Abstract
Hereditary hemochromatosis (HH) type I is a disorder of iron metabolism caused by a mutation in the HFE gene. Whereas the prevalence of the mutation is very high, its penetrance seems very low. The goal of our study was to determine whether hepcidin, a recently identified iron-regulatory peptide, could be a genetic modifier contributing to the HH phenotype. In mice, deficiency of either HFE (Hfe-/-) or hepcidin (Usf2-/-) is associated with the same pattern of iron overload observed in patients with HH. We intercrossed Hfe-/- and Usf2+/- mice and asked whether hepcidin deficiency increased the iron burden in Hfe-/- mice. Our results showed that, indeed, liver iron accumulation was greater in the Hfe-/-Usf2+/- mice than in mice lacking Hfe alone. This result, in agreement with recent findings in humans, provides a genetic explanation for some variability of the HH phenotype. (Blood. 2004;103: 2841-2843)
Introduction
Hereditary hemochromatosis (HH) is a genetic disorder of iron metabolism resulting in a chronic increase in intestinal iron absorption. As the disease progresses, patients develop toxic iron overload and complications of tissue damage including liver cirrhosis, hepatocarcinoma, and heart disease. Most patients with HH are homozygous for a missense mutation (C282Y) that disrupts the conformation of HFE, an atypical major histocompatibility class I molecule.1 Similar to human patients, mice lacking the Hfe protein2-4 or producing a mutated protein analogous to the human C282Y protein3 develop increased hepatic iron levels and elevated transferrin saturation. Whereas HH is among the most prevalent genetic disorders (about 5 people in 1000 are homozygotes for the C282Y mutation), the clinical penetrance of the mutation is low, suggesting that the HFE C282Y mutation is a necessary but not sufficient cause of clinical HH.5 Variable penetrance may be due to epigenetic, environmental, and genetic factors.
In mice, there is a marked difference in hepatic iron loading between the C57BL/6 and DBA/2 Hfe-/- strains, indicating that other genes modify the murine HH phenotype.6 Several candidate modifier genes have been investigated in mice and humans.7,8 Here, we evaluated the iron regulatory peptide, hepcidin,9-12 as a potential modifier of iron loading. Hepcidin is believed to act as a negative regulator of iron release from absorptive enterocytes and from macrophages that mediate iron recycling from senescent red cells. Its complete absence in hepcidin-deficient mice (the Usf2-/- model12 ) leads to iron accumulation in parenchymal cells.12 Accordingly, homozygous mutations in HAMP, the gene encoding human hepcidin, have been identified in 2 families with severe juvenile hemochromatosis.13 Whereas hepcidin synthesis is physiologically increased by dietary iron to avoid excess iron accumulation,10 this response is defective in patients homozygous for the C282Y mutation in HFE14 and in Hfe-/- mice.14-17 Furthermore, forced expression of hepcidin prevented iron overload in Hfe-/- mice.17 These data support the hypothesis that inappropriate hepcidin regulation contributes to the iron loading phenotype. In the present study, we asked whether haploinsufficiency for hepcidin would exacerbate the phenotype of Hfe-/- mice.
Study design
Animals
Hfe-/- mice and hepcidin-deficient Usf2-/- mice were described previously.3,12 It is noteworthy that the Usf2-/- mice (originally mixed 129/Sv-C57BL/6) were bred to uniformity (8 backcrosses) on a 129/Sv genetic background. Hfe genotype analysis was performed on tail DNA using primers specific for the wild-type Hfe allele (forward: 5′-TTCTTTAGATAGCCTCTCAC-3′, and reverse: 5′-GTGGCGAGTCACTTTCACCA-3′) and the targeted Hfe allele (forward: 5′-AGTTGGGAGTGGTGTCCGA-3′, and reverse: 5′-CTAGCTTCGGCCGTGACG-3′), resulting in 502-base pair (bp) and 190-bp products, respectively. Usf2 genotype analysis was performed as previously described.12 Hfe-/- and Usf2+/- mice were interbred and resulting double heterozygous progeny were crossed with Hfe-/-Usf2+/+ mice. Due to the poor viability of Usf2-/- mice,12 Hfe-/-Usf2-/- animals could not be investigated.
Liver iron determination
Liver nonheme iron content was determined as described previously.18
RNA isolation and PCR analysis
Total RNA was isolated and double-stranded cDNA was synthesized as described previously.12 Real-time quantification of transcripts was performed on 25-μL samples in an ABI PRISM 7700 Sequence Detector (Applied Biosystems, Foster City, CA) using SYBR Green polymerase chain reaction (PCR) master mix (Applied Biosystems). Sequences of the primers were: hepc1, forward: 5′-CCTATCTCCATCAACAGATG-3′ and reverse: 5′-AACAGATACCACACTGGGAA-3′; gapdh, forward: 5′-TGCACCACCAACTGCTTAG-3′ and reverse: 5′-GGATGCAGGGATGATGTTC-3′.
Statistical analysis
Results are expressed as mean ± SD for n animals, and statistical analysis was performed using Student t test (unpaired, 2 tailed). All statistical analyses were performed using StatView version 5.0 (SAS Institute, Cary, NC). To study the effects of Hfe and Usf2 genotypes on hepatic iron content and hepc1 gene expression, a Kruskal-Wallis test was performed.
Results and discussion
We intercrossed Hfe-/- mice and Usf2+/- mice to further investigate the role of hepcidin in the pathogenesis of liver iron overload in HH. Usf2+/- mice carry only one functional allele for both hepc1 and hepc2, the gene products resulting from a tandem duplication of the hepcidin gene in mice, due to insertional disruption of one allele of Usf2, a gene that lies in close proximity to the murine hepcidin locus. Although the roles of these proteins are incompletely understood, hepc1, the peptide most closely resembling human hepcidin, is known to be important in iron metabolism.19 Compound mutant mice were killed at 4 or 8 weeks of age to determine liver iron content. The abundance of hepc1 transcripts was assessed by real-time PCR quantification.
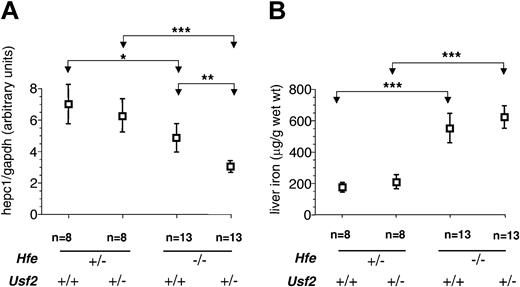
Figure 1A shows the relationship between genotype and liver hepc1 expression. In agreement with previously reported data,15-17 we found a 40% reduction in hepc1 expression in Hfe-/-Usf2+/+ mice as compared to Hfe+/-Usf2+/+ mice. This reduction in the level of hepc1 was greater (75%) in Hfe-/-Usf2+/- mice, indicating that haploinsufficiency for hepcidin expression results from inactivation of one allele. Figure 1B shows, as described previously, that Hfe-/-Usf2+/+ mice accumulate 3 times more iron than Hfe+/-Usf2+/+ mice.17 They also accumulate more iron than doubly heterozygous Hfe+/-Usf2+/- mice. However, there was no difference in iron loading observed when comparing Hfe-/-Usf2+/+ and Hfe-/-Usf2+/- mice at 4 weeks of age, despite decreased hepc1 expression in the Hfe-/-Usf2+/- group.
Hepc1 quantification and nonheme iron concentration in livers of 4-week-old mice.Hepc1 amount was quantified by real-time PCR and normalized to the amount of gapdh as described in “Study design.” Results are expressed as mean ± SD for n animals. *P < .01; **P < .001; ***P < .0001. The Kruskal-Wallis test was P < .0001 (mean rank: Hfe-/-Usf2+/-, 10.75; Hfe-/-Usf2+/+, 21.35; Hfe+/-Usf2+/-, 30.50; Hfe+/-Usf2+/+, 34.25). (B) Nonheme iron concentrations in liver samples from 4-week old mice with different Hfe and Usf2 genotypes. Iron content was measured in 2 pieces of liver for each mouse. Results are expressed as mean ± SD for n animals. The Kruskal-Wallis test was P < .0001 (mean rank: Hfe-/-Usf2+/-, 30.29; Hfe-/-Usf2+/+, 29.70; Hfe+/-Usf2+/-, 11.50; Hfe+/-Usf2+/+, 5.50).
Hepc1 quantification and nonheme iron concentration in livers of 4-week-old mice.Hepc1 amount was quantified by real-time PCR and normalized to the amount of gapdh as described in “Study design.” Results are expressed as mean ± SD for n animals. *P < .01; **P < .001; ***P < .0001. The Kruskal-Wallis test was P < .0001 (mean rank: Hfe-/-Usf2+/-, 10.75; Hfe-/-Usf2+/+, 21.35; Hfe+/-Usf2+/-, 30.50; Hfe+/-Usf2+/+, 34.25). (B) Nonheme iron concentrations in liver samples from 4-week old mice with different Hfe and Usf2 genotypes. Iron content was measured in 2 pieces of liver for each mouse. Results are expressed as mean ± SD for n animals. The Kruskal-Wallis test was P < .0001 (mean rank: Hfe-/-Usf2+/-, 30.29; Hfe-/-Usf2+/+, 29.70; Hfe+/-Usf2+/-, 11.50; Hfe+/-Usf2+/+, 5.50).
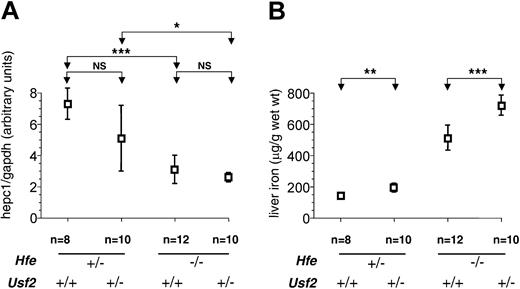
We hypothesized that the physiologic effects of decreased hepcidin synthesis might become more pronounced with increasing age, as a cumulative effect on intestinal iron absorption. To further investigate this possibility, we analyzed a second group of mice at 8 weeks of age. In contrast to 4-week-old animals, we observed a sexual dimorphism in iron loading, as previously reported,20 with females accumulating more iron than males (data not shown). At 8 weeks we found that haploinsufficiency for hepcidin was associated with increased liver iron overload (Figure 2B). The increase was approximately 40% for both sexes (females: 721 ± 86 μg/g in Hfe-/-Usf2+/- [n = 5] versus 514 ± 72 μg/g in Hfe-/-Usf2+/+ [n = 6], P < .01; males: 502 ± 32 μg/g in Hfe-/-Usf2+/- [n = 5] versus 354 ± 65 μg/g in Hfe-/-Usf2+/+ [n = 6], P < .01). These data strongly support the interpretation that hepcidin deficiency contributes to iron overload in Hfe-/- mice.
Hepc1quantification and nonheme iron concentration in livers of 8-week-old mice.Hepc1 quantification (A) and nonheme iron concentration (B) are as in Figure 1 but in liver samples from 8-week-old mice. Results are expressed as mean ± SD for n animals (NS indicates nonsignificant). Results in panel B were normalized according to the sex. The sex ratio (female-male) for hepatic iron was 1.44 for Hfe-/-Usf2+/- mice, 1.45 for Hfe-/-Usf2+/+ mice, 1.28 for Hfe+/-Usf2+/- mice, and 1.24 for Hfe+/-Usf2+/+ mice. The Kruskal-Wallis test for hepc1 quantification was P < .01 (mean rank: Hfe-/-Usf2+/-, 11.55; Hfe-/-Usf2+/+, 13.67; Hfe+/-Usf2+/-, 23.64; Hfe+/-Usf2+/+, 31.57) and for nonheme iron concentration P < .0001 (mean rank: Hfe-/-Usf2+/-, 29.80; Hfe-/-Usf2+/+, 21.92; Hfe+/-Usf2+/-, 10.71; Hfe+/-Usf2+/+, 4.29). *P < .01; **P < .001; ***P < .0001.
Hepc1quantification and nonheme iron concentration in livers of 8-week-old mice.Hepc1 quantification (A) and nonheme iron concentration (B) are as in Figure 1 but in liver samples from 8-week-old mice. Results are expressed as mean ± SD for n animals (NS indicates nonsignificant). Results in panel B were normalized according to the sex. The sex ratio (female-male) for hepatic iron was 1.44 for Hfe-/-Usf2+/- mice, 1.45 for Hfe-/-Usf2+/+ mice, 1.28 for Hfe+/-Usf2+/- mice, and 1.24 for Hfe+/-Usf2+/+ mice. The Kruskal-Wallis test for hepc1 quantification was P < .01 (mean rank: Hfe-/-Usf2+/-, 11.55; Hfe-/-Usf2+/+, 13.67; Hfe+/-Usf2+/-, 23.64; Hfe+/-Usf2+/+, 31.57) and for nonheme iron concentration P < .0001 (mean rank: Hfe-/-Usf2+/-, 29.80; Hfe-/-Usf2+/+, 21.92; Hfe+/-Usf2+/-, 10.71; Hfe+/-Usf2+/+, 4.29). *P < .01; **P < .001; ***P < .0001.
Our observations suggest that differences in basal hepcidin expression might contribute to phenotypic variability in clinical expression of HH in human patients. A similar conclusion was reached by Merryweather-Clarke et al,21 who identified heterozygous hepcidin mutations in 2 HH families. They noted a correlation between the severity of the iron overload and the presence of hepcidin (HAMP) mutations in individuals with HFE C282Y mutations. In the accompanying paper, Jacolot et al extend those results, providing further evidence that coexistence of mutations in HFE and HAMP genes may lead to new forms of hemochromatosis (see accompanying article by Jacolot et al,22 beginning on page 2835).
It is worth noting that at 8 weeks, the amount of hepc1 remained low in the Hfe-/-Usf2+/- mice as compared to the other groups (Figure 2A). This difference, however, did not reach statistical significance most likely due to the wide range in hepc1 expression in the Hfe-/-Usf2+/+ group.
In conclusion, our result confirms the key role of hepcidin in the control of iron homeostasis and further highlight that decreased hepcidin contributes to the iron homeostasis abnormalities characteristic of HH. Although the mechanism of action of hepcidin has not yet been established, we speculate that hepcidin or its analogs may prove to be useful in the treatment of hemochromatosis.
Prepublished online as Blood First Edition Paper, December 4, 2003; DOI 10.1182/blood-2003-09-3358.
An Inside Blood analysis of this article appears in the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Bernard Grandchamp for support, discussion, and advice. We acknowledge the technical assistance of Jacqueline Bauchet, Nicolas Sorhaindo, Azhour Lahdar and Houda Beltaif (Centre d'Explorations Fonctionnelles Intégré, Faculté de Médecine Xavier Bichat, Paris) and Laurent Gouya for helpful discussion.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal