTGCCATCAGGCTTGAATGTGTC-3′. The HA-tagged WT- or KN-CKIδ and -CKIϵ cDNA fragments were inserted into the HpaI site of the retroviral vector pDON-AI containing a Neo marker gene or into the SwaI site of the adenoviral vector pAxCAwt (Takara, Tokyo, Japan). Recombinant CKIδ or CKIϵ expressed transiently in 293T cells by pAxCAwt was purified with an anti-HA monoclonal antibody, followed by in vitro kinase assays using casein as a substrate.10 The kinase activities of KN-CKIδ and -CKIϵ were successfully destroyed as predicted (data not shown). Recombinant retroviruses were prepared by transfection of the vector plasmids into ψ CRIP-P131 packaging cells.
32D cells were transduced by the recombinant retrovirus in a RetroNectin-coated 6-well plate (Takara) for 24 hours at 37°C. Then, cells were transferred to a 96-well plate to select G418 (Life Technologies)-resistant cells. As control cells, a retrovirus containing only Neo gene was infected into 32D cells.
RNA blot analysis
A human multiple tissue Northern blot (MTN blot) was purchased from Clontech Laboratories (Palo Alto, CA). Total RNAs of human blood cells were isolated by the acid guanidinium thiocyanate–phenol chloroform method.39 RNA blots were hybridized with 32P-labeled human CKIδ- and CKIϵ-specific cDNA probes, which were prepared from the 3′-noncoding (nucleotides 1543-1906) and C-terminal coding (nucleotides 920-1302) fragments, respectively.
Antibodies
Biotinylated anti–mouse CD11b (Mac-1) monoclonal antibody was purchased from Caltag Laboratories (San Francisco, CA). CKIδ- and CKIϵ-specific polyclonal antisera were obtained after immunization of healthy rabbits with 2 peptides, PGRVASSGLQSVVHR and IPASQTSVPFDHLGK, coupled to keyhole limpet hemocyanin by using glutaraldehyde, respectively. Antibodies to total and tyrosine 705–phosphorylated STAT3 (P(Y)-STAT3), serine 727–phosphorylated STAT3 (P(S)-STAT3), tyrosine 694–phosphorylated STAT5 (P(Y)-STAT5), β-catenin, and horseradish peroxidase (HRP)–conjugated anti–rabbit and anti–mouse IgG were purchased from New England Biolabs (Beverly, MA). Antibodies to STAT5 (N-20), SOCS3 (M-20), CIS (N-19), β-actin (H-196), and HRP-conjugated anti–goat IgG were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-HA monoclonal antibody, 12CA5, was purchased from Roche Diagnostics (Indianapolis, IN).
Immunoblot analysis
Cells were lysed in lysis buffer containing 50 mM Tris (tris(hydroxymethyl)aminomethane)-HCl, pH 7.5, 50 mM NaCl, 2 mM EDTA (ethylenediaminetetraacetic acid), 1% Triton X-100, 10 mM sodium pyrophosphate, 1 mM sodium vanadate, 50 mM sodium fluoride, 1.0 μg/mL leupeptin, 1.0 μg/mL aprotinin, 1.0 μg/mL pepstatin, and 1 mM phenylmethylsulfonyl fluoride with or without 10 μg/mL MG132. For immunoprecipitation, total cell lysates were incubated with specific antibodies, followed by the addition of protein G-sepharose (Amersham Pharmacia Biotech) on a rotating shaker for 4 hours at 4°C. The immunoprecipitates were washed 3 times with lysis buffer and 3 times with phosphate-buffered saline.
Cell lysates were separated on 12% to 15% sodium dodecyl sulfate–polyacrylamide gels (SDS-PAGE) and transferred onto nitrocellulose membranes (Portran; Schleicher & Schuell, Dassel, Germany). The membranes were first incubated with TBST (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.1% Tween 20) containing 5% nonfat dried milk and probed with specific antibodies. Immune complexes were detected by chemiluminescence (SuperSignal West Pico Chemiluminescent Substrate; Pierce, Rockford, IL).
Statistical analysis
Student t test was used for the comparison of numerical values in the cell growth assay. The nonparametric Mann-Whitney U test was applied to evaluate differences in the amount of the Mac-1 expression on each 32D cell line.
Results
Isoform-specific expression of CKIδ and CKIϵ in human peripheral leukocytes
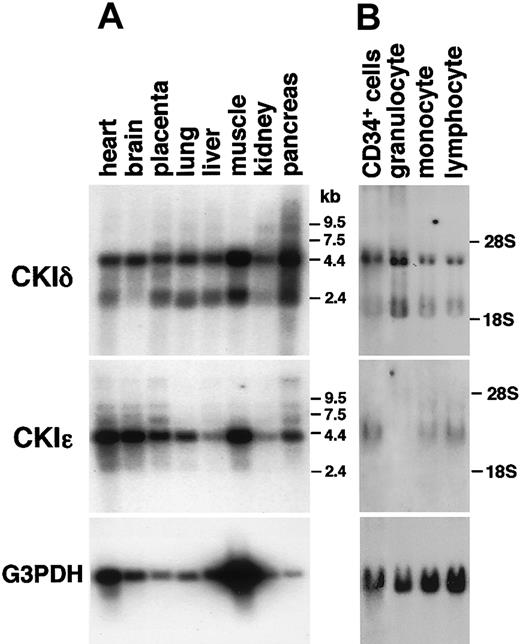
By using isoform-specific cDNA probes, RNA blot analysis of various human tissues was performed to assess the expression profiles of the CKIδ and CKIϵgenes. Because their amino acid sequences were 98% identical between their kinase domains, full-length cDNA probes cross-hybridized with both transcripts.33 Here, we employed human CKIδ- and CKIϵ-specific probes derived from the 3′-noncoding or C-terminal-coding sequences that encode only 53% identical amino acid sequences. Both CKIδ and CKIϵ mRNAs were ubiquitously expressed in all human tissues, such as the brain, heart, lung, liver, pancreas, kidney, placenta, and skeletal muscles (Figure 1A).
Various expressions of human CKIδ and CKIϵ mRNAs. RNA blot analysis of various normal human tissues (A) and human CD34+ hematopoietic stem cells and normal peripheral leukocytes (B) by using CKIδ- and CKIϵ-specific cDNA probes. Granulocytes, monocytes, and lymphocytes were prepared as PMNs, adherent-, and nonadherent MNC fractions, respectively. The signal that hybridized with the glyceraldehyde-3-phosphte dehydrogenase (G3PDH) cDNA probe is shown as a control.
Various expressions of human CKIδ and CKIϵ mRNAs. RNA blot analysis of various normal human tissues (A) and human CD34+ hematopoietic stem cells and normal peripheral leukocytes (B) by using CKIδ- and CKIϵ-specific cDNA probes. Granulocytes, monocytes, and lymphocytes were prepared as PMNs, adherent-, and nonadherent MNC fractions, respectively. The signal that hybridized with the glyceraldehyde-3-phosphte dehydrogenase (G3PDH) cDNA probe is shown as a control.
In human blood cells, CKIδ mRNA was highly expressed in CD34+ hematopoietic cells and PMN (mature granulocytes) (Figure 1B), as in other human tissues described above. It also was detectable in MNCs (monocytes and lymphocytes). On rehybridization of the same RNA blot with human CKIϵ-specific cDNA probe, CD34+ cells highly expressed CKIϵ. The expression of CKIϵ mRNA also was detectable in monocytes and lymphocytes, but not in granulocytes. These results suggested that there was isoform-specific expression of CKIδ and CKIϵ in human leukocytes, and CKIϵ was remarkably down-regulated along with granulocytic differentiation.
Down-regulation of CKIϵ, but not CKIδ, in granulocytic differentiation
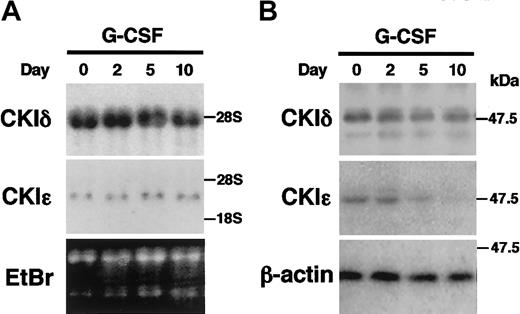
IL-3–dependent murine myeloid progenitor 32D cells undergo granulocytic differentiation in response to G-CSF. To study the regulation of the CKIϵ expression in granulocytic differentiation, 32D cells were treated with G-CSF in a differentiation medium. The mRNA expression of CKIδ and CKIϵ did not change significantly during the 10-day culture in the presence of G-CSF (Figure 2A). To examine the expression of CKIδ and CKIϵ at the protein levels, cell lysates were subjected to immunoblot analysis by using the CKI isoform-specific antibodies (Figure 2B). The expression of CKIδ protein was constitutively detectable at similar levels. However, the expression of CKIϵ protein decreased gradually by G-CSF treatment and then was almost undetectable in 10 days. These observations indicated that there was a regulatory mechanism to down-regulate the CKIϵ expression in accordance with granulocytic differentiation.
Decreased expression of CKIϵ in G-CSF–induced granulocytic differentiation of 32D cells. 32D cells were treated with 10 ng/mL G-CSF in a differentiation medium. Total RNAs and cell lysates extracted at days 0, 2, 5, and 10 were subjected to RNA blot analysis by using CKIδ- and CKIϵ-specific cDNA probes (A), or immunoblot analysis using anti-CKIδ, CKIϵ, and β-actin (control) polyclonal antibodies (B). Ethidium bromide (EtBr) staining indicates the amount of RNAs loaded in each lane.
Decreased expression of CKIϵ in G-CSF–induced granulocytic differentiation of 32D cells. 32D cells were treated with 10 ng/mL G-CSF in a differentiation medium. Total RNAs and cell lysates extracted at days 0, 2, 5, and 10 were subjected to RNA blot analysis by using CKIδ- and CKIϵ-specific cDNA probes (A), or immunoblot analysis using anti-CKIδ, CKIϵ, and β-actin (control) polyclonal antibodies (B). Ethidium bromide (EtBr) staining indicates the amount of RNAs loaded in each lane.
CKIδ and CKIϵ in cytokine-dependent cell growth
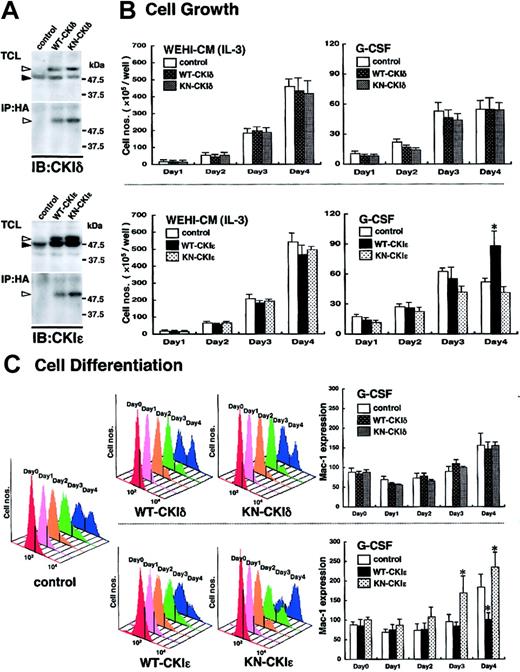
To elucidate the functional significance of the CKIϵ down-regulation in granulocytic differentiation, WT- or KN-CKIδ and -CKIϵ cDNA was introduced into 32D cells. To confirm the expression of exogenous human CKIδ and CKIϵ in 32D cells, the immunoprecipitates with anti-HA antibody were subjected to immunoblot analysis by using each CKI isoform-specific antibody. WT- and KN-CKIδ– and -CKIϵ–infected, but not control virus–infected, 32D cells produced HA-tagged human CKI protein in addition to mouse endogenous 49-kDa CKIδ and 47-kDa CKIϵ, respectively (Figure 3A).
Involvement of exogenous CKIϵ in G-CSF–induced cell differentiation. (A) Expression of WT- or KN-CKI. Total cell lysates (TCLs) extracted from recombinant virus–infected 32D cell lines and immunoprecipitates (IPs) with anti-HA antibody were subjected to immunoblot analysis (IB) by using anti-CKIδ and CKIϵ antibodies. Black arrowheads indicate the position of endogenous 49-kDa CKIδ or 47-kDa CKIϵ. White arrowheads indicate HA-tagged recombinant CKIδ or CKIϵ. (B) Cell growth of virus-infected 32D cell lines in the presence of WEHI-CM (IL-3) or G-CSF. Each cell line was subcultured at 5 × 105 cells/well in a medium containing 15% WEHI-CM (IL-3) and in a differentiation medium with 10 ng/mL G-CSF on day 0. Viable cells after staining with trypan blue were counted at days 1, 2, 3, and 4. Values are the means of cell numbers/well ± SE of four samples in 5 independent experiments (*P < .05 vs control cells). (C) Cell differentiation induced by 10 ng/mL G-CSF was assessed daily by flow cytometry (FACScan; Becton Dickinson, San Jose, CA) by using Mac-1 antibody with phycoerythrin-conjugated streptavidin (Becton Dickinson) (upper panels: WT-CKIδ- and KN-CKIδ-32D cells; lower panels: WT-CKIϵ- and KN-CKIϵ-32D cells). Daily histogram analysis of Mac-1 expression is a representative 1 of 5 similar results. Data represent the mean ± SE of Mac-1 expression of 5 independent experiments (*P < .05 vs control cells).
Involvement of exogenous CKIϵ in G-CSF–induced cell differentiation. (A) Expression of WT- or KN-CKI. Total cell lysates (TCLs) extracted from recombinant virus–infected 32D cell lines and immunoprecipitates (IPs) with anti-HA antibody were subjected to immunoblot analysis (IB) by using anti-CKIδ and CKIϵ antibodies. Black arrowheads indicate the position of endogenous 49-kDa CKIδ or 47-kDa CKIϵ. White arrowheads indicate HA-tagged recombinant CKIδ or CKIϵ. (B) Cell growth of virus-infected 32D cell lines in the presence of WEHI-CM (IL-3) or G-CSF. Each cell line was subcultured at 5 × 105 cells/well in a medium containing 15% WEHI-CM (IL-3) and in a differentiation medium with 10 ng/mL G-CSF on day 0. Viable cells after staining with trypan blue were counted at days 1, 2, 3, and 4. Values are the means of cell numbers/well ± SE of four samples in 5 independent experiments (*P < .05 vs control cells). (C) Cell differentiation induced by 10 ng/mL G-CSF was assessed daily by flow cytometry (FACScan; Becton Dickinson, San Jose, CA) by using Mac-1 antibody with phycoerythrin-conjugated streptavidin (Becton Dickinson) (upper panels: WT-CKIδ- and KN-CKIδ-32D cells; lower panels: WT-CKIϵ- and KN-CKIϵ-32D cells). Daily histogram analysis of Mac-1 expression is a representative 1 of 5 similar results. Data represent the mean ± SE of Mac-1 expression of 5 independent experiments (*P < .05 vs control cells).
Because mutant hrr25, the homolog of human CKIδ and CKIϵ, was previously reported to inhibit the cell growth of budding yeast,11 we first examined the effects of the exogenous CKIδ and CKIϵ on cell growth. As shown in Figure 3B, in the presence of 15% WEHI-CM, no significant difference in cell growth was found among any 32D cell lines infected control (Neo gene alone), WT- and KN-CKIδ and -CKIϵ viruses. These results suggested that neither CKIδ nor CKIϵ was involved in IL-3–dependent cell growth.
G-CSF stimulated the cell growth of control virus–infected 32D cells significantly for the first 3 days in a differentiation medium. However, the cell growth in this differentiation medium was very slow, compared with that in the presence of enough IL-3 (15% WEHI-CM). Moreover, the G-CSF–induced cell growth ceased at day 4 (Figure 3B). Such a growth arrest in the presence of G-CSF also was seen in KN-CKIϵ-32D cells (Figure 3B), but not in WT-CKIϵ-32D cells (Figure 3B). The latter cells continued to proliferate significantly even at day 4. In contrast to CKIϵ, both WT- and KN-CKIδ did not influence G-CSF–induced cell growth significantly (Figure 3B). These results suggested that G-CSF–, but not IL-3–induced cell growth was influenced by the exogenous CKIϵ specifically.
Involvement of CKIϵ in G-CSF–induced cell differentiation
We next examined whether CKIϵ is functionally involved in cytokine-induced granulocytic differentiation by using the 32D cell lines. The expression of a differentiation antigen, Mac-1 (CD11b), of WT- and KN-CKIϵ-32D cells was similar to that of control virus–infected 32D cells in the medium containing 15% WEHI-CM (shown as day 0 in Figure 3C). In control cells, G-CSF gradually increased the Mac-1 expression day by day. On day 4 after G-CSF addition, 2 peaks indicating low and high Mac-1 expressions appeared in the histogram (Figure 3C). G-CSF increased the cell population expressing high levels of Mac-1 in both WT- and KN-CKIϵ-32D cells. Compared with control cells, the increase of Mac-1 expression was clearly suppressed in WT-CKIϵ cells (Figure 3C). In contrast, it was promoted in KN-CKIϵ cells during the 4 days of culture. Even at day 3, the Mac-1 expression was significantly increased in KN-CKIϵ cells. These results showed that KN-CKIϵ, which could inhibit kinase activity of endogenous CKIϵ in a dominant-negative manner, accelerated granulocytic differentiation. Moreover, the overexpression of wild-type CKIϵ inhibited the differentiation.
In contrast to CKIϵ, there was no apparent difference in the profiles of G-CSF–induced Mac-1 expression between WT- and KN-CKIδ cells (Figure 3C).
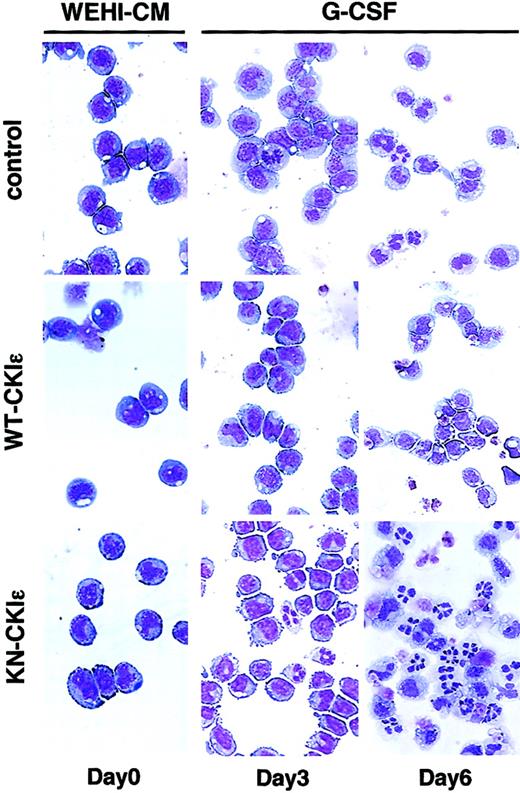
We further examined whether the phenotypic differentiation influenced by CKIϵ accompanied morphological changes to granulocytes. Criteria for granulocytic differentiation in 32D cells include nuclear segmentation, an increased cytoplasm/nucleus ratio, and increased eosinophilia and granularity in cytoplasm (Figure 4). Most WT-CKIϵ cells retained undifferentiated blastoid morphology at day 3, and about half of the cells remained undifferentiated 6 days after G-CSF treatment. In contrast, KN-CKIϵ cells started to exhibit differentiated morphology and lost cytoplasmic basophilia by day 3. Most KN-CKIϵ cells terminally differentiated like mature granulocytes at day 6. These results further confirmed that the overexpression of CKIϵ inhibited G-CSF–induced granulocytic differentiation not only phenotypically but also morphologically.
Effect of exogenous CKIϵ on morphological differentiation. Photomicrographs of 3 types of cell lines Wright-stained and cultured in a medium containing 15% WEHI-CM (day 0) and then in a differentiation medium with 10 ng/mL G-CSF for 3 and 6 days are shown (original magnification, × 400).
Effect of exogenous CKIϵ on morphological differentiation. Photomicrographs of 3 types of cell lines Wright-stained and cultured in a medium containing 15% WEHI-CM (day 0) and then in a differentiation medium with 10 ng/mL G-CSF for 3 and 6 days are shown (original magnification, × 400).
Involvement of CKIϵ in STAT3, but not STAT5, activation
In the cytokine-induced intracellular signal transduction cascades, the activation of signal transducers and activators of transcription (STATs) is essential for the cell growth and differentiation of hematopoietic cells. Especially, STAT3 and STAT5 activations are reported to be critical for G-CSF and IL-3 signalings, respectively.40 Thus, we examined whether CKIϵ is involved in cytokine-induced STATs activation by immunoblot analysis. IL-3 stimulation lead to a robust activation of STAT5, but not STAT3, protein via tyrosine phosphorylation, in control as well as WT- and KN-CKIϵ cells within 5 minutes (Figure 5A). The phosphorylated STAT5 (P(Y)-STAT5) increased in a time-dependent manner until 60 minutes. There is no difference in the time courses of IL-3–induced STAT5 activation among the 3 cell lines.
Regulation of STAT3 activation by CKIϵ. 32D transfectants were stimulated with 5 ng/mL IL-3 (A) or 10 ng/mL G-CSF (B) for the indicated periods (minutes). Total cell lysates were subjected to immunoblot to analyze the expression levels of the total and tyrosine 705– and serine 727–phosphorylated STAT3 (P(Y)-STAT3 and P(S)-STAT3, respectively) and the total and tyrosine 694–phosphorylated STAT5 (P(Y)-STAT5) by using their specific antibodies. Anti-STAT3 polyclonal antibody recognizes STAT3α (92 kDa) as well as STAT3β (83 kDa). Anti-STAT5 antibody also recognizes STAT5a (96 kDa) as well as STAT5b (94 kDa). These data represent 3 independent experiments.
Regulation of STAT3 activation by CKIϵ. 32D transfectants were stimulated with 5 ng/mL IL-3 (A) or 10 ng/mL G-CSF (B) for the indicated periods (minutes). Total cell lysates were subjected to immunoblot to analyze the expression levels of the total and tyrosine 705– and serine 727–phosphorylated STAT3 (P(Y)-STAT3 and P(S)-STAT3, respectively) and the total and tyrosine 694–phosphorylated STAT5 (P(Y)-STAT5) by using their specific antibodies. Anti-STAT3 polyclonal antibody recognizes STAT3α (92 kDa) as well as STAT3β (83 kDa). Anti-STAT5 antibody also recognizes STAT5a (96 kDa) as well as STAT5b (94 kDa). These data represent 3 independent experiments.
On the other hand, both STAT3 and STAT5 were tyrosine-phosphorylated in response to G-CSF. However, significant differences among the 3 cell lines were observed in the STAT3 activation (Figure 5B). In control cells, tyrosine-phosphorylated STAT3 (P(Y)-STAT3) was induced within 5 minutes after the G-CSF addition, reached the maximum levels at 15 minutes, and decreased gradually. In WT-CKIϵ cells, P(Y)-STAT3 also was detectable in 5 minutes, but decreased rapidly. It was almost undetectable by 30 minutes. In contrast, the high level of P(Y)-STAT3 was sustained for 60 minutes in KN-CKIϵ cells. The serine residue at 727 of STAT3 in each cell line also was phosphorylated (P(S)-STAT3) with a similar time course of P(Y)-STAT3.
Although P(Y)-STAT5 in response to G-CSF was weaker than that induced by IL-3, no significant difference among the 3 cell lines was observed in the expression levels of P(Y)-STAT5. Neither IL-3 nor G-CSF changed the expression levels of STAT3 and STAT5 proteins in any of the cell lines (Figure 5A-B). These results suggested that CKIϵ was specifically involved in the G-CSF–induced STAT3 activation.
Stabilization of SOCS3 protein by CKIϵ
A family of suppressors of cytokine signaling (SOCS), including cytokine-inducible SH2 proteins (CIS), is a novel negative regulator of Janus kinases (JAKs)–STAT pathway in cytokine signaling.40,41 Among them, SOCS3 and CIS are reported to be involved in STAT3 activation.40-44 Thus, we further examined whether CKIϵ regulates the expression of these proteins. Figure 6A shows immunoblots of SOCS3 and β-actin. In response to IL-3, SOCS3 protein was induced within 1 hour and then down-regulated in a cell line–dependent manner. Compared with control cells, the expression of SOCS3 protein in KN-CKIϵ cells was significantly weak and rapidly disappeared by 6 hours after IL-3 treatment. In contrast, the expression of SOCS3 in WT-CKIϵ cells was prolonged for 9 hours. As well, the expression levels of SOCS3 induced by G-CSF varied on the cell lines. After 3 hours of treatment with G-CSF, SOCS3 protein was barely detectable only in WT-CKIϵ cells. In the 9 hours' treatment with G-CSF, SOCS3 was faintly detectable in control cells, but not in KN-CKIϵ cells. The expression level of SOCS3 in each cell line was consistent with the activation level of STAT3, because SOCS3 negatively regulates the activation.
Stabilization of SOCS3 and β-catenin by CKIϵ. 32D transfectants were treated with 5 ng/mL IL-3 or 10 ng/mL G-CSF in the absence or presence of MG132 (20 or 40 μM) for the indicated times. Then, total cell lysates were subjected to immunoblot analysis (IB) by using anti-SOCS3 and β-actin (*) (A), anti-CIS (B), and anti–β-catenin (C) polyclonal antibodies. To detect CIS protein in the absence of MG132, the immunoblot filter was exposed for a long time. Equal loading of cell lysates in each lane was probed with an anti–β-actin antibody. The same lysates were employed in IB for CIS and β-catenin.
Stabilization of SOCS3 and β-catenin by CKIϵ. 32D transfectants were treated with 5 ng/mL IL-3 or 10 ng/mL G-CSF in the absence or presence of MG132 (20 or 40 μM) for the indicated times. Then, total cell lysates were subjected to immunoblot analysis (IB) by using anti-SOCS3 and β-actin (*) (A), anti-CIS (B), and anti–β-catenin (C) polyclonal antibodies. To detect CIS protein in the absence of MG132, the immunoblot filter was exposed for a long time. Equal loading of cell lysates in each lane was probed with an anti–β-actin antibody. The same lysates were employed in IB for CIS and β-catenin.
Recently, proteasome was reported to play a major role in the rapid degradation of the CIS/SOCS proteins.45,46 Thus, we employed a proteasome inhibitor, MG132, to confirm the actual induction levels of these proteins by cytokines. In the presence of 20 μM MG132, the expression levels of SOCS3 protein induced by IL-3 of 3 cell lines became almost similar. These results indicated that CKIϵ had inhibited the proteasomal degradation of SOCS3. The same dose of MG132 made the expression patterns of SOCS3 clearly distinguishable in each cell line treated with G-CSF. While WT-CKIϵ increased SOCS3 protein compared with control cells, KN-CKIϵ rapidly decreased it. A higher dose of MG132 (40 μM) stabilized SOCS3 more with little difference in the expression profiles of SOCS3 protein among the 3 cell lines. In all the cell lines, SOCS3 protein was rapidly induced in 1 hour and decreased with similar time courses. The down-regulation of SOCS3 protein seemed to be caused by its negative feedback mechanism in the G-CSF signaling.
In response to IL-3, CIS was also induced within 1 hour. However, no significant difference in CIS expression was observed among 3 cell lines (Figure 6B). MG132 also sustained an increase in the CIS protein levels until 9 hours in all the cell lines. Contrary to IL-3, G-CSF did not increase the expression of CIS protein. The CIS signals were barely detectable by a long exposure to films and completely down-regulated by 3 hours in all the cell lines. Even in the presence of MG132, CIS was not induced by G-CSF, and the time-dependent decrease of CIS protein was clearly confirmed. These results indicate that CKIϵ specifically stabilized SOCS3 but not CIS protein.
β-catenin, a downstream component of Wnt signaling, is reportedly stabilized by CKIϵ. The unstabilized β-catenin directs its degradation by an ubiquitin-proteasome system.47 We also examined the expression of β-catenin protein induced by cytokines (Figure 6C). Although β-catenin was undetectable until 9 hours after IL-3 stimulation in the absence of MG132, the addition of MG132 made the IL-3–induced expression of β-catenin of control cells clearly detectable in 3 hours. Overexpression of WT-CKIϵ stabilized the β-catenin expression and increased high molecular weight of β-catenin possibly ubiquitinated. Contrary to the case of WT-CKIϵ, KN-CKIϵ destabilized it, as described previously.31,32,48,49
Taken together, these observations suggested that CKIϵ specifically stabilized SOCS3 as well as β-catenin by preventing proteasomal degradation.
Specific association of CKIϵ with SOCS3
We finally examined the interaction of CKIϵ with STAT3 and SOCS3 protein by using their specific antibodies. As previously reported,40,50 STAT3 was coimmunoprecipitated with anti-SOCS3 antibody from the control, WT- and KN-CKIϵ cell lysates (Figure 7A). However, STAT3 could not be coimmunoprecipitated with the CKIϵ antibody (Figure 7A), nor could CKIϵ be detectable in the immunoprecipitates of STAT3-specific antibody (Figure 7B). In contrast, CKIϵ was coimmunoprecipitated with SOCS3-specific antibody (Figure 7B). SOCS3 associated not only with endogenous CKIϵ (indicated by a black arrowhead) but also with exogenous CKIϵ (indicated by a white arrowhead). However, CKIδ was not coimmunoprecipitated with anti-SOCS3 as well as anti-STAT3 antibodies. These results demonstrate that CKIϵ, but not CKIδ, specifically associates with SOCS3 protein, possibly to stabilize this protein.
Association of CKIϵ with SOCS3. 32D transfectants were treated with 10 ng/mL G-CSF. After 3 hours, total cell lysates (TCLs) were immunoprecipitated (IP) with anti-CKIϵ, anti-STAT3, or anti-SOCS3 polyclonal antibodies. Each blot was probed by using anti-STAT3 (A), or anti-CKIδ or CKIϵ (B) antibodies. Black and white arrowheads indicate the position of endogenous CKIϵ and HA-tagged recombinant CKIϵ, respectively. An asterisk (*) indicates the position of endogenous CKIδ.
Association of CKIϵ with SOCS3. 32D transfectants were treated with 10 ng/mL G-CSF. After 3 hours, total cell lysates (TCLs) were immunoprecipitated (IP) with anti-CKIϵ, anti-STAT3, or anti-SOCS3 polyclonal antibodies. Each blot was probed by using anti-STAT3 (A), or anti-CKIδ or CKIϵ (B) antibodies. Black and white arrowheads indicate the position of endogenous CKIϵ and HA-tagged recombinant CKIϵ, respectively. An asterisk (*) indicates the position of endogenous CKIδ.
Discussion
The 2 closest CKI isoforms, CKIδ and CKIϵ, expressed ubiquitously in many human tissues, have been thought to possess similar functions not only in vitro but also in vivo.34,51 Here, we showed that the expression of CKIϵ, but not CKIδ, is regulated specifically along with granulocytic differentiation and is critical for G-CSF–induced cell differentiation. There are some mechanisms that down-regulate CKIϵ expression at transcriptional and/or translational levels. The kinase activity of CKIϵ and granulocytic differentiation are decreased in all mechanisms.
CKIϵ, but not CKIδ, inhibited G-CSF–induced phenotypic as well as morphologic differentiation in a kinase-dependent manner. However, it did not significantly affect IL-3–dependent cell growth of 32D cells. The cytokine signaling pathway is one of the critical systems for regulating the cell growth, differentiation, and survival of various hematopoietic cells.52-54 Many different cytokines use several common signal transduction cascades including the JAK-STAT pathway,40 but each cytokine shows various cell-specific biologic functions. In JAK-STAT pathways, STAT's activation appears to orchestrate the downstream events propagated by cytokine stimulations.40 Recent studies using dominant-negative STATs have shown that STAT3 activation plays a key role in the differentiation responses to G-CSF,55 whereas STAT5 appears essential for proliferative responses to IL-3 and G-CSF.56,57 In the present study, we demonstrated that CKIϵ suppressed G-CSF–induced STAT3 activation but not IL-3– or G-CSF–induced STAT5 activation. Therefore, the activation status of STAT3 regulated by CKIϵ in WT- and KN-CKIϵ cells was consistent with the biologic effect of G-CSF on the cell differentiation. These results indicate that the expression level of functional CKIϵ defines the stage-specific responsiveness for G-CSF in myeloid cell differentiation. Moreover, the modification of intracellular signaling from cytokine receptors by CKIϵ might explain the cell-specific biologic responses of given cytokines in each cell type.
The stabilization of target substrates is one of the novel molecular functions of CKIϵ. In response to DNA damage, p53 is stabilized through the phosphorylation of threonine 18 by CKIϵ, followed by the prevention of Mdm2-dependent degradation.25 Moreover, β-catenin is also stabilized by CKIϵ in Wnt signaling as described below.31,32,48,49 Here, we showed that CKIϵ is involved in the stabilization of SOCS3 as well. The duration and intensity of cytokine signaling from cytokine receptors are tightly regulated, in part, by CIS/SOCS.40,41 They are up-regulated in cytokine stimulation and inhibit JAK-STAT pathways. In this study, there was no significant difference among the 3 cell lines in the IL-3–induced expression of CIS protein. It is consistent with the fact that STAT5 had been activated equivalently by IL-3 in all the cell lines as shown in Figure 5A. On the contrary, the expression levels of SOCS3 protein in the cell lines were different. SOCS3 is known to regulate G-CSF signaling negatively via STAT3 activation directly or indirectly.40,41,58 SOCS3 protein was induced by both IL-3 and G-CSF stimulations in 32D cells. However, it was highly unstable and rapidly down-regulated by proteasomal degradation. The rapid degradation of SOCS3 was enhanced in KN-CKIϵ cells, whereas the overexpression of wild-type CKIϵ stabilized the SOCS3 protein. Therefore, SOCS3 induced by G-CSF was less expressed in KN-CKIϵ cells, followed by the sustained activation of STAT3. In addition, we also confirmed the physical association of CKIϵ with SOCS3. Taken together, these results suggested that CKIϵ is directly involved in the stabilization of SOCS3 protein and consequently inhibits the G-CSF–induced STAT3 activation. Indeed, there are several serine/threonine residues that possibly are phosphorylated by CKIϵ in the SOCS3 amino acid sequences, including the kinase inhibitory region.59
SOCS3 also was reported to inhibit the STAT5 activation by binding to JAK2 or activated G-CSF receptor.58,60 JAK2 is an early downstream component of both IL-3 and G-CSF receptor signalings. However, both IL-3– and G-CSF–induced STAT5 phosphorylation were not affected in 32D cells overexpressing WT- and KN-CKIϵ. This may be explained by the previous reports that SOCS3 binds to JAK2 with a low affinity and is a significantly less efficient inhibitor.61-64 The inhibition of STAT5-mediated gene expression by SOCS3 was shown by ectopic overexpression of G-CSF receptors and SOCS3.57,58 The regulatory kinetics of CIS/SOCS proteins are complicated by a variety of factors such as the kind of cytokine stimulations, cell types, and their own expression levels. Although ectopic overexpression studies reveal the potential of SOCS family members to inhibit multiple cytokine-induced signaling pathways, these studies could overestimate the SOCS action.40,41,46 For instance, the dual conflicting effects of SOCS2 on growth hormone signaling depend on its expression levels.65 In this study, the expression levels of G-CSF receptor, JAK, STAT, and SOCS are within physiological ranges. Moreover, the transcriptional activity of STAT5 is possibly regulated not only by its tyrosine phosphorylation but also by its subcellular localization. For example, another signaling pathway, such as phosphatidylinositol 3-kinase activation, is reported to be involved in the nuclear localization of STAT5.66 These might be the reasons the STAT5 phosphorylation was not affected by recombinant CKIϵ in 32D cells.
In addition, negative feedback components induced by IL-3 might affect its signal transduction little in IL-3–dependent 32D cells. However, SOCS3 induced by IL-3 might influence another cytokine signaling, such as G-CSF signaling, to inhibit cell differentiation. Denson et al recently reported such a cross-talk between IL-6 and hepatic growth hormone signalings via up-regulation of CIS and SOCS3.44
A Wnt signaling pathway was reported recently to be critical for hematopoietic cell self-renewal.67 β-catenin, or other components of this signal pathway, also are stabilized by CKIϵ directly or indirectly from proteasomal degradation.31,32,47-49 Here, we first demonstrated that IL-3 induced the expression of β-catenin in the murine hematopoietic progenitor cells. We also confirmed that CKIϵ could stabilize β-catenin as well as SOCS3 in hematopoietic cells. These observations suggest the possibility that Wnt signaling for hematopoietic cell self-renewal could be amplified through the induction of β-catenin by IL-3 or its stabilization by CKIϵ. Moreover, both the negative regulation of STAT3 via SOCS3 and the stabilization of β-catenin by CKIϵ are likely to inhibit hematopoietic cell differentiation in collaboration. In other words, CKIϵ might integrate several intracellular signalings and lead hematopoietic cells to a common biologic response such as self-renewal and inhibition of cell differentiation through the stabilization of several signaling molecules. Further studies are necessary to clarify how CKIϵ specifically contributes to the hematopoietic cell growth and differentiation in vivo.
In conclusion, we provide the first evidence that the regulatory mechanism of CKIϵ expression is essential for hematopoietic granulocytic differentiation. In particular, CKIϵ is directly involved in the G-CSF–induced differentiation signal in a kinase-dependent manner as a negative regulator. CKIϵ regulates the G-CSF–induced STAT3 activation through the stabilization of SOCS3 as well as β-catenin induced by IL-3. Further analyses of CKIϵ in cytokine signalings might help our understanding of the molecular mechanisms of not only hematopoiesis but also hematological disorders with cell differentiation, such as myelodysplastic syndrome and leukemia.
Prepublished online as Blood First Edition Paper, January 8, 2004; DOI 10.1182/blood-2003-08-2768.
Supported in part by Grants-in-Aid for scientific research from the Ministry of Education, Culture, Sports, and Technology of Japan.
An Inside Blood analysis of this article appears in the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We would like to thank Kaori Tsujimoto for her technical assistance.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal