Abstract
The first congenital defect of hypoxia-sensing homozygosity for VHL 598C>T mutation was recently identified in Chuvash polycythemia. Subsequently, we found this mutation in 11 unrelated individuals of diverse ethnic backgrounds. To address the question of whether the VHL 598C>T substitution occurred in a single founder or resulted from recurrent mutational events in human evolution, we performed haplotype analysis of 8 polymorphic markers covering 340 kb spanning the VHL gene on 101 subjects bearing the VHL 598C>T mutation, including 72 homozygotes (61 Chuvash and 11 non-Chuvash) and 29 heterozygotes (11 Chuvash and 18 non-Chuvash), and 447 healthy unrelated individuals from Chuvash and other ethnic groups. The differences in allele frequencies for each of the 8 markers between 447 healthy controls (598C) and 101 subjects bearing the 598T allele (P < 10–7) showed strong linkage disequilibrium. Haplotype analysis indicated a founder effect. We conclude that the VHL 598C>T mutation, the most common defect of congenital polycythemia yet found, was spread from a single founder 14 000 to 62 000 years ago.
Introduction
Diverse physiologic and pathophysiologic processes are regulated by tissue hypoxia.1,2 Hypoxia inducible factor 1α (HIF-1α) is a component of HIF, the “master” transcription regulator of the genes that respond to hypoxia.3 The HIF-1α protein is stable under hypoxia but is rapidly degraded in normoxia via a pathway that includes binding to the von Hippel-Lindau (VHL) protein.4 Chuvash polycythemia (CP) is a congenital disorder of augmented hypoxia sensing due to a homozygous mutation of the VHL gene.5,6 Subsequent to its original description in the Chuvash population isolate, the same mutation was detected in 11 unrelated individuals belonging to the diverse ethnic groups of Asian, white, and African-American origin.7-9
The worldwide distribution of the VHL 598C>T mutation raised the question of whether the VHL 598C>T substitution occurred in a single founder or resulted from recurrent mutational events. To address this issue, we characterized haplotypes of Chuvash and non-Chuvash subjects bearing the VHL 598C>T mutation and healthy unrelated individuals from diverse ethnic groups.
Study design
Subjects
We obtained 150 Chuvash DNA samples (61 with CP and 89 controls) and 35 non-Chuvash samples (29/35 non-Chuvash samples had VHL mutation, 6/35 non-Chuvash samples are their healthy relatives that were used as the controls).7-9 We also examined 363 unrelated healthy individuals: Southeast Asian (n = 76), white (n = 101), African American (n = 88), and Hispanic (n = 98).
VHL 598C>T mutation screen
Haplotype analysis of the VHL gene
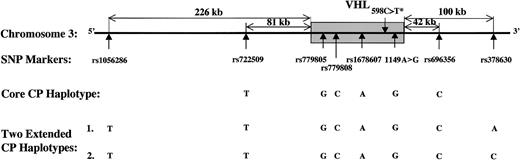
After screening 41 reported single-nucleotide polymorphisms (SNPs) spanning the VHL gene, we found 8 polymorphisms, illustrated in Figure 1 and Table 1, to be highly informative in the Chuvash population.
VHL haplotype system. The shaded area is the VHL gene (14 543 bp). The asterisk (*) indicates that this mutation is associated with Chuvash polycythemia.
VHL haplotype system. The shaded area is the VHL gene (14 543 bp). The asterisk (*) indicates that this mutation is associated with Chuvash polycythemia.
Allele-specific restriction analyses of 8 SNPs
SNPs and primers . | Primer sequence (5′-3′) . | Annealing temperature, °C . | DMSO, % . | PCR product, bp . | Enzyme . | Gel analysis . | Fragment sizes, bp . |
|---|---|---|---|---|---|---|---|
| rs1056286 | 65 | 7.5 | 161 | TaqαI | 2% agarose gel | T = 161 | |
| C = 105 + 56 | |||||||
| F | GTC TGC TGC GGG CGC TGG AC | ||||||
| R | ACT GCG GTG GAG CTC TGC TC | ||||||
| rs722509 | 57 | — | 151 | NsiI | 3% Nusieve | A = 151 | |
| 3:1 agarose gel | T = 129 + 22 | ||||||
| F | GGA ACC CTG ATT CTT TTA GA | ||||||
| R | AAC AAG GTG AGG GGA AAT AC | ||||||
| rs779805 (19A>G)10 | 63 | — | 276 | HaeIII | 4% Nusieve | A = 186 + 90 | |
| 3:1 agarose gel | G = 186 + 45 + 45 | ||||||
| F | GAA ATA CAG TAA CGA GTT GGC CTA GC | ||||||
| R | ACC TCG GCC TCG TCC CAG T | ||||||
| rs779808 | 67 | — | 286 | BstNI | 3% Nusieve | T = 199 + 68 + 19 | |
| 3:1 agarose gel | C = 152 + 68 + 47 + 19 | ||||||
| F | CAT TGC AAC ATC TGC CAC CT | ||||||
| R | GCG ACA GAT CCA GAC TCC AA | ||||||
| rs1678607 | 60 | — | 208 | HpyCH4III | 3% Nusieve | A = 208 | |
| 3:1 agarose gel | C = 147 + 61 | ||||||
| F | CGT ACA AAT ACA TCA CTT CC | ||||||
| R | TGA CGG ACA GCC TAT TTT TG | ||||||
| 1149A>G11 | 50 | — | 110 | AccI | 3% Nusieve | A = 110 | |
| 3:1 agarose gel | G = 89 + 21 | ||||||
| F | CTG CCC ATT AGA GAA GTA TTT | ||||||
| R | AAT TCC CAC TGA ATT ACG TAT A | ||||||
| rs696356 | 58 | 10 | 201 | AvaII | 2% agarose gel | A = 109 + 80 + 12 | |
| C = 189 + 12 | |||||||
| F | CAG TTT CTC TCC AAC AAG GG | ||||||
| R | CCA GGC TGG TCC AGA ACT CC | ||||||
| rs378630 | 67 | — | 225 | BanI | 2% agarose gel | A = 225 | |
| C = 155 + 70 | |||||||
| F | TTA CAG CTC TCC ACT CAC TG | ||||||
| R | GGA GCT CTC TGC AGG GTG AG |
SNPs and primers . | Primer sequence (5′-3′) . | Annealing temperature, °C . | DMSO, % . | PCR product, bp . | Enzyme . | Gel analysis . | Fragment sizes, bp . |
|---|---|---|---|---|---|---|---|
| rs1056286 | 65 | 7.5 | 161 | TaqαI | 2% agarose gel | T = 161 | |
| C = 105 + 56 | |||||||
| F | GTC TGC TGC GGG CGC TGG AC | ||||||
| R | ACT GCG GTG GAG CTC TGC TC | ||||||
| rs722509 | 57 | — | 151 | NsiI | 3% Nusieve | A = 151 | |
| 3:1 agarose gel | T = 129 + 22 | ||||||
| F | GGA ACC CTG ATT CTT TTA GA | ||||||
| R | AAC AAG GTG AGG GGA AAT AC | ||||||
| rs779805 (19A>G)10 | 63 | — | 276 | HaeIII | 4% Nusieve | A = 186 + 90 | |
| 3:1 agarose gel | G = 186 + 45 + 45 | ||||||
| F | GAA ATA CAG TAA CGA GTT GGC CTA GC | ||||||
| R | ACC TCG GCC TCG TCC CAG T | ||||||
| rs779808 | 67 | — | 286 | BstNI | 3% Nusieve | T = 199 + 68 + 19 | |
| 3:1 agarose gel | C = 152 + 68 + 47 + 19 | ||||||
| F | CAT TGC AAC ATC TGC CAC CT | ||||||
| R | GCG ACA GAT CCA GAC TCC AA | ||||||
| rs1678607 | 60 | — | 208 | HpyCH4III | 3% Nusieve | A = 208 | |
| 3:1 agarose gel | C = 147 + 61 | ||||||
| F | CGT ACA AAT ACA TCA CTT CC | ||||||
| R | TGA CGG ACA GCC TAT TTT TG | ||||||
| 1149A>G11 | 50 | — | 110 | AccI | 3% Nusieve | A = 110 | |
| 3:1 agarose gel | G = 89 + 21 | ||||||
| F | CTG CCC ATT AGA GAA GTA TTT | ||||||
| R | AAT TCC CAC TGA ATT ACG TAT A | ||||||
| rs696356 | 58 | 10 | 201 | AvaII | 2% agarose gel | A = 109 + 80 + 12 | |
| C = 189 + 12 | |||||||
| F | CAG TTT CTC TCC AAC AAG GG | ||||||
| R | CCA GGC TGG TCC AGA ACT CC | ||||||
| rs378630 | 67 | — | 225 | BanI | 2% agarose gel | A = 225 | |
| C = 155 + 70 | |||||||
| F | TTA CAG CTC TCC ACT CAC TG | ||||||
| R | GGA GCT CTC TGC AGG GTG AG |
Nusieve 3:1 agarose gel was made by Cambrex BioScience Rockland (Rockland, ME). DMSO indicates dimethyl sulfoxide; F, forward primer; and R, reverse primer.
Estimation of haplotype frequencies and determination of statistical significance
We used the expectation maximization (EM) algorithm, which handles uncertainty in phase determination, to estimate haplotype frequencies12 as implemented by Mander.13 Log-linear model was used to test for linkage disequilibrium and disease association.13 To evaluate the haplotypes in pedigrees we used GeneHunter.14 Since GeneHunter (Whitehead Institute, Cambridge, MA) does not allow for linkage disequilibrium among the markers, we rejected any haplotypes that failed to preserve strong associations among the marker alleles.
Phase determination
Long and accurate polymerase chain reaction (LA PCR; Serologicals, Norcross, GA) was employed to determine haplotype phase of the VHL 598C>T alleles from individuals no. 1 and no. 2, who did not have extensive family members available for phasing the haplotypes. The primers were VHL433F (5′-AAA AAA CAC CAA ACC TTA GAG GGG TG-3′) and VHL9246R (5′-CCC AAA GCA GGA GGC AGA CAA GTC ACC-3′). The 8.8-kb gel-purified amplicon covering the markers rs779805 through 1149A>G11 was cloned into the pCR-XL-TOPO vector (Invitrogen, Carlsbad, CA) and sequenced.
Dating the origin of VHL 598C>T mutation
We calculated the time of origin of the VHL 598C>T mutation using a formula that relates the proportion of disease-bearing chromosomes not bearing the ancestral haplotype to the number of generations15 (g) since the mutation event according to the formula g = log[(1–Q)/(1–PN)]/log(1–θ), where Q is the proportion of disease-bearing chromosomes not bearing the ancestral haplotype, PN is the frequency of the disease allele in the population, and θ is the recombination distance assuming that 1% corresponds to 1 megabase (Mb).
Results and discussion
The VHL 598C>T missense mutation was found in Chuvashians, African Americans, Bangladeshi, Punjabi, English, and whites.5,7-9 This finding suggests either that the VHL 598C>T mutation occurred several times in human evolution or that it originated once in some common founder.
We compared the allele frequencies of 8 polymorphic markers in 447 healthy controls (VHL 598C) and 101 subjects bearing the VHL 598C>T mutation that implied 173 598C>T chromosomes from 72 homozygotes (61 Chuvash and 11 non-Chuvash) and 29 heterozygotes (11 Chuvash and 18 non-Chuvash). In the 144 chromosomes from the 72 homozygotes for the VHL 598T genotype, the frequency of one of the alleles for each SNP (see underlined text in Table 2) ranged from 0.73 to 1.00, whereas in 894 chromosomes from the 447 healthy controls (VHL 598C), the frequency was only 0.13 to 0.85. The linkage disequilibrium between VHL 598T mutation and the polymorphic markers was complete for 5 SNPs (rs722509, rs779805, rs779808, rs1678607, 1149A>G) and extensive for 3 SNPs (rs1056286, rs696356, rs378630) (Table 2). Based on χ2 analysis, the differences in allele frequencies for each marker between 447 healthy controls (598C) and 101 subjects bearing 598T were highly significant (P < 10–7; Table 2). This degree of linkage disequilibrium indicates that the VHL 598C>T mutation occurred only once on the founder haplotype-rs1056286T-rs722509T-rs779805G-rs779808C-rs1678607A-1149G-rs696356C-rs378630A (Table 2).
Allele frequencies of 8 SNPs of VHL haplotype
. | . | Chuvash . | . | . | Non-Chuvash . | . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNPs and alleles . | Distance from 598C> T, kb . | Homozygotes, n = 61 . | Heterozygotes, n = 11 . | Healthy individuals, n = 78 . | Homozygotes, n = 11 . | Heterozygotes, n = 18 . | Healthy individuals, n = 369 . | Founder haplotype* . | ||||
| rs1056286, upstream | ||||||||||||
| T | 234.8 | 0.97 | 0.41 | 0.46 | 1.00 | 0.58 | 0.36 | T | ||||
| C | 234.8 | 0.03 | 0.59 | 0.54 | 0.00 | 0.42 | 0.64 | T | ||||
| rs722509, upstream | ||||||||||||
| A | 89.8 | 0.00 | 0.09 | 0.15 | 0.00 | 0.17 | 0.20 | T | ||||
| T | 89.8 | 1.00 | 0.91 | 0.85 | 1.00 | 0.83 | 0.80 | T | ||||
| rs779805, 5′-UTR of VHL | ||||||||||||
| A | 8.3 | 0.00 | 0.59 | 0.67 | 0.00 | 0.28 | 0.61 | G | ||||
| G | 8.3 | 1.00 | 0.41 | 0.33 | 1.00 | 0.72 | 0.39 | G | ||||
| rs779808, intron 1 of VHL | ||||||||||||
| T | 6.6 | 0.00 | 0.59 | 0.67 | 0.00 | 0.25 | 0.60 | C | ||||
| c | 6.6 | 1.00 | 0.41 | 0.33 | 1.00 | 0.75 | 0.40 | C | ||||
| rs1678607, intron 2 of VHL | ||||||||||||
| A | 3.2 | 1.00 | 0.32 | 0.15 | 1.00 | 0.56 | 0.13 | A | ||||
| C | 3.2 | 0.00 | 0.68 | 0.85 | 0.00 | 0.44 | 0.87 | A | ||||
| 1149A>G, 3′-UTR of VHL | ||||||||||||
| A | 0.3 | 0.00 | 0.55 | 0.68 | 0.00 | 0.31 | 0.61 | G | ||||
| G | 0.3 | 1.00 | 0.45 | 0.32 | 1.00 | 0.69 | 0.39 | G | ||||
| rs696356, downstream | ||||||||||||
| A | 47.7 | 0.01 | 0.32 | 0.31 | 0.00 | 0.22 | 0.33 | C | ||||
| C | 47.7 | 0.99 | 0.68 | 0.69 | 1.00 | 0.78 | 0.67 | C | ||||
| rs378630, downstream | ||||||||||||
| A | 105.7 | 0.73 | 0.36 | 0.37 | 0.73 | 0.33 | 0.34 | A | ||||
| C | 105.7 | 0.27 | 0.64 | 0.63 | 0.27 | 0.67 | 0.66 | A | ||||
. | . | Chuvash . | . | . | Non-Chuvash . | . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNPs and alleles . | Distance from 598C> T, kb . | Homozygotes, n = 61 . | Heterozygotes, n = 11 . | Healthy individuals, n = 78 . | Homozygotes, n = 11 . | Heterozygotes, n = 18 . | Healthy individuals, n = 369 . | Founder haplotype* . | ||||
| rs1056286, upstream | ||||||||||||
| T | 234.8 | 0.97 | 0.41 | 0.46 | 1.00 | 0.58 | 0.36 | T | ||||
| C | 234.8 | 0.03 | 0.59 | 0.54 | 0.00 | 0.42 | 0.64 | T | ||||
| rs722509, upstream | ||||||||||||
| A | 89.8 | 0.00 | 0.09 | 0.15 | 0.00 | 0.17 | 0.20 | T | ||||
| T | 89.8 | 1.00 | 0.91 | 0.85 | 1.00 | 0.83 | 0.80 | T | ||||
| rs779805, 5′-UTR of VHL | ||||||||||||
| A | 8.3 | 0.00 | 0.59 | 0.67 | 0.00 | 0.28 | 0.61 | G | ||||
| G | 8.3 | 1.00 | 0.41 | 0.33 | 1.00 | 0.72 | 0.39 | G | ||||
| rs779808, intron 1 of VHL | ||||||||||||
| T | 6.6 | 0.00 | 0.59 | 0.67 | 0.00 | 0.25 | 0.60 | C | ||||
| c | 6.6 | 1.00 | 0.41 | 0.33 | 1.00 | 0.75 | 0.40 | C | ||||
| rs1678607, intron 2 of VHL | ||||||||||||
| A | 3.2 | 1.00 | 0.32 | 0.15 | 1.00 | 0.56 | 0.13 | A | ||||
| C | 3.2 | 0.00 | 0.68 | 0.85 | 0.00 | 0.44 | 0.87 | A | ||||
| 1149A>G, 3′-UTR of VHL | ||||||||||||
| A | 0.3 | 0.00 | 0.55 | 0.68 | 0.00 | 0.31 | 0.61 | G | ||||
| G | 0.3 | 1.00 | 0.45 | 0.32 | 1.00 | 0.69 | 0.39 | G | ||||
| rs696356, downstream | ||||||||||||
| A | 47.7 | 0.01 | 0.32 | 0.31 | 0.00 | 0.22 | 0.33 | C | ||||
| C | 47.7 | 0.99 | 0.68 | 0.69 | 1.00 | 0.78 | 0.67 | C | ||||
| rs378630, downstream | ||||||||||||
| A | 105.7 | 0.73 | 0.36 | 0.37 | 0.73 | 0.33 | 0.34 | A | ||||
| C | 105.7 | 0.27 | 0.64 | 0.63 | 0.27 | 0.67 | 0.66 | A | ||||
n indicates number of people.
P < 10-7 for linkage disequilibrium for each SNP with VHL 598C> T mutation.
We have detected the VHL 598C>T mutation in 11 unrelated non-Chuvash families or individuals.7-9 Haplotype analysis demonstrated that 5 individuals (nos. 3-7) homozygous for 598C>T shared the same founder haplotype as the polycythemia patients from Chuvashia and thus are of common ancestry. Six non-Chuvash individuals (nos. 1, 2, and 8-11) had the VHL 598C>T mutation on a single allele. Phase determination on individuals no. 1 and no. 2 showed that the VHL 598C>T allele contained the identical SNP pattern, as did the VHL 598C>T allele from Chuvash patients.
The analyses of the haplotypes observed in Chuvash and non-Chuvash subjects with VHL 598C>T mutation can be further subdivided into the core CP haplotype (covering 6 SNPs) and 2 extended CP haplotypes (covering 8 SNPs) (Figure 1). The 2 extended CP haplotypes TTGCAT*GCA (haplotype 1) and TTGCAT*GCC (haplotype 2) cover 340 kb containing the VHL 598C>T mutation (T*) (Figure 1). In these analyses, 99.2% Chuvash and non-Chuvash individuals homozygous for the VHL 598C>T mutation share the core CP haplotype, while 96.5% share the 2 extended CP haplotypes (72.2% haplotype 1 and 24.3% haplotype 2). The analyses of the 6 heterozygous non-Chuvash individuals reveal that 3 of 6 individuals (nos. 1, 2, and 8) share the core CP haplotype. Of those without the exact CP haplotype (individuals nos. 9-11), individual no. 9 has a haplotype that may have arisen from a crossover between SNPs rs779808 and rs1678607 within the VHL gene, individual no. 10 has a C to A transition/crossover at rs696356, and individual no. 11 has the A to C transition at the SNP rs1678607. Statistical differences in founder haplotype were verified between normal alleles and alleles bearing the VHL 598T mutation. Out of 144 VHL 598T alleles derived from homozygotes, 72.2% bore the founder haplotype, TTGCAGCA, which was in striking contrast to 0.9% of 894 VHL 598C alleles derived from healthy controls without VHL 598C>T mutation (P < .000 001). These data strongly support the conclusion that the VHL 598C>T missense mutation arose from a single founder.
We estimated the time of origin of the VHL 598C>T mutation using an approach suggested by Risch et al.15 In the 11 non-Chuvash families or individuals that we studied, we observed 6 TTGCAT*GCC haplotypes, 1 CCGCCT*GCC haplotype, 2 CTGCAT*GCC haplotypes (African American and white), 6 TTGCAT*GCA haplotypes (Bengali and Punjabi origin), and 1 TTGCCT*GCC haplotype (English) among 5 families having affected individuals homozygous for the Chuvash mutation, 5 patients heterozygous with the Chuvash mutation, and 1 African American heterozygous for the Chuvash mutation who was otherwise healthy. We assumed the absence of back mutations. For these calculations, we excluded Chuvash individuals, since their mutations are likely due to founder effect. Among the remaining controls, 1 of 363 individuals (excluding 6 nonpolycythemic relatives who were ascertained because of polycythemic probands) showed a VHL 598C>T mutation (individual no. 1), yielding an estimated population frequency of the disease allele (PN) of 0.001377. Given the genomic distance between VHL 598T and rs1056286 of 234.8 kb, 715 generations of 20 years or 14 293 years are required to account for the data. For the next closest 5′ marker, rs722509, which was 89.8 kb from the VHL 598C>T mutation, the mutation is estimated to have occurred 3110 generations or 62 188 years ago. In the 3′ region, the distance from the marker rs378630 to the VHL 598C>T mutation was 105.7 kb and the estimated number of generations is 930 or 18 593 years ago. The wide range of estimated dates for the origin of the 598C>T mutation reflects the study of a limited number of haplotypes in non-Chuvash subjects. In addition, because the Bengali and the Punjabi families share a unique common haplotype in the 3′ region, the estimated time could be biased by sampling several patients from this region. The calculations concerning generation time are highly influenced by the mutation frequency in the controls, but the rarity of this mutation in controls precludes an accurate estimate of the age of the mutation. The mutation could be much older than our estimate because we only observed a single African-American control with a mutation and our calculations were all performed with whites, for whom the estimated control mutation frequency is 0. We conclude that the VHL 598C>T mutation arose in a single ancestor at least 14 000 to 62 000 years ago. This age estimate and the fact that the mutation is found in different races and ethnic groups suggest that this mutation occurred early in the development of the human race.
We conclude that homozygosity for the VHL 598C>T mutation, the first disorder of augmented hypoxia sensing and the most common genetic defect of congenital polycythemia, originated from a single founder. It is possible that this wide dissemination from the original founder may be associated with some survival advantages. Such an advantage might be related to subtle improvement of iron metabolism, erythropoiesis, embryonic development, energy metabolism,1,16 or some other yet unknown effect. Another intriguing possibility is the recent demonstration of the protective role of HIF-1α in regulating vascular endothelial growth factor (VEGF) in pre-eclampsia,16,17 which is the leading cause of maternal and fetal mortality worldwide.18 However, at this time this is only speculation and might prompt further in vitro and in vivo studies.
Prepublished online as Blood First Edition Paper, November 6, 2003; DOI 10.1182/blood-2003-07-2550.
Supported by grants from the National Heart, Lung, and Blood Institute (HL66333 and UH1HL03679; J.T.P.); the Howard University General Clinical Research Center grant from the National Institutes of Health (2MO1 RR10284; V.G.); a grant from the National Human Genome Research Institute (R01HG02275; C.I.A.); and the Northern Ireland Leukaemia Research Fund (T.R.J.L.).
E.L. and M.J.P. contributed equally to this study.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We are grateful to Baylor Human Polymorphism Resource for providing us with the DNA samples of 4 ethnic groups and to Xiangjun Gu and Wei V. Chen at the UT MD Anderson Cancer Center for their assistance in data analysis.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal