Abstract
For the clinical evaluation of the efficacy of cellular immunotherapy it is necessary to analyze the effector functions of T cells against primary leukemic target cell populations which are usually considerably heterogeneous caused by differential maturation stages of the leukemic cells. An appropriate assay should not only allow the quantitative analysis of rapid cell death induction as measured by the conventional 51Cr release assay but also of the more slowly executing pathways of T-cell-induced apoptosis occurring within days instead of hours which cannot be measured using this method. Furthermore, it should dissect the differential susceptibility to T-cell-induced cell death of various target cell subpopulations and characterize the malignant precursor cells capable of producing malignant progeny. To fulfill these requirements we developed a new assay based on carboxyfluorescein diacetate succinimidyl ester (CFSE) labeling of the target cell population combined with antibody staining of specific cell populations and addition of fluorescent microbeads to quantitatively monitor target cell death occurring within a longer time frame up to at least 5 days. This new assay facilitates the analysis of differential recognition of distinct cell types within a heterogeneous target cell population and allows simultaneously evaluation of the proliferative status of surviving target cells in response to relevant cytokines. (Blood. 2004;103: 2677-2682)
Introduction
Cellular immunotherapy of relapsed leukemia after allogeneic stem cell transplantation using donor lymphocyte infusions (DLIs)1,2 or leukemia-reactive cytotoxic T lymphocytes (CTLs)3 has been found to be effective in a limited number of patients. Especially, chronic myeloid leukemia (CML) was found to be a suitable target for immunotherapy. CTLs mediate tumor cell death by various effector mechanisms, including the induction of rapid target cell lysis by way of the perforin/granzyme pathway occurring already within minutes up to a few hours after interaction with the T cell, or a more slow induction of target cell apoptosis through the interaction of Fas ligand (FasL) to the Fas receptor or tumor necrosis factor α (TNF-α) to the TNF receptor,4 which may take 12 to 24 hours to occur.5,6 The contribution of each of these effector mechanisms in clinically relevant CTL-induced cell death is still unclear.
For the clinical evaluation of immunotherapeutic interventions in patients with leukemia it is necessary to determine the efficacy and the specificity of isolated T cells for the leukemia. Not only the ability of these T cells to induce rapid perforin-mediated cell death but also their capacity to execute apoptosis by way of the slower pathways may be of importance. In addition, it is essential to determine the specific toxicity of T cells against different specific subpopulations within a cell suspension. Primary leukemic cell populations are usually considerably heterogeneous, consisting of cells at differential maturation stages, with only a minority of cells being capable of sustained proliferation and production of malignant progeny. The fate of the subpopulation of CD34+ leukemic progenitor cells within a leukemic cell population may play an important role in the evaluation of the probability of success of the treatment. We have previously demonstrated that following DLI for CML after allogeneic stem cell transplantation (SCT) the response correlated with the frequency of T cells that were capable of recognizing the leukemic progenitor cells within the target cell population and illustrated that these T cells were capable of inhibition of leukemic precursor cells capable of cytokine-induced proliferation in CML.2,7 We recently further demonstrated the relevance of the cell cycle status of leukemic cells for their sensitivity to Fas-mediated cell death, which was found to be specific for cells in activated G1 phase of the cell cycle.6 Leukemic (precursor) cells in noncycling dormant G0 phase of the cell cycle as well as leukemic cells in cycling S or G2/M phase of the cell cycle were relatively resistant against this pathway of cell death induction. Therefore, the susceptibility to T-cell-mediated cell death of the subpopulation of leukemic (precursor) cells in dormancy as compared with those in active phases of the cell cycle needs to be investigated in detail.
Thus, the requirements for an appropriate in vitro assay for the clinical evaluation of the effectiveness of immunotherapeutic interventions are not limited to high sensitivity and the possibility of quantitative analysis, but in addition the assay should facilitate the analysis of the more slowly executing pathways of target cell death induction occurring within days instead of hours. Furthermore, it should dissect the relative susceptibility of distinct cell types within a heterogeneous cell population to T-cell recognition and should give information on the recognition of malignant precursor cells capable of producing progeny.
The most widely used assay for quantification of CTL-induced target cell death is the conventional 51Cr release assay.8 Although this assay has been proven to be a reliable and reproducible assay for the analysis of rapid target cell lysis occurring within 4 hours, the increasing spontaneous release of 51Cr by many primary cell types in assays longer than 4 to 8 hours disables the analysis of the more slowly executing effector mechanisms of T-cell-induced cell death. The 51Cr release assay does not allow the evaluation of susceptibility to lysis of distinct cell types within the target cell population. In addition, radioactivity of the labels hampers the use in certain laboratories, although it may be replaced by europium (Eu)9 or lactate dehydrogenase (LDH).10 However, all these assays are still not suitable for the detection of differential recognition and kill of different cell types within a target cell population, and they do not supply information on the proliferative capacity of the target cells. To fulfill the requirements for a more optimal assay for the clinical evaluation of the efficacy of immunotherapeutic interventions we developed a new type of cytotoxicity assay based on staining with carboxyfluorescein diacetate succinimidyl ester (CFSE) and flow cytometry as read out.
CFSE is a dye that spontaneously and irreversibly binds to intracellular proteins. On cell division the CFSE labeling is equally distributed among the daughter cells, which, therefore, contain half the fluorescent dye compared with the parental cells.11,12 On the basis of the CFSE fluorescence the number of cell divisions can be followed. Recently, CFSE staining has been described for its use in functional assays for the monitoring of immunologic processes.13-15 In these assays CFSE was used primarily as a dye to stain either the effector cells or the target population, enabling discrimination of these cell types in a mixed lymphocyte reaction (MLR) rather than supplying information on the specific proliferation within the CFSE-labeled population.
We developed a novel cytotoxicity assay using CFSE labeling not only as a dye to specifically stain the target cell population but also as a dye allowing the analysis of the number of individual cell divisions occurring within the target cell populations, combined with monoclonal antibodies to stain for specific cell types within a heterogeneous (target) cell population and fluorescent beads to enable quantitative analysis of the number of CFSE-labeled target cells that underwent cell death. Not only does this assay function as a replacement for the conventional 51Cr release assay, but also the stability of the CFSE signal in nondividing target cells allows the analysis of the more slowly executing CTL effector mechanisms in a time frame of days instead of hours. Specific staining of distinct cell types, including the leukemic precursor cells within the target cell population, facilitated the analysis of differential susceptibility to lysis of these cells. In addition, on the basis of the distribution of the CFSE fluorescence following cell divisions, the specific proliferation of all cell types within the target cell population in response to specific cytokines can be followed in time. By combining these technical features this assay fulfills the requirements for adequate clinical monitoring of the effectiveness of immunotherapeutic interventions.
Materials and methods
Target cell populations
Peripheral blood of a patient with acute myeloid leukemia (AML) French-American-British (FAB) type M2 and from 3 patients with chronic-phase CML was obtained after informed consent. Ficoll density separation was performed, and mononuclear cells were cryopreserved. After thawing, the cells were resuspended in assay medium consisting of Iscove modified Dulbecco medium (IMDM; BioWhittaker, Verviers, Belgium) supplemented with 3 mM l-glutamine (BioWhittaker), antibiotics, 10% human serum, 20 μg/mL transferrin (Serva, Heidelberg, Germany), and 5 × 10-5 M β-mercaptoethanol (Sigma-Aldrich, Zwijndrecht, the Netherlands). The cells were cultured at 37°C and 5% CO2. Proliferation was induced by the addition of 50 ng/mL interleukin-3 (IL-3; Novartis, Bern, Switzerland), 100 ng/mL granulocyte-macrophage colony-stimulating factor (GM-CSF; Novartis), 100 ng/mL granulocyte colony-stimulating factor (G-CSF; Neupogen; Amgen Europe, Breda, the Netherlands), and 25 ng/mL stem cell factor (SCF; Amgen). For the CML cells, 2 U/mL recombinant human erythropoietin (r-HuEPO; Eprex; Janssen-Cilag, Tilburg, the Netherlands) was also added.
The growth factor-dependent AML cell line AML-19316 was obtained from American Type Culture Collection (ATCC; Rockville, MD). The cells were cultured in serum-free medium, consisting of IMDM (BioWhittaker) supplemented with 3 mM l-glutamine (BioWhittaker), antibiotics, 0.4% human serum albumin (HSA) (wt/vol) (CLB, Amsterdam, the Netherlands), 20 μg/mL cholesterol (Sigma-Aldrich), 20 μg/mL transferrin (Serva), 5 × 10-5 M β-mercaptoethanol (Sigma-Aldrich), and 10 μg/mL insulin (Sigma-Aldrich). The cells were cultured at 37°C and 5% CO2 in serum-free medium in the presence of 20 ng/mL recombinant human (rh) GM-CSF (Novartis).
Effector T-cell clones
The CD8+ CTL clones H and G directed against the AML-193 cell line were isolated by limiting dilution assay from an in vitro-generated immune response by using peripheral blood of an unrelated partially HLA-matched healthy donor as responder population and the AML-193 cell line as stimulator population.
The CD8+ HA-1-specific T-cell clone was isolated from a patient with HLA-A2/HA-1-positive CML after treatment with DLI from an HLA-matched HA-1-negative donor by using the interferon γ (IFN-γ) secretion assay (Miltenyi Biotec, Bergisch Gladbach, Germany), following stimulation of the peripheral blood T cells with pre-SCT bone marrow of the patient.
The CD8+ HLA-A1-restricted anti-HY CTL clone was generated as described previously.17
51Cr release assay
The conventional 51Cr release assay was performed as described previously.6 In short, target cells were labeled with 100 μCi (3.7 MBq) Na251CrO4 (Amersham Pharmacia Biotech Benelux, Roosendaal, The Netherlands) for 1 hour at 37°C. After 3 washes the 51Cr-labeled targets were plated in a 96-well U-bottom microtiter plate in 100 μL IMDM plus 10% human serum at 5 × 104 cells/mL. CTLs were added at different effector-target ratios. Plates were incubated in a humidified atmosphere of 5% CO2 and 37°C. After 4 hours 25 μL supernatant was harvested on solid scintillator-coated lumaplates (Packard, Groningen, the Netherlands) and counted in an automatic gamma counter (Topcount, Packard). For each target cell type, spontaneous release and maximal release were determined. The ratio of maximal to spontaneous release always exceeded 5. Percentage of lysis was calculated as follows: % specific cytotoxicity = [experimental release (cpm) - spontaneous release (cpm)]/[maximal release (cpm) - spontaneous release (cpm)] × 100.
CFSE-based cytotoxicity assay
After washing with phosphate-buffered saline (PBS) the target cell suspensions were resuspended at 20 × 106 cells/mL and labeled with 10 μM CFSE (Molecular Probes Europe, Leiden, the Netherlands) for 10 minutes at 37°C. The reaction was stopped by the addition of an equal volume of fetal calf serum (FCS), followed by a 2-minute incubation at room temperature. After 2 washes the CFSE-labeled target cells were resuspended in assay medium and either directly used or cultured for 1 to 3 days at 37°C and 5% CO2 in the presence or absence of cytokines. The cell concentration was adjusted to 5 × 104 cells/mL, and 100 μL/well was plated in 96-well microtiter plates. CTLs were added at different effector-target ratios. Plates were incubated in a humidified atmosphere of 5% CO2 and 37°C. After 4 hours, 24 hours, 48 hours, and 72 hours relevant antibodies to stain for specific subpopulations were added and incubated at 4°C for at least 15 minutes. Phycoerythrin (PE)-labeled mouse CD3, CD33, and CD34 antibodies were purchased from Becton Dickinson (BD; San Jose, CA). PE-Cy5 CD3 antibodies were obtained from Immunotech (Beckman Coulter, Mijdrecht, the Netherlands). The wells were harvested, and to allow quantitative analysis of the cell populations, 10 000 Flow-Count Fluorospheres (Coulter Corporation, Miami, FL) were added. This was done just prior to the analysis to avoid the formation of complexes between the cells and the beads. To stain for dead cells, propidium iodide (1 μg/mL) was added, and samples were mixed properly and directly analyzed by flow cytometry. For each sample 5000 microbeads were acquired, facilitating the calculation of absolute numbers of target cells. The absolute number of surviving cells was determined at each time point by calculation of the ratio between the number of cells and the number of beads. Because a fixed amount of beads (10 000) was always added and acquired (5000), the absolute number of cells could be extrapolated at every time point x. The absolute number of cells at the moment of T-cell addition (t = 0) is the number of target cells added to the wells (5000). The percentage of survival was calculated as follows: % survival = [absolute no. viable CFSE+ target cells (t = x)]/[absolute no. viable CFSE+ target cells (t = 0)] × 100.
To analyze the HLA class I or II restriction of the target cell lysis, blocking studies were performed. Target cells were incubated with saturated concentrations of anti-HLA class I or anti-HLA class II monoclonal antibodies (W6/32 and PdV5.2, respectively) for 30 minutes before effector and target cells were cocultured.
Results
Correlation between the CFSE-based cytotoxicity assay and the conventional 51Cr release assay
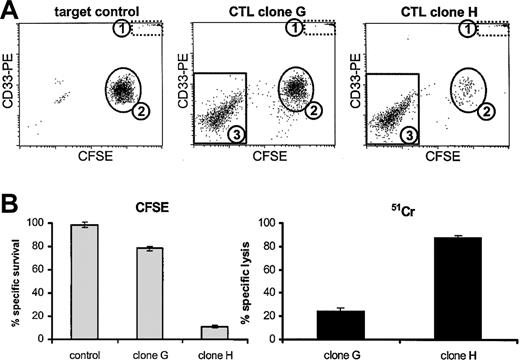
To first determine whether the results obtained in a standard 51Cr release assay could be mimicked by the CFSE-based cytotoxicity assay, we performed these assays in parallel. CD33+ AML-193 cells were labeled either with CFSE or with 51Cr and incubated with 2 specific CTL clones at a 1:1 E/T ratio for 4 hours. After 4 hours, the wells were harvested and analyzed according to the assay protocols described. Figure 1A demonstrates the design of the CFSE-based cytotoxicity assay. Dot plots are shown of the viable cells within the life gate. In the target control, 2 populations could be discriminated, comprising the fluorescent microbeads (population 1) used for quantification of the absolute number of viable cells in each sample, which could be discriminated as a distinct population both on the basis of their scattering pattern as well as by their high fl-1, fl-2, and fl-3 fluorescence, and also as a population of CD33+ CFSE-positive AML-193 cells (population 2). After 4 hours of incubation with the nonpotently killing CTL clone G, a small decrease in the number of AML-193 cells was visible. The effector T cells were visible as a population of CFSE-negative CD33- cells (population 3). Incubation for 4 hours with the potent CTL clone H resulted in a strong induction of target cell death, resulting in almost complete deletion of population 2. The percentage-specific survival of AML-193 cells was calculated relative to the target control incubated in the absence of CTLs using the ratio between the number of viable AML-193 cells and the number of fluorescent beads. Figure 1B demonstrates the results obtained with the CFSE-based cytotoxicity which revealed that 78% ± 2% of the AML-193 cells survived after 4 hours of incubation with CTL clone G. This correlated well with the 24% ± 3% lysis which was observed in the 51Cr release assay. Incubation with the more potent CTL clone H resulted in 11% ± 2% survival and 88% ± 2% lysis as measured with the CFSE assay and the 51Cr release assay, respectively. In conclusion, in 4 hours of assays the results obtained with the CFSE assay correlated with the results obtained in the 51Cr release assay, showing identical sensitivity after 4 hours of effector target cell incubation.
Correlation between the CFSE-based cytotoxicity assay and the conventional 51Cr release assay. (A) Dot plots of AML-193 cells labeled with CFSE and incubated for 4 hours in the absence of T cells (target control) or in the presence of 2 specific CTL clones (E/T ratio, 1:1). Population 1 contained the fluorescent microspheres which could be discriminated on the basis of their scattering pattern and their fluorescence. Population 2 contained the CFSE and CD33+ AML-193 cells. The effector T cells added could be discriminated as a population of CFSE-negative CD33- cells (population 3). (B) The percentages of surviving AML-193 cells (± SD) as measured by the CFSE-based cytotoxicity assay (▦) and the percentages of lysed AML-193 cells as measured by the 51Cr release assay (▪) after 4 hours of incubation in the absence of CTLs (control) or after incubation with CTL clone G or CTL clone H (n = 3).
Correlation between the CFSE-based cytotoxicity assay and the conventional 51Cr release assay. (A) Dot plots of AML-193 cells labeled with CFSE and incubated for 4 hours in the absence of T cells (target control) or in the presence of 2 specific CTL clones (E/T ratio, 1:1). Population 1 contained the fluorescent microspheres which could be discriminated on the basis of their scattering pattern and their fluorescence. Population 2 contained the CFSE and CD33+ AML-193 cells. The effector T cells added could be discriminated as a population of CFSE-negative CD33- cells (population 3). (B) The percentages of surviving AML-193 cells (± SD) as measured by the CFSE-based cytotoxicity assay (▦) and the percentages of lysed AML-193 cells as measured by the 51Cr release assay (▪) after 4 hours of incubation in the absence of CTLs (control) or after incubation with CTL clone G or CTL clone H (n = 3).
Induction and analysis of leukemic cell proliferation using the CFSE assay
Labeling of the target cell population with CFSE allows not only the discrimination between (CFSE-labeled) target cells and the effector T cells but also information on the proliferative status of the target cells. Staining with specific markers facilitates detailed analysis of specific cell types within a heterogeneous target population like the mononuclear fraction of the peripheral blood of a patient with leukemia, which contains not only a large fraction of leukemic cells but also may, for instance, contain a percentage of contaminating healthy nonleukemic cells.
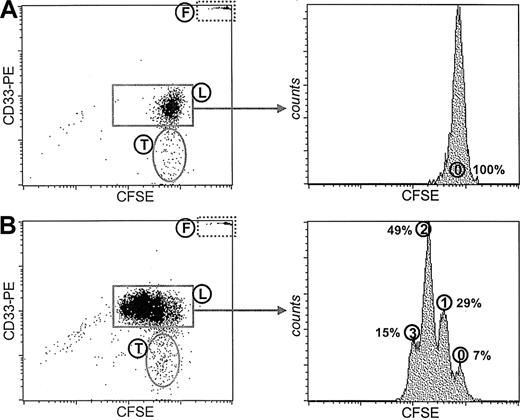
Figure 2 demonstrates the CFSE patterns of the mononuclear cell fraction isolated from the peripheral blood of a patient with AML which was cultured for 3 days either in the absence of cytokines (Figure 2A) or in the presence of a mixture of leukemic cell proliferation inducing cytokines, including GM-CSF, IL-3, G-CSF, and SCF (Figure 2B). CD33 was used as a specific staining for the leukemic cells, facilitating the discrimination between the population of leukemic cells (population L) and the population of nonleukemic cells (population T) which appeared to be T cells after staining with CD3 (data not shown). On the basis of the CFSE fluorescence demonstrated as histograms specifically for the population of CFSE-positive CD33+ cells the number of cell divisions that the leukemic cells underwent could be visualized. In the absence of cytokines only the undivided CFSE peak was visible, demonstrating that these AML blasts do not divide in the absence of cytokines. The addition of the cytokine mixture resulted in clear induction of proliferation of the leukemic cells. Only a small number (7%) of leukemic cells resided in the undivided CFSE peak (peak 0), whereas the majority of the leukemic cells underwent 1, 2, or 3 cell divisions, visualized by the respective CFSE peaks.
Manipulation of the proliferative status of primary leukemic cells by cytokines. Leukemic cells within the mononuclear cell (MNC) fraction isolated from the peripheral blood of a patient with AML are visualized by CD33 staining. Plotted are the propidium iodide (PI)-negative events in the life gate. Population F represents the fluorescent microspheres added for quantification. Population T represents a population of healthy patient T cells. The CFSE histograms are shown of the viable CD33+ leukemic cells (population L) cultured for 3 days either in the absence of cytokines (A) or in the presence of GM-CSF, G-CSF, IL-3, and SCF (B). The number of cell divisions is marked as well as the percentages of cells in every fraction.
Manipulation of the proliferative status of primary leukemic cells by cytokines. Leukemic cells within the mononuclear cell (MNC) fraction isolated from the peripheral blood of a patient with AML are visualized by CD33 staining. Plotted are the propidium iodide (PI)-negative events in the life gate. Population F represents the fluorescent microspheres added for quantification. Population T represents a population of healthy patient T cells. The CFSE histograms are shown of the viable CD33+ leukemic cells (population L) cultured for 3 days either in the absence of cytokines (A) or in the presence of GM-CSF, G-CSF, IL-3, and SCF (B). The number of cell divisions is marked as well as the percentages of cells in every fraction.
Analysis of CTL-induced cytotoxicity in a heterogeneous primary cell population
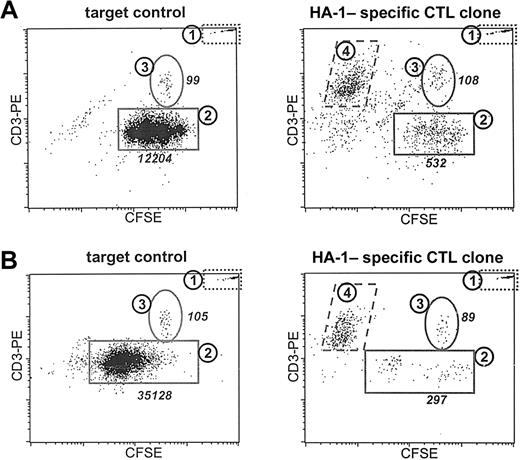
To demonstrate the analysis of T-cell-induced cytotoxicity in a heterogeneous primary target cell population using the CFSE-based cytotoxicity assay we labeled the MNC fraction of an HLA-A2 minor histocompatibility antigen HA-1-positive female patient with AML, which contained 84% CD33+ leukemic blasts. T cells and leukemic cells were specifically stained with CD3-PE/CY5 and CD33-PE, respectively. Depending on the underlying research question this analysis can be performed in 2 ways. Figure 3A demonstrates an experiment in which malignant progeny was induced by 3 days of culture on a cytokine mixture containing GM-CSF, G-CSF, IL-3, and SCF. At day 3 the effector T cells were added to investigate whether they were able to kill the malignant progeny. Viable cells were gated on the basis of a fsc/ssc-based life gate and PI negativity.
Analysis of the specific cytotoxicity induced by a T-cell clone in different cell types within a heterogeneous target cell population. The MNC fraction isolated from a female patient with AML positive for HLA-A2 was stained with CFSE. (A) Malignant progeny were induced by culturing the target population in the presence of GM-CSF, IL-3, G-CSF, and SCF for 3 days (target control). The effect of 24 hours of incubation of the induced malignant progeny with an HA-1-specific T-cell clone in a 1:1 E/T ratio is demonstrated. (B) The capacity of leukemic cells to produce malignant progeny after the simultaneous addition of cytokines and the cytotoxic T-cell clone (E/T ratio, 1:1) is demonstrated. Demonstrated are the target control after 5 days of culture and the surviving cells after 5 days of exposure to cytokines and CTLs. Plotted are the PI-negative events in the life gate counterstained with CD3-PE. Population 1 represented the population of fluorescent microspheres. Population 2 represented the leukemic cells. Populations 3 and 4 contained the healthy patient T cells present and the added CFSE-negative effector T cells, respectively. The absolute numbers are listed for populations 2 and 3.
Analysis of the specific cytotoxicity induced by a T-cell clone in different cell types within a heterogeneous target cell population. The MNC fraction isolated from a female patient with AML positive for HLA-A2 was stained with CFSE. (A) Malignant progeny were induced by culturing the target population in the presence of GM-CSF, IL-3, G-CSF, and SCF for 3 days (target control). The effect of 24 hours of incubation of the induced malignant progeny with an HA-1-specific T-cell clone in a 1:1 E/T ratio is demonstrated. (B) The capacity of leukemic cells to produce malignant progeny after the simultaneous addition of cytokines and the cytotoxic T-cell clone (E/T ratio, 1:1) is demonstrated. Demonstrated are the target control after 5 days of culture and the surviving cells after 5 days of exposure to cytokines and CTLs. Plotted are the PI-negative events in the life gate counterstained with CD3-PE. Population 1 represented the population of fluorescent microspheres. Population 2 represented the leukemic cells. Populations 3 and 4 contained the healthy patient T cells present and the added CFSE-negative effector T cells, respectively. The absolute numbers are listed for populations 2 and 3.
Figure 3A demonstrates the CFSE/CD3 dot plot of the target cell population. Population 1 again represents the fixed amount of fluorescent microbeads added to calculate the absolute number of viable cells within each sample. The leukemic blasts (population 2) could be distinguished on the basis of the CD33 staining. Population 3 represents the population of residing healthy patient T cells in the sample. After 24 hours of exposure of the induced malignant progeny to an HA-1-specific T-cell clone at a 0.5:1 E/T ratio (Figure 3B) a decrease of the absolute number of CFSE-positive/CD33+ leukemic blasts was observed. The effector T cells could be distinguished as a CFSE-negative, CD3+ population (population 4). Calculation of the ratio between the number of acquired viable leukemic cells and the fixed number of acquired fluorescent beads made it possible to calculate absolute numbers and to quantify the cytotoxicity. The leukemic progeny within this heterogeneous target cell population appeared to be significantly more sensitive to cell death induction induced by the T-cell clone compared with the population of nonleukemic healthy T cells. In Figure 3B an experiment is described in which we analyzed whether malignant progeny could be formed following exposure to the cytotoxic T-cell clone. Therefore, the leukemic blasts were exposed to the effector T cells at day 0 and simultaneously cytokines were added. Demonstrated are the target control cultured for 5 days in the presence of cytokines and the small population of residual leukemic cells after 5 days of exposure to the effector T-cell clone. Similar populations can be discriminated as described for Figure 3A.
In summary, the CFSE-based cytotoxicity assay not only facilitates the visualization of different cell types within a heterogeneous target cell population, but in addition supplies information on the proliferative status of these populations and their differential susceptibility to T-cell-induced cell death.
Evaluation of the HLA restriction of target cell lysis using anti-HLA class I-and class II-blocking antibodies in the CFSE-based cytotoxicity assay
To demonstrate the HLA restriction of CTL-induced lysis of target cells in the CFSE-based cytotoxicity assay blocking studies were performed. A representative result of this analysis is shown in Figure 4.
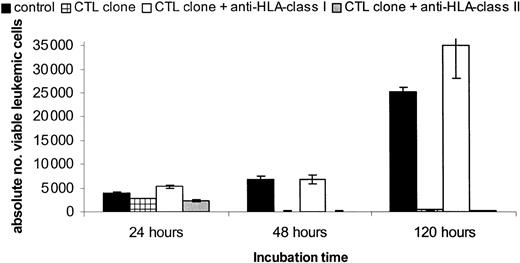
Analysis of the HLA restriction of CTL-induced target cell killing using the CFSE-based cytotoxicity assay. The MNC fraction of a patient with AML positive for HLA-A2/HA-1 was stained with CFSE at day -1. At day 0, GM-CSF, IL-3, and G-CSF were added. The target population was incubated in the presence of absence of saturating concentrations of blocking HLA class I or II antibodies before the HLA-A2-restricted HA-1-specific CTL clone was added at a 0.5:1 E/T ratio. Plotted are the calculated absolute numbers of viable CD33+ leukemic cells (± SD) after 24, 48, and 120 hours of CTL incubation.
Analysis of the HLA restriction of CTL-induced target cell killing using the CFSE-based cytotoxicity assay. The MNC fraction of a patient with AML positive for HLA-A2/HA-1 was stained with CFSE at day -1. At day 0, GM-CSF, IL-3, and G-CSF were added. The target population was incubated in the presence of absence of saturating concentrations of blocking HLA class I or II antibodies before the HLA-A2-restricted HA-1-specific CTL clone was added at a 0.5:1 E/T ratio. Plotted are the calculated absolute numbers of viable CD33+ leukemic cells (± SD) after 24, 48, and 120 hours of CTL incubation.
Peripheral blood of the HLA-A2/HA-1-positive female patient with AML was labeled with CFSE at day -1. At day 0 a mixture of cytokines containing GM-CSF, IL-3, and G-CSF was added to the cells, and the cells were incubated in the absence or presence of blocking anti-HLA class I or class II antibodies before the effector and target cells were cocultured in the assay plates. As effector cells, the HLA-A2-restricted HA-1-specific T-cell clone was used at an 0.5:1 E/T ratio. The absolute numbers of viable leukemic cells after 24, 48, and 120 hours of CTL incubation are shown. The specific lysis induced by the HLA-A2-restricted HA-1-specific CTL clone was completely abrogated by preincubation of the target cells with blocking anti-HLA class I antibodies even after 120 hours of CTL incubation. The addition of control HLA class II antibodies did not affect the lysis. These results demonstrate the feasibility of analyzing HLA restriction of the recognition by T-cell clones and lines using the CFSE-based cytotoxicity assay.
Specific analysis of CTL-induced cell death of CML precursor cells
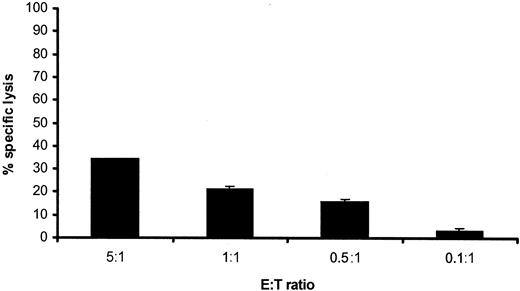
To investigate whether it was possible to demonstrate specific killing of CD34+ CML precursor cells by specific T cells, we labeled the MNC fraction of 3 HLA-A1-positive male patients with chronic-phase CML, in which the CD34 percentages ranged from 5% to 25% (data not shown) with CFSE. Proliferation was induced by the addition of a specific cytokine mixture containing GM-CSF, G-CSF, IL-3, SCF, and r-huEPO. An HLA-A1-restricted anti-HY CTL clone was added at different E/T ratios. Representative results for one of the patients are shown in Figures 5 and 6. The conventional 4-hour 51Cr release assay showed 34% specific lysis at the highest E/T ratio tested (Figure 5).
Susceptibility to CTL-induced lysis of chronic-phase CML. A conventional 4-hour 51Cr release assay was performed by using the MNCs derived from the peripheral blood of a male patient with chronic-phase CML positive for HLA-A1 as target cell population. HLA-A1-restricted CTLs directed against the HY antigen were added at different E/T ratios. Data shown are as mean percentages of specific lysis induced ± SD (n = 3).
Susceptibility to CTL-induced lysis of chronic-phase CML. A conventional 4-hour 51Cr release assay was performed by using the MNCs derived from the peripheral blood of a male patient with chronic-phase CML positive for HLA-A1 as target cell population. HLA-A1-restricted CTLs directed against the HY antigen were added at different E/T ratios. Data shown are as mean percentages of specific lysis induced ± SD (n = 3).
Differential recognition of proliferating CD34+ CML precursors. Plotted are the CFSE histograms of the viable CD34+ target cells after 4, 24, and 48 hours of incubation in the absence (A; target control) or presence (B) of HLA-A1-restricted HY-specific CTLs at a 1:1 E/T ratio. The absolute numbers of viable CD34+ cells are indicated. (C) The percentages of cells within the CD34+ fraction and the total CFSE+ target cell population which survived after 4, 24, and 48 hours of T-cell exposure (solid lines, ▪), or after culture for the same periods of time in the absence of T cells (target control; dashed lines, •).
Differential recognition of proliferating CD34+ CML precursors. Plotted are the CFSE histograms of the viable CD34+ target cells after 4, 24, and 48 hours of incubation in the absence (A; target control) or presence (B) of HLA-A1-restricted HY-specific CTLs at a 1:1 E/T ratio. The absolute numbers of viable CD34+ cells are indicated. (C) The percentages of cells within the CD34+ fraction and the total CFSE+ target cell population which survived after 4, 24, and 48 hours of T-cell exposure (solid lines, ▪), or after culture for the same periods of time in the absence of T cells (target control; dashed lines, •).
To investigate whether this lysis was only represented by the death of mature CML blasts, we investigated the fate of the CD34+ CML precursor cells specifically in the CFSE assay. Figure 6A demonstrates the proliferation of the CD34+ CML precursor cells in the target cell control after 4, 24, and 48 hours of culture represented by CFSE histograms of the gated CFSE-positive CD34+ population. The measured absolute numbers of CD34+ cells are shown in the figures. The majority of the CD34 precursors started to proliferate on the cytokine stimulation. However, a small but significant fraction of the CD34+ cells did not divide on the addition of cytokines and resided in the undivided CFSE peak. These cells probably reflect a population of precursor cells in dormancy. Figure 6B demonstrates the CFSE histograms and the absolute numbers of viable CD34+ target cells after 4, 24, and 48 hours of incubation with the HY-A1-specific CTL clone. The absolute number of CD34+ cells was significantly decreased compared with the target control already after 4 hours of CTL incubation and further decreased in time. The CFSE histograms demonstrated a relative specific deletion of proliferating CD34+ precursor cells compared with a subpopulation of undivided dormant CD34+ cells which remained present even after 48 hours of CTL incubation, probably because of relative resistance to T-cell-induced cell death.
Figure 6C represents the percentages of survival of the CD34+ CFSE-positive cells and the total CFSE-positive cells in the absence (dotted lines) and presence (solid lines) of the T-cell clone after 4, 24, and 48 hours of incubation. The highest relative increase in absolute cell numbers is observed in the CD34+ fraction (> 500% after 48 hours). The total number of CFSE-positive cells underwent a 2-fold increase in 48 hours. The most profound effect of the CTL addition is observed in the CD34+ population of which the majority was deleted by 48 hours of exposure to T cells resulting in 25% survival. Of the total CFSE-positive population 39% survived after 48 hours of T-cell exposure. In conclusion, this assay provides quantitative, phenotypical, and functional information on distinct cell types within a target cell population rather than only on the population as a whole.
Discussion
Immunotherapeutic interventions by adoptive transfer of donor T cells have been found to be an effective approach in patients with relapsed leukemia after allogeneic stem cell transplantation.1,2 The clinical response is most likely to be dependent on the ability of these T cells to specifically kill or inhibit the proliferation of leukemic (precursor) cells.1,7 For the clinical evaluation of immunotherapeutic interventions in patients with leukemia in vitro assays to determine the efficacy and the specificity of T cells for distinct cellular subsets within a heterogeneous leukemic target cell population are essential.
On the basis of previous findings that the fate of specific leukemic subpopulations like the CD34+ leukemic progenitor cells in CML determines the outcome of immunotherapeutic interventions, and on the basis of recent studies in which we analyzed the importance of the proliferative status of leukemic cells on their susceptibility to both chemotherapy and Fas-mediated apoptosis,6,18 we designed a new assay for the detailed monitoring of T-cell-mediated cell death. By combining CFSE labeling of the target cell population and staining with malignancy or cell lineage specific markers like CD34, CD33, or CD3 this assay not only facilitated within a time frame of days the quantitative analysis of susceptibility to T-cell-mediated lysis of specific cell types within a heterogeneous target cell population, but also it enabled the detailed analysis of individual cell divisions occurring within specific cell populations. Thus, this assay fulfills many requirements of an appropriate assay for the clinical evaluation of the efficacy of immunotherapeutic interventions.
The advantage of this assay is that it is easy to perform, and within one single assay a variety of simultaneous information becomes available which otherwise would require the performance of multiple assays. The CFSE-based cytotoxicity assay can be used for the assessment of multiple immune-mediated target cell read-outs, including the determination of the specificity of CTLs for defined cell types within a heterogeneous target cell population, their ability to either kill or inhibit the proliferation of these cell types, the effect of the proliferative status of specific cell types within a target cell population on the sensitivity to T cells, and the analysis of the different effector pathways comprising not only the rapid induction of perforin-mediated target cell lysis which is measured in the conventional 4-hour 51Cr release assay but also the more slowly executing receptor-mediated pathways of target cell apoptosis. Furthermore, it may be possible to combine this assay with specific intracellular staining for cytokines like IFN-γ produced by the T cells.
Although the assay is easy to perform, the flow cytometry read-out requiring individual acquisition and analysis of each sample makes it laborious compared with the conventional 51Cr release assay. Therefore, whether the CFSE assay is useful to perform depends on the type of information needed. If homogeneous target cell populations like cell lines are used as target populations, and only information on the ability of T cells to exert rapid toxicity against these cells is required, the 51Cr release assay may be easier to perform. However, if heterogeneous (primary) target cell populations are studied, or more detailed information is required also on the long-term survival of specific subsets within this target cell population, the CFSE-based cytotoxicity assay may be indispensable.
In addition to the detailed information on distinct cellular subtypes within a heterogeneous population our new CFSE-based cytotoxicity assay has other advantages compared with the functional assays for the analysis of T-cell-mediated cell death described until now. The assay not only supplies information on the population of cells that is killed but also allows analysis of the characteristics and the ability to proliferate of the surviving cells. As compared with other functional assays designed to evaluate the effect of T cells on the proliferation of leukemic (precursor) cells, like the Progenitor Inhibition Assay (PIA)2,19 and the colony-forming assays using culture in methylcellulose,20 the CFSE staining is not toxic to the cells and enables the isolation and further culture of the surviving target cell populations from the suspension. In contrast to the PIA it is not necessary to irradiate the effector T-cell population in the CFSE assay, which facilitates the analysis of not only the short-term interactions between (irradiated) T cells and target cells but also of long-term interactions between target cells and proliferating T cells.
In conclusion, we here present a new assay to simultaneously determine quantitative target cell death induction by cytotoxic T cells, differential recognition of distinct cell types within a target cell population, and the proliferative capacity of the surviving target cells in a single assay within a time frame of days, allowing detailed monitoring of immunotherapeutic interventions.
Prepublished online as Blood First Edition Paper, November 20, 2003; DOI 10.1182/blood-2003-06-2070.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal