Abstract
Hereditary hemochromatosis is a genetically heterogeneous disease of iron metabolism. The most common form of the disorder is an adult-onset form that has mainly been associated with the HFE pC282Y/pC282Y genotype. The phenotypic expression of this genotype is very heterogeneous and could be modulated by both environmental factors and modifier genes. The non-HFE hereditary hemochromatosis forms include a juvenile onset form associated with mutations in HAMP. From a cohort of 392 C282Y homozygous patients, we found 5 carriers of an additional HAMP mutation at the heterozygous state (pR59G, pG71D, or pR56X). We found that iron indices of these 5 patients were among the most elevated of the cohort. Moreover, we specified that the HAMP mutations were not detected in 300 control subjects. These results revealed that mutations in HAMP might increase the phenotypic expression of the pC282Y/pC282Y genotype. From a cohort of 31 patients with at least one chromosome lacking an HFE mutation, we further identified 4 males carrying a heterozygous HAMP mutation (pR59G or pG71D). Based on a digenic model of inheritance, these data suggest that the association of heterozygous mutations in the HFE and HAMP genes could lead, at least in some cases, to an adult-onset form of primary iron overload. (Blood. 2004;103:2835-2840)
Introduction
Hereditary hemochromatosis (HH) is an inherited disorder of iron metabolism characterized by progressive accumulation of iron in tissues. Such an iron overload can lead to cirrhosis, diabetes mellitus, arthropathy, endocrine abnormalities, hepatocellular carcinoma, and cardiomyopathy.1 The most common form of the disorder, called hereditary hemochromatosis of type I (Online Mendelian Inheritance in Man [OMIM] no. 235200), is an adult-onset form associated with duodenal iron hyperabsorption. It is inherited in an autosomal recessive pattern and has, since 1996, been linked mainly with the pC282Y HFE.2 In addition to the pC282Y mutation, which has been reported to be homozygous in 64% to 100% of HH patients of European origin,3 18 other mutations have now been reported.2,4-19 The pH63D and pS65C substitutions have been particularly associated with milder iron overload phenotypes.6,20
Non-HFE-related forms of primary iron overload have been documented in recent years. These include an autosomal recessive form caused by mutations in the TfR2 gene (HH type III; OMIM no. 604250), an autosomal dominant form due to mutations in the SLC11A3 gene (HH type IV; OMIM no. 606069), another autosomal dominant form due to a mutation in the iron-responsive element of H ferritin mRNA (HH type V; OMIM no. 134770), and an autosomal recessive juvenile-onset form associated with mutations in either the very recently identified 1q-related gene (HH type IIA; OMIM no. 602390)21 or in the HAMP gene (HH type IIB; OMIM no. 606464).
The HAMP gene (GenBank no. AJ277280) has very recently been implicated in a juvenile form of hereditary hemochromatosis on the basis of the description of 2 mutations, found in the homozygous state, in 2 families: the deletion of guanine 93 (nucleotide 1 is the adenine in the initiator methionine), which results in a frameshift, and the g166C>T transition, which changes amino acid 56 from arginine to a stop codon (pR56X).22
Since the identification of the HFE gene (GenBank no. Z92910), several studies have reported that the phenotypic expression of the pC282Y/pC282Y genotype is very heterogenous.23,24 The influence of environmental factors, such as an excessive alcohol consumption,25 surely explains one aspect of this phenotypic heterogeneity. As recognized by different authors,26-28 the influence of other genes, which act via the same molecular pathways as HFE (SwissProt no. Q30201), is an attractive additional explanation.
The main objective of the present study was to determine if mutations in the HAMP gene were associated with more severe iron overload phenotypes in pC282Y homozygous patients. In 5 individuals we identified a HAMP gene mutation in the heterozygous state: either the pR59G and pG71D missense mutations, which are novel, or the previously described pR56X nonsense mutation. We argue that iron parameters of the 5 pC282Y homozygous patients with an additional HAMP mutation were effectively among the most elevated.
On the basis of these findings, we also searched for HAMP mutations in 31 HH patients with at least one chromosome without an HFE mutation. In 1 pC282Y, 2 pS65C, and 1 pH63D carrier, the pR59G or the pG71D HAMP mutation could be detected. A family study, from the pC282Y/pR59G proband, further confirmed that mutations in both the HAMP and HFE genes can lead to an iron overload phenotype, whereas normal iron parameters were found in subjects heterozygous for a mutation in either gene. These additional data led us to propose that, at least in some cases, the addition of a heterozygous mutation at both the HFE and HAMP loci could be responsible for an adult-onset iron overload phenotype.
Patients, materials, and methods
Patients and controls
Informed consent was obtained from all subjects including controls for DNA studies.
We first studied the HAMP coding region in 392 pC282Y homozygous patients that had a transferrin saturation level greater than or equal to 45%. At the first visit of these patients to a blood center of the western part of France, a clinical questionnaire was completed by the specialized physician. Information contained in this questionnaire was previously described in detail.25 It notably provided information regarding sociodemographic characteristics of patients, their age at onset, the circumstances of HH discovery, the biochemical parameters, and the clinical signs associated at the time of onset. This questionnaire also included data related to the treatment, such as the number and quantity of phlebotomies needed to reach depletion and the quantity of iron extracted.
We also selected 31 HH patients, from a cohort of 450 treated by phlebotomy, on the basis of 3 criteria: (1) a transferrin saturation level above the threshold (ie, ≥ 60% in males and ≥ 50% in females), (2) at least one chromosome without an HFE mutation (this point was checked by a complete denaturing high performance liquid chromatography [DHPLC] scanning of the HFE gene, as previously described29 ), and (3) exclusion of secondary known causes of iron overload. There was no history of excess alcohol intake (< 60 g daily), hematologic disease, blood transfusions, or excess oral iron intake. Moreover, serologic testing for hepatitis C and B were negative.
Included in the study as controls were 300 bone marrow donors with normal iron indices from the same geographic area.
HAMP mutations analysis
Sequence information and polymerase chain reaction (PCR). For the DHPLC scanning of the whole HAMP coding sequence we designed 2 sets of primers based on the GenBank AJ277280HAMP gene sequence: exon 1 was amplified using the forward primer 5′-GCCCCATAAAAGCGACTGTC-3′ and the reverse primer 5′-CATCCCTGCTGCTGCCCTGCTA-3′, while exons 2 and 3 were coamplified using the forward primer 5′-TCTCAGAGGTCCACTGGGC-3′ and the reverse primer 5′-GACACTCGGCAGAGAGAAAG-3′. These primers were also used for the sequencing analysis.
PCR reactions were carried out using the GeneAmp PCR system 9700 (Applied Biosystems, Foster City, CA). After a first denaturation step of 94°C for 5 minutes, PCR cycles (N/40) were as follows: 94°C for 30 seconds, 57°C or 59°C (exon 1 and exons 2-3, respectively) for 30 seconds, and 72°C for 30 seconds.
To promote heteroduplex formation the PCR product was heated to 95°C for 5 minutes and then cooled to 68°C over 15 minutes.
DHPLC analysis and sequencing. DHPLC analysis was carried out using the WAVE DNA Fragment Analysis System (Transgenomic, San Jose, CA). The melting profile and analytical conditions for each DNA fragment (exon 1 and exons 2-3 of the HAMP gene) were first predicted using WaveMaker software (Transgenomic), and then analytical conditions were established based on experimentally determined melting curves. DHPLC conditions are shown in Table 1. Sequencing of the DHPLC-positive samples was performed using a fluorescent-tagged dideoxy chain termination method with an ABI Prism Genetic Analyser (Applied Biosystems).
DHPLC temperature and gradient conditions for HAMP coding fragments, with a flow rate of 0.9 mL/min and buffer B increased at 2% per minute
Exon . | Temperature, °C . | Acetonitrile gradient, buffer B, % . | Retention time, min . |
|---|---|---|---|
| 1 | 64 | 53-59 | 4.1 |
| 2-3 | 60 | 58-64 | 4.1 |
| 2-3 | 64 | 56-62 | 4.1 |
Exon . | Temperature, °C . | Acetonitrile gradient, buffer B, % . | Retention time, min . |
|---|---|---|---|
| 1 | 64 | 53-59 | 4.1 |
| 2-3 | 60 | 58-64 | 4.1 |
| 2-3 | 64 | 56-62 | 4.1 |
Results
HAMP analysis in 392 pC282Y homozygous HH patients and 300 control subjects
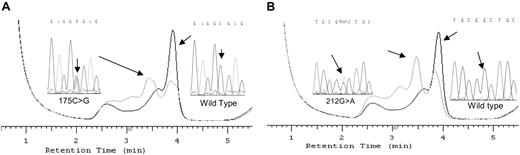
DHPLC analysis and subsequent DNA sequence analysis provide a sensitive means of identifying novel mutations in scanned genes. We used this method to analyze the HAMP coding region from 392 pC282Y homozygous HH patients with mild to severe iron overloads. DHPLC analysis revealed that 2 siblings (a male and a female) and 3 nonrelated females were heterozygotes for a variation in the region encompassing exons 2 to 3 of the HAMP gene. Subsequent sequencing analysis identified a cytosine to guanine transition at position 175 (nucleotide 1 is the adenine in the initiator methionine) in 2 nonrelated females, which changes amino acid 59 from arginine to glycine (pR59G; Figure 1A), a guanine to adenine transition at position 212 in another female, which changes amino acid 71 from glycine to aspartic acid (pG71D; Figure 1B), and the previously described pR56X HAMP mutation in the 2 siblings.
DHPLC elution profiles for the HAMP g175G>C sequence alteration and g212G>A sequence variation compared with wild-type homozygous DNA and the respective sequences. Panel A displays the overlay of chromatograms showing HAMP g175G>C sequence alteration and wild type and the respective sequences. Panel B shows the second alteration profile of HAMP g212G>A and sequences.
DHPLC elution profiles for the HAMP g175G>C sequence alteration and g212G>A sequence variation compared with wild-type homozygous DNA and the respective sequences. Panel A displays the overlay of chromatograms showing HAMP g175G>C sequence alteration and wild type and the respective sequences. Panel B shows the second alteration profile of HAMP g212G>A and sequences.
None of the 3 pR56X, pR59G, and pG71D HAMP mutations was detected in a control group of 300 individuals from the same geographic area.
Positions of the pR59G and pG71D mutations on the HAMP gene product (hepcidin)
HAMP encodes for a propeptide of 84 amino acids (SwissProt no. P81172), called the hepcidin precursor protein, that undergoes enzymatic cleavage into mature peptides of 20, 22, and 25 amino acids. As illustrated in Figure 2, arginine 59 is at the critical position of a penta-arginine basic domain (residues 55-59) from which the cleavage processing occurs (the furin prohormone convertase is probably implicated in this biologic activity30 ). Glycine 71 is located between 4 of the 8 cysteines that, based on the structure recently defined by Hunter et al using proton nuclear magnetic resonance (H MNR) spectroscopy,31 form intramolecular bonds and stabilize the β-sheet structure of the 25-amino acid mature peptide.
Peptide sequence of the hepcidin precursor protein and the position of the changed amino acids.
Peptide sequence of the hepcidin precursor protein and the position of the changed amino acids.
Iron overload phenotypes of the 5 pC282Y homozygous HH probands who have inherited an additional mutation in the HAMP gene
The male who carried the HAMP pR56X mutation presented with a severe iron overload phenotype as indicated by a transferrin saturation of 89%, a serum ferritin of 2242 μg/L, and a quantity of iron removed by phlebotomy of 14.6 g. Despite this significant iron reduction, normal iron indices have not been reached at the time of this report. His transferrin saturation level is still elevated (56%), while serum ferritin is normal (38 μg/L).
As shown in Table 2, iron overload phenotypes observed in the 4 females appeared to be more severe than those found in pC282Y homozygous females of similar age ranges. In particular we found that the mean transferrin saturation level of the 4 females was significantly different from that of pC282Y homozygous females (age range at diagnosis, 45 to 75 years) without a HAMP gene mutation (92.5% [σ = 5.4] vs 74.3% [σ = 16.7]; P = .0326, Student t test). A similar statistical analysis was performed on the serum ferritin means, using a logarithmic transformation in order to normalize the values, but no significant difference was obtained (644.0 [σ = 259.3] vs 661.8 [σ = 1098.3] μg/L; P = .3530, Student t test).
Comparative study of iron indices parameters of pC282Y homozygous females
Cohort of the pC282Y homozygous females* . | . | . | . | The 4 females with a HAMP gene mutation . | . | . | . | . | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Age range, y . | No. of patients . | Means of SF, μg/L . | Means of TS, % . | HAMP mutation . | Age-D, y . | SF, μg/L . | TS, % . | Phlebotomy IR, g . | |||||||
| 45-55 | 37 | 670 ± 1307 | 75.5 ± 19.4 | pR59G | 50 | 719 | 97 | 10.2 | |||||||
| 55-65 | 39 | 719 ± 1070 | 72.9 ± 15.7 | pG71D | 61 | 852 | 86 | 6.5 | |||||||
| pR56X | 62 | 265 | 97 | 3.4 | |||||||||||
| 65-75 | 16 | 502 ± 528 | 74.8 ± 12.4 | pG71D | 71 | 740 | 90 | 4.9 | |||||||
Cohort of the pC282Y homozygous females* . | . | . | . | The 4 females with a HAMP gene mutation . | . | . | . | . | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Age range, y . | No. of patients . | Means of SF, μg/L . | Means of TS, % . | HAMP mutation . | Age-D, y . | SF, μg/L . | TS, % . | Phlebotomy IR, g . | |||||||
| 45-55 | 37 | 670 ± 1307 | 75.5 ± 19.4 | pR59G | 50 | 719 | 97 | 10.2 | |||||||
| 55-65 | 39 | 719 ± 1070 | 72.9 ± 15.7 | pG71D | 61 | 852 | 86 | 6.5 | |||||||
| pR56X | 62 | 265 | 97 | 3.4 | |||||||||||
| 65-75 | 16 | 502 ± 528 | 74.8 ± 12.4 | pG71D | 71 | 740 | 90 | 4.9 | |||||||
SF indicates serum ferritin; TS, transferrin saturation; Age-D, age at diagnosis; and phlebotomy IR, phlebotomy iron removed.
Excluding those with a HAMP gene mutation.
These results led us to take into account the fact that HAMP gene mutations could also contribute to the expression of an iron overload phenotype in HH patients with at least one chromosome without an HFE mutation.
HAMP DHPLC in HH patients with at least one chromosome without an HFE mutation
The entire coding region of the HAMP gene was screened in 31 selected HH patients with at least 1 chromosome lacking an HFE mutation (details of the selection criteria are provided in “Patients, materials, and methods”). In 4 male carriers of one mutation at the HFE locus (either the pC282Y, pH63D, or pS65C) the pR59G or pG71D missense mutations could be detected at the HAMP locus. Details of patient genotypes as well as of their biochemical iron parameters assayed at the time of diagnosis are provided in Table 3.
Genotypes and biochemical iron parameters from 4 patients with HFE and HAMP mutations
Patients . | HFE mutation . | . | . | HAMP mutation . | Biochemical parameters . | . | |||
|---|---|---|---|---|---|---|---|---|---|
| Age-D, y . | pC282Y . | pH63D . | pS65C . | . | SF, μg/L . | TS, % . | |||
| 44 | −/− | −/− | +/− | pR59G | 716 | 75 | |||
| 50 | −/− | −/− | +/− | pR59G | 1052 | 61 | |||
| 43 | +/− | −/− | −/− | pR59G | 849 | 70 | |||
| 34 | −/− | +/− | −/− | pG71D | 275 | 85 | |||
Patients . | HFE mutation . | . | . | HAMP mutation . | Biochemical parameters . | . | |||
|---|---|---|---|---|---|---|---|---|---|
| Age-D, y . | pC282Y . | pH63D . | pS65C . | . | SF, μg/L . | TS, % . | |||
| 44 | −/− | −/− | +/− | pR59G | 716 | 75 | |||
| 50 | −/− | −/− | +/− | pR59G | 1052 | 61 | |||
| 43 | +/− | −/− | −/− | pR59G | 849 | 70 | |||
| 34 | −/− | +/− | −/− | pG71D | 275 | 85 | |||
All patients were male.
SF indicates serum ferritin; TS, transferrin saturation; and Age-D, age at diagnosis.
Venesections were instituted in the 4 HH patients. An equivalent of 7.4 g iron, sufficient to restore normal iron status, was removed in one pS65C/pR59G carrier (the 50-year-old man), whereas an equivalent of 9 g iron was removed in the pH63D/pG71D carrier. In the other 2 men the curative part of the venesection process has begun more recently and is still in course. Consequently, quantities of iron removed are not yet informative.
Family study
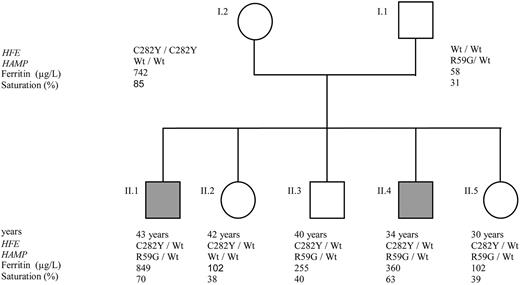
For the pC282Y/pR59G mutations carrier, a rapid family study was feasible. The family tree is present in Figure 3.
Family tree of the pC282Y/pR59G proband. Wt indicates wild-type allele; grey square or circle, patients with a HH phenotype; SF, serum ferritin; and TS, transferrin saturation.
Family tree of the pC282Y/pR59G proband. Wt indicates wild-type allele; grey square or circle, patients with a HH phenotype; SF, serum ferritin; and TS, transferrin saturation.
The proband (number II.1) was a 43-year-old man with a transferrin saturation of 70% and serum ferritin of 849 μg/L. Interestingly, his mother was already known to display the pC282Y homozygous genotype. She was included in a venesections protocol at our center during which an equivalent of 8 g iron was removed in order to restore normal iron indices. The proband's father was found to be heterozygous for the pR59G HAMP mutation and had normal iron parameters.
The proband has 2 brothers and 2 sisters. The younger brother (II.4) had an identical genotype with a transferrin saturation of 63% and serum ferritin of 360 μg/L. As for the proband, he has no other known cause of iron disorder. The second brother (II.3) and one sister (II.5) were found to be heterozygous for the pC282Y HFE and pR59G HAMP mutations without evidence of an iron overload. It must be emphasized that the woman gave birth 3 months prior to our determination of iron parameters and that, before this event, she had a transferrin saturation of 63% (data collected from another biologic laboratory). The second sister (II.2) was heterozygous for the pC282Y HFE mutation with normal transferrin saturation and serum ferritin levels.
Discussion
The phenotypic expression of the pC282Y homozygous genotype is very heterogeneous and could be modulated by both environmental factors and modifier genes.23,24,26-28 To address the question of whether HAMP could be one such gene, we performed DHPLC scanning of the HAMP coding region, with subsequent sequence analysis, in 392 pC282Y homozygous patients. In 5 of these patients, 1 male and 4 females, we identified 3 HAMP mutations in the heterozygous state. Of these mutations, 2 were novel, the pR59G and pG71D missense mutations,32,33 and 1 was previously described, the pR56X nonsense mutation.
Positional study of the pR59G amino acid change revealed that it occurred in a penta-arginine basic domain (residues 55-59) in which arginine 59 is at the critical position for activity of prohormone convertases and, particularly, for furin, which processes substrates having an RX(K/R)R sequence.34 Functionally, one may thus consider that the effect of this mutation is to prevent the formation of the 25-amino acid mature peptide. The example of mutations that change the last arginine of the fibroblast growth factor 23 (FGF23) RXXR processing site, which are thought to prevent a proteolytic cleavage activity and result in the autosomal dominant hypophosphatemic rickets (ADHR; OMIM no. 193100) disorder, argue for this assumption.35,36 To date, the functional relevance of the pG71D amino acid substitution is less clear. However, it must be emphasized that this missense mutation is located between 4 of the 8 structural cysteines of the 25-amino acid mature peptide,31 and that the change of a neutral amino acid to an acidic residue frequently leads to crucial protein structure modifications.
The comparative study presented here of pC282Y homozygous females of similar age ranges has shown that iron indices of the 4 females with a HAMP mutation were among the most elevated. The results were also significant (P = .0326, Student t test) for the transferrin saturation levels. The male who carried the HAMP pR56X mutation harbored a more severe iron overload phenotype (transferrin saturation of 89%, serum ferritin of 2242 μg/L, and a quantity of iron removed by phlebotomy of 14.6 g), and, while his serum ferritin concentration is now normal (38 μg/L), in spite of 55 phlebotomies, his transferrin saturation level is still clearly above the normal range (56%). These data led us to propose that, even if they are not frequent,37-39 HAMP mutations could effectively explain one part of the pC282Y/pC282Y-related phenotypic heterogeneity by accentuating the iron burden.
Based on these findings, another objective of our study was to determine if HAMP mutations could also contribute to the expression of an iron overload phenotype in a group of 31 adult-onset patients with at least one chromosome lacking an HFE mutation. These patients were identified in a screening study on the basis of an elevated transferrin saturation level (≥ 60% in males and ≥ 50% in females) in the absence of other known causes of iron disorder. In 4 of the 31 HH selected patients (ie, 13%), we identified the 2 novel pR59G and pG71D missense mutations in the heterozygous state.
We further performed a family study in relatives of the pC282Y and pR59G mutations carrier. The proband had a transferrin saturation level of 70% and a serum ferritin level of 849 μg/L at diagnosis. His younger brother (II.4), who is also a carrier of the pC282Y and pR59G mutations, had a transferrin saturation level of 63% and a serum ferritin level (360 μg/L) that is clearly above the normal range for a 34-year-old man. The proband's father and one of his sisters, who are heterozygous for the pR59G and pC282Y mutations, respectively, had normal transferrin saturation and serum ferritin levels.
Taken together, these data confirm that the combination of a genetic alteration at both the HFE and HAMP loci could lead to an adult-onset phenotype of hereditary hemochromatosis that is not apparent when an individual carries only one of these gene alterations. A similar situation has very recently been reported by Merryweather-Clarke et al who have partially described the pedigree of a proband heterozygous for the C282Y HFE mutation and for a HAMP deletion that removes the last 3 nucleotides of exon 2 and the first one of intron 2.33 Thus, on the basis of the example of several other genetic disorders,40 it is tempting to propose that, at least in some cases, a digenic model of inheritance might be responsible for an adult-onset iron overload phenotype.
Observation that the older brother of the proband (II.3) had normal iron parameters led us to postulate that the penetrance of the pC282Y/pR59G genotype might, however, not be complete. This might be related to the recognition of nonexpressing elderly pC282Y homozygous individuals, even with relatives of iron overload pC282Y/pC282Y patients.41,42 On the other hand, one can also consider a more complex genetic situation implicating a third mutation that might be located either in noncoding regions of the HFE and HAMP genes or in another iron metabolism gene. Considering the familial tree of the pC282Y/pR59G proband, this third mutation could be detected in the proband (II.1), his younger brother (II.4), and his younger sister (II.5), but not in his older brother (II.3). Today, because nongenetic factors account for part of the iron overload processing, this later subject must be considered as carrying mutations that may lead to hemochromatosis but not yet having the disease. He needs to be followed carefully.
Our results shed new light on the molecular basis of primary iron overloads. They are in accordance with the demonstration by Nicolas et al of an increased hepatic iron loading in Hfe-/- mice with an additional HAMP deficient allele (see accompanying article by Nicholas et al,43 beginning on page 2841). They are also in accordance with several studies that, during the past 2 years, have concluded that the HAMP gene product, called hepcidin, might act through the same regulatory pathways as HFE to control the amount of iron in the body.28,44-47 In the case of an excess of iron in the body, the control process should include a form of action on the reticulo-endothelial macrophages, by promoting inhibition of iron release from senescent red blood cells,30,48 and an action on duodenal cells, by promoting inhibition of alimentary iron absorption.30,49,50 Even if the relationship between HFE and HAMP has still to be clearly defined, it is interesting to note that the first demonstration of a relation between the 2 proteins has very recently been forthcoming from 3 studies that have highlighted that HFE plays an important role in the regulation of hepatic HAMP gene expression.45-47
Prepublished online as Blood First Edition Paper, December 11, 2003; DOI 10.1182/blood-2003-10-3366.
Supported by grants from the Programme Hospitalier de Recherches Clinique “Mise en place d'un diagnostic phenotypique des surcharges en fer primaire” and the Conseil Scientifique de l'Etablissement Français du Sang (projet 2003.19).
S.J. and G.L.G. contributed equally to this study.
An Inside Blood analysis of this article appears in the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal