Abstract
Although it has been suggested that REL is the critical target gene of 2p12-16 amplification in diffuse large B-cell lymphoma (DLBCL), little experimental evidence supports this notion. In the present study, we sought to evaluate the relationship between REL amplification and REL function in a panel of 46 newly diagnosed DLBCLs and to correlate with DLBCL subgroups as identified by gene expression profiles and clinical features. The results indicate that amplification of the REL locus is not associated with accumulation of the active form of REL, as evaluated by immunofluorescence analysis. Upon subgrouping of the DLBCL cases based on gene expression signatures, REL amplification was detected in all subgroups, while high levels of nuclear-located REL were more frequently detected in activated B-cell–like DLBCL. Correlative analyses of REL copy number and REL nuclear accumulation with clinical parameters did not reveal any significant associations. Together these results indicate that 2p12-16 amplification does not lead to abnormal REL activation, suggesting that REL may not be the functional target of the amplification event. Nonetheless, these data indicate that DLBCLs are heterogeneous with respect to REL and thus nuclear factor–κB (NF-κB) activity.
Introduction
Diffuse large B-cell lymphoma (DLBCL) is an aggressive malignancy that comprises approximately one third of all non-Hodgkin lymphoma (NHL).1 Despite its potential curability and high response rates with standard chemotherapy, approximately half of the patients ultimately die of their disease.1 DLBCL typically displays immunoglobulin (IG) gene rearrangement and somatic hypermutation that reflect their mature B-cell origin and play pivotal roles in transformation.2 Underlying chromosomal translocations that often involve IG gene loci cause deregulated expression of the BCL2, MYC, and BCL6 genes and occur in approximately 50% of cases.2 Genomic amplification as an additional mechanism leading to deregulated gene expression in NHL has gone largely unrecognized. In solid tumors, amplification of genes regulating cell proliferation or drug resistance represents an important pathway to aggressive behavior.3 Application of comparative genomic hybridization (CGH)4 for screening tumor genomes for DNA sequence gains and losses has now identified a number of chromosomal sites and candidate target genes amplified in DLBCL, associated with advanced-stage disease.5,6 Amplification and gain of the 2p12-16 region has frequently been described in NHL and Hodgkin disease (HD) with frequencies as high as 50%.5-8 Candidate genes mapped within this CGH amplicon include REL and BCL11A, with REL being the most frequently amplified gene (more than 25%).7,9-11 The functionally relevant overexpressed amplified target gene(s), however, has not as yet been identified.
The candidate gene REL encodes a member of the REL/nuclear factor–κB (REL/NF-κB) family of transcription factors that play an important and complex role in cell growth, differentiation, and survival regulation, in particular, in the hematopoietic lineages.12,13 REL/NF-κB complexes are normally bound to inhibitory IκB proteins in the cytoplasm.14 Upon activation by diverse stimuli such as pathogens, cytokines, and chemotherapeutic agents, they translocate to the nucleus, promoting the expression of many immunomodulatory target genes.14 Aberrant constitutive NF-κB activity has been reported in both lymphoid malignancies and solid tumors, with the suggestion that active REL/NF-κB generates cell survival signals preventing apoptosis.15-18 Recently, a requirement for active NF-κB was demonstrated for the survival of a subgroup of DLBCL cell lines that exhibit expression of transcripts common to activated peripheral blood B cells.19 These activated B-cell–like (ABL) DLCBL cell lines displayed a higher NF-κB activity than another subgroup of DLBCL cell lines that expressed transcripts common to normal germinal center (GC) B cells.19 Molecular classification of DLBCL specimens based on gene expression signatures using the Lymphochip cDNA array indicated that patients with germinal center B-cell–like (GCBL) DLBCL had a higher overall survival than those with ABL DLBCL.20 This correlation was not confirmed in another gene expression profiling study of DLBCL utilizing oligonucleotide arrays.21 More recently, using the 100 most differentially expressed genes between the 2 subgroups, a third subgroup has been identified (Type 3) that does not express either set of genes at a higher level yet exhibits a poor clinical outcome.22 REL was one of the discriminatory genes with higher expression in GCBL DLBCL.22 In addition, genomic amplification of REL was found to occur exclusively within GCBL DLBCL, a subgroup not requiring active NF-κB for survival.22
Together, these results point to REL as an important player in DLBCL biology but do not provide information on its functional status in these tumors. In particular, it is not known whether amplification of the REL locus is associated with expression of nuclear REL, the transcriptionally active form of this NF-κB factor. Thus, in the present study, we sought to evaluate the relationship between REL amplification and REL function in a panel of newly diagnosed DLBCLs with relation to DLBCL expression signature subgroup and clinical features. Upon molecular classification of the DLBCL specimens as GCBL, ABL, or Type 3 DLBCL, a correlation between subgroup assignment and clinical outcome of DLBCL could not be confirmed, and REL amplification was found to occur in all subgroups. No clear correlation was evident between REL amplification and REL nuclear accumulation as evaluated by immunofluorescence, suggesting that REL is not the functional target of the 2p12-16 amplicon in DLBCL. Higher nuclear accumulation of REL was more frequent in ABL DLBCL, but a significant in vivo transcript expression signature was not found to be associated with nuclear REL. Correlation of REL copy number and REL nuclear accumulation with clinical parameters did not reveal any significant associations. Overall, these results indicate that while REL may not be the critical target gene of 2p12-16 amplification, DLBCLs are heterogeneous with respect to REL function.
Materials and methods
Tumor specimens
The study cohort comprised 46 newly diagnosed DLBCL cases ascertained at the Memorial Sloan-Kettering Cancer Center (MSKCC) between March 1985 and October 1998. The original diagnostic material of all patients was reviewed to confirm histology according to the World Health Organization (WHO) classification.23 Percentage of malignant cells was evaluated for each case as an approximate estimation of tumor or large cell lymphoma observed over the total tissue section, including general lymphoma, fibrosis, adipose tissue, fibrovascular tissue, and/or glandular or epithelial tissue. As controls, 2 reactive lymph node specimens resected from patients with no history of lymphoma were included in the study. The following categories of patients were excluded from this study: those with primary mediastinal DLBCL, evidence of histologic transformation from a low-grade lymphoma, human immunodeficiency virus (HIV)–associated lymphoma, and posttransplantation lymphoproliferative disorder. The medical records of all patients were reviewed to obtain relevant clinical information: lactate dehydrogenase (LDH) levels, extent of disease/stage at diagnosis, presence of pure extranodal disease (disease confined to extralymphatic organs), International Prognostic Index (IPI)24 score, treatment, response, time to treatment failure (TTF), and overall survival (OS). OS and TTF were calculated for the 39 patients who received an anthracycline-containing chemotherapy regimen, using a log-rank test using the SAS statistical software program.25 The median follow-up was 46 months, and OS significantly correlated with IPI score (P = .005). The Fisher exact test was used to test for association between variables. The study was conducted according to the principles of the Declaration of Helsinki and the Institutional Review Board of the Memorial Sloan-Kettering Cancer Center.
Karyotype analysis and IGH-BCL2 rearrangement by real-time PCR
G-banded karyotype analysis was attempted for 42 cases, of which 28 were found to be abnormal, 3 were normal, and 11 were failures. Real-time polymerase chain reaction (PCR) analysis was performed on available tumor DNAs for detection of t(14;18)(q32;q21) rearrangements between IGH and the major breakpoint cluster region (MBR) of BCL2 as described.26
RNA extraction and expression profiling
Total RNA was extracted from snap-frozen or optimal cutting temperature medium (OCT)–embedded fresh tumor or reactive lymph node specimens using an RNeasy kit (Qiagen, Valencia, CA) and the quality assessed by spectrophotometry and agarose gel electrophoresis.27 Total RNA was subjected to expression profiling as previously described by us.27 Briefly, first- and second-strand cDNA synthesis was performed using a cDNA synthesis kit (Invitrogen, Carlsbad, CA), and biotinylated cRNA was prepared according to manufacturer's recommendations (Enzo, Farmingdale, NY). The cRNA was fragmented and 15 μg hybridized to Human Genome 95A Version 2 oligonucleotide arrays (HG-U95Av2: Affymetrix, Santa Clara, CA), followed by rinsing, staining, and scanning all according to the manufacturer's recommended conditions.
Expression data analysis
Expression signals were quantitated using the Affymetrix Microarray Suite 5.0, with the mean hybridization intensity (signal) of each array scaled to 2500 (Additional Data A—all additional data are provided at http://www.mskcc.org/GCL/DLBCL1/). The signal data were transformed using the logarithm base 2. Prior to clustering, the transcripts were standardized by subtracting off the mean and dividing by the standard deviation. Molecular classification of the DLBCL into GCBL/ABL/Type 3 subgroups was performed utilizing the entire cohort of 46 newly diagnosed DLBCL specimens. Of the 100 genes reported to distinguish GCBL/ABL DLBCL at the significance level of P values less than .001,22 81 had available GenBank accession numbers that could be assigned to UniGene clusters. Of these, 62 were present on the HG-U95Av2 array, being represented by 99 probe pair sets (Additional Data B). The 46 DLBCL specimens were submitted to hierarchic agglomerative clustering using the average linkage method with one minus the correlation as the distance metric for the 99 probe pair sets.28
Differentially expressed transcripts between DLBCL with high and low percent REL-positive nuclei were determined using the Wilcoxon rank sum test on a transcript-by-transcript basis. A permutation method was used to determine whether the number of significant transcripts found by our criterion was more than expected. In this method, the array labels were permuted and the number of significant transcripts computed for this new set of identifiers. The permutation process was repeated 1000 times. A P value was calculated as the proportion of times the number of significant transcripts for the real data were greater than or equal to the number of significant transcripts for the permuted data.
Determination of REL copy number by Southern blot analysis and validation by fluorescence in situ hybridization (FISH)
The gene copy number of REL was determined relative to a restriction fragment length polymorphism probe (D2S48) mapped to 2p21-23 by quantitative Southern blotting of restricted genomic DNAs isolated from 10 snap-frozen or OCT-embedded fresh tumor specimens as previously described.5,9 Within the present study group, the REL copy number had previously been evaluated in 33 cases.5,9 A gene copy of 4 or more was considered indicative of regional gene amplification. The REL copy number as evaluated by Southern blotting was validated by fluorescence in situ hybridization (FISH) analysis using a REL-containing BAC (RP11-373L24)29 on 1 independent DLBCL. In this case with a REL copy number of 7, FISH analysis detected on average 6 copies of the REL locus per malignant cell (data not shown). Fixed cells were not available for the panel of 46 cases. A test based on the Kendall τ statistic was used to examine the association between REL copy number and REL transcript levels.
Analysis of nuclear REL accumulation by immunofluorescence staining
For REL immunofluorescence analysis, a primary rabbit polyclonal REL antiserum (265) raised against the C-terminal 15 amino acids of human REL developed at the National Cancer Institute (NCI)–Frederick Cancer Research and Development Center was utilized (gift from N. Rice, NCI-Frederick Cancer Research and Development Center).30 In Western blotting analysis of a panel of NHL cell lines (OCI-LY series from the Ontario Cancer Institute, gifts from M. Messner, University of Toronto, Canada), this antibody detected a single band of approximately 80 kDa (data not shown). While validation of nuclear REL staining by this antibody had previously been reported for immunohistochemical staining,30 it was also validated in an in vitro system for immunofluorescence analysis. B-cell lymphoma cell lines were treated with CD40L to induce nuclear translocation of REL and activation of NF-κB activity. Comparison with mock-treated cells indicated nuclear REL only in cells with demonstrated increase in NF-κB activity evaluated by electrophoretic mobility shift assays (data not shown). Twomicrometer paraffin-embedded tumor sections were stained with the primary antibody and then with a secondary goat antirabbit biotin-conjugated antibody (Jackson Immunoresearch, West Grove, PA). Finally, the slides were counterstained with avidin–fluorescein isothiocyanate (avidin-FITC) and DAPI (4′,6-diamidino-2-phenylindole) and viewed with a Nikon epifluorescence microscope (Nikon). A total of 200 cells per section were scored to obtain percent of malignant cells with REL-positive nuclei. Nuclei were considered positive when staining was observed over the entire nuclear area (defined by DNAcounterstain) irrespective of cytoplasmic staining. In tumor sections, cytoplasmic-specific REL staining was routinely detected in small lymphocytes (mostly resting B cells), and nuclear and cytoplasmic REL staining was detected in occasional residual germinal centers (data not shown).
Results
Frequency of REL amplification in DLBCL expression signature subgroups
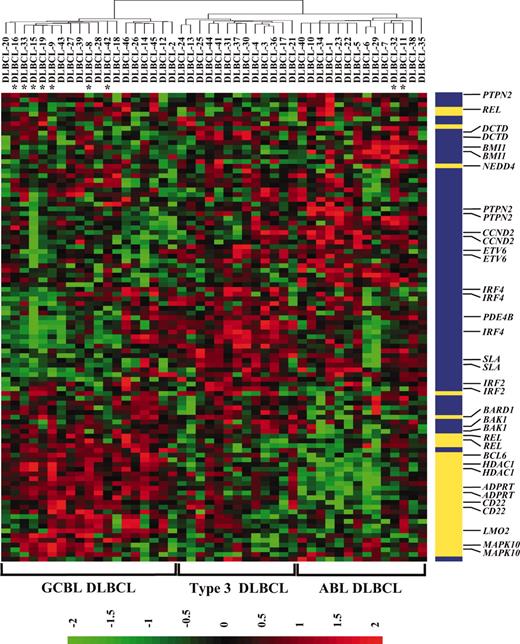
The REL locus is frequently amplified in DLBCL,5,6,9 and it has previously been reported that amplification occurs exclusively within GCBL DLBCL.22 To validate these findings, a panel of 46 newly diagnosed DLBCLs with known clinical parameters was initially submitted to oligonucleotide expression profiling for expression signature classification. The specimens were assigned as GCBL, ABL, or Type 3 DLBCL on the basis of hierarchic clustering using a subset of 62 (99 probe sets) of the 100 discriminatory genes that were represented on the oligonucleotide array used in the present study.22 Reclustering of the published specimens22 using only this subset of 62 genes revealed the 3 expected subgroups, with the expected survival correlation between the groups (P < .001), indicating that the 62 genes were sufficient for subgroup clustering. For the 46 specimens in the present study, hierarchic agglomerative clustering resulted in a dendrogram with 2 main branches (Figure 1 and Additional Data B). One branch comprised 19 specimens that more highly expressed the GCBL signature genes. The other branch, comprising 27 specimens, broadly expressed the ABL signature genes at higher levels, although they could be further subdivided into 2 branches. A subgroup of ABL DLBCL comprising 14 specimens exhibited a marked decrease in the expression of the GCBL signature genes, not evident in the other subgroup of 13 specimens (Type 3). Such clustering is in keeping with those previously published, and the assignments for each DLBCL are listed in Table 1. The REL copy number was previously known or determined for 43 of the 46 specimens (Table 1). Nine specimens (20.9%) exhibited an increased REL copy number (4 or more), consistent with the frequency previously reported by us.9 To examine the association between REL copy number and array-based REL transcript expression, a test based on the Kendall τ statistic was applied for each of 3 probe sets representing REL on the array. For 2 of these, a low and insignificant negative correlation estimate (–0.05, P = .64 to .66) was found, while for the third a low but significant positive correlation estimate (0.23, P = .03) was obtained. Thus, no consistent significant correlation was found between REL copy number and REL transcript level. REL amplification was detected in all 3 DLBCL subgroups (Table 2), in contrast with the previous report.22 It has also been reported that t(14;18)(q32;q21) occurs exclusively among GCBL DLBCL when evaluated by FISH or by PCR of the MBR of BCL2.22,31 In the present study, abnormal karyotypes were obtained for 28 of the specimens (Additional Data C) and IGH-BCL2 MBR rearrangement was evaluated by real-time PCR in 33 specimens (Table 1). For the 22 specimens studied by both assays, concordance between the assays was evident for 20. For specimen DLBCL-42, rearrangement was detected by PCR but not by G-banding analysis where only 1 karyotypically abnormal cell with a marker chromosome was encountered, accounting for the discrepancy between the assays. DLBCL-11 exhibited a t(14; 18)(q32;q21) in 53% of abnormal cells evaluated, and no rearrangement was detected by PCR. This could be explained by a breakpoint occurring within the minor breakpoint cluster region (MCR) or elsewhere in BCL2 undetected by PCR analysis, but observed by FISH, as previously reported.31 The frequency of t(14;18)(q32;q21) in the 3 expression subgroups was evaluated combining the results from both assays (39 specimens) and considering DLBCL-11 and -42 as positive. The translocation was detected in both GCBL and ABL subgroups (Table 2), with most in the former (7 of 9 with translocation). For each subgroup, the (14;18)(q32;q21)–containing specimens were noted to cluster within 1 arm of the subgroup (Figure 1). Overall then, 2 genetic abnormalities (REL amplification and t(14;18)(q32;q21)) previously reported to occur exclusively among GCBL DLBCL22,32 have not been confirmed in the present study.
Clustering of DLBCL into GCBL, ABL, and type 3 DLBCL. The 46 DLBCLs were hierarchically clustered using the average linkage method for 99 probe pair sets that represented 62 genes of the 100 reported to distinguish GCBL/ABL DLBCL at the significance level of P < .001.22 Of the 62 genes, 23 (31 probe pair sets, yellow) were represented within the GCBL DLBCL expression signature and 39 (68 probe pair sets, blue) within the ABL DLBCL expression signature. Select transcripts are listed, with a full list given in Additional Data B. Red indicates high expression and green, low expression, with the color scale at the bottom showing the relative expression in standard deviations from the mean. *Specimens with t(14;18)(q32;q21) translocation.
Clustering of DLBCL into GCBL, ABL, and type 3 DLBCL. The 46 DLBCLs were hierarchically clustered using the average linkage method for 99 probe pair sets that represented 62 genes of the 100 reported to distinguish GCBL/ABL DLBCL at the significance level of P < .001.22 Of the 62 genes, 23 (31 probe pair sets, yellow) were represented within the GCBL DLBCL expression signature and 39 (68 probe pair sets, blue) within the ABL DLBCL expression signature. Select transcripts are listed, with a full list given in Additional Data B. Red indicates high expression and green, low expression, with the color scale at the bottom showing the relative expression in standard deviations from the mean. *Specimens with t(14;18)(q32;q21) translocation.
Molecular classification of 46 newly diagnosed DLBCLs
DLBCL . | GCBL/Type 3/ABL . | % malignant cells . | t(14;18): karyotype . | t(14;18): PCR . | REL copy no. . | % REL-positive nuclei . |
|---|---|---|---|---|---|---|
| 1 | ABL | 90 | - | - | 2 | 12 |
| 2 | GCBL | 85 | - | - | 3 | 15 |
| 3 | Type 3 | 90 | - | - | 2 | NE |
| 4 | Type 3 | 70 | NE | NE | 2 | 2 |
| 5 | ABL | 90 | N | - | 1 | 90 |
| 6 | ABL | 50 | - | - | 2 | 80 |
| 7 | ABL | 90 | NE | NE | 3 | NE |
| 8 | GCBL | 90 | F | + | 2 | 85 |
| 9 | GCBL | 80 | + | + | 2 | 50 |
| 10 | ABL | 50 | - | NE | 4 | 95 |
| 11 | ABL | 90 | + | - | 2 | 90 |
| 12 | GCBL | 50 | F | - | 5 | 20 |
| 13 | Type 3 | 90 | F | - | 2 | 40 |
| 14 | GCBL | 95 | - | NE | 2 | 40 |
| 15 | GCBL | 95 | + | + | 2 | 5 |
| 16 | GCBL | 90 | + | + | 1 | 5 |
| 17 | Type 3 | 85 | - | - | 5 | 85 |
| 18 | GCBL | 60 | - | NE | 2 | 90 |
| 19 | GCBL | 75 | + | + | 1 | NE |
| 20 | GCBL | 90 | - | NE | 5 | 10 |
| 21 | Type 3 | 95 | F | NE | 1 | 20 |
| 22 | ABL | 40 | - | - | 1 | 70 |
| 23 | ABL | 30 | N | - | 2 | 25 |
| 24 | Type 3 | 85 | - | - | 1 | NE |
| 25 | Type 3 | 90 | - | - | 3 | 2 |
| 26 | GCBL | 95 | F | - | 2 | 80 |
| 27 | GCBL | 85 | - | - | 4 | 90 |
| 28 | GCBL | 90 | - | - | 3 | 95 |
| 29 | ABL | 30 | F | NE | NE | 90 |
| 30 | Type 3 | 60 | F | - | 5 | NE |
| 31 | Type 3 | 65 | F | - | 1 | 5 |
| 32 | ABL | 85 | + | + | 2 | NE |
| 33 | GCBL | 30 | + | NE | 1 | 5 |
| 34 | ABL | 95 | - | NE | 4 | NE |
| 35 | ABL | 40 | - | - | 2 | 10 |
| 36 | Type 3 | 95 | - | - | 2 | 90 |
| 37 | Type 3 | 90 | F | - | 2 | 95 |
| 38 | ABL | 95 | - | - | 2 | 50 |
| 39 | GCBL | 95 | NE | NE | NE | 15 |
| 40 | ABL | 90 | NE | NE | NE | 90 |
| 41 | Type 3 | 90 | F | - | 1 | 85 |
| 42 | GCBL | 90 | - | + | 4 | 15 |
| 43 | GCBL | 95 | N | NE | 2 | 5 |
| 44 | Type 3 | 50 | F | - | 2 | 20 |
| 45 | GCBL | 95 | - | - | 1 | 85 |
| 46 | GCBL | 80 | - | - | 141 | 1 |
DLBCL . | GCBL/Type 3/ABL . | % malignant cells . | t(14;18): karyotype . | t(14;18): PCR . | REL copy no. . | % REL-positive nuclei . |
|---|---|---|---|---|---|---|
| 1 | ABL | 90 | - | - | 2 | 12 |
| 2 | GCBL | 85 | - | - | 3 | 15 |
| 3 | Type 3 | 90 | - | - | 2 | NE |
| 4 | Type 3 | 70 | NE | NE | 2 | 2 |
| 5 | ABL | 90 | N | - | 1 | 90 |
| 6 | ABL | 50 | - | - | 2 | 80 |
| 7 | ABL | 90 | NE | NE | 3 | NE |
| 8 | GCBL | 90 | F | + | 2 | 85 |
| 9 | GCBL | 80 | + | + | 2 | 50 |
| 10 | ABL | 50 | - | NE | 4 | 95 |
| 11 | ABL | 90 | + | - | 2 | 90 |
| 12 | GCBL | 50 | F | - | 5 | 20 |
| 13 | Type 3 | 90 | F | - | 2 | 40 |
| 14 | GCBL | 95 | - | NE | 2 | 40 |
| 15 | GCBL | 95 | + | + | 2 | 5 |
| 16 | GCBL | 90 | + | + | 1 | 5 |
| 17 | Type 3 | 85 | - | - | 5 | 85 |
| 18 | GCBL | 60 | - | NE | 2 | 90 |
| 19 | GCBL | 75 | + | + | 1 | NE |
| 20 | GCBL | 90 | - | NE | 5 | 10 |
| 21 | Type 3 | 95 | F | NE | 1 | 20 |
| 22 | ABL | 40 | - | - | 1 | 70 |
| 23 | ABL | 30 | N | - | 2 | 25 |
| 24 | Type 3 | 85 | - | - | 1 | NE |
| 25 | Type 3 | 90 | - | - | 3 | 2 |
| 26 | GCBL | 95 | F | - | 2 | 80 |
| 27 | GCBL | 85 | - | - | 4 | 90 |
| 28 | GCBL | 90 | - | - | 3 | 95 |
| 29 | ABL | 30 | F | NE | NE | 90 |
| 30 | Type 3 | 60 | F | - | 5 | NE |
| 31 | Type 3 | 65 | F | - | 1 | 5 |
| 32 | ABL | 85 | + | + | 2 | NE |
| 33 | GCBL | 30 | + | NE | 1 | 5 |
| 34 | ABL | 95 | - | NE | 4 | NE |
| 35 | ABL | 40 | - | - | 2 | 10 |
| 36 | Type 3 | 95 | - | - | 2 | 90 |
| 37 | Type 3 | 90 | F | - | 2 | 95 |
| 38 | ABL | 95 | - | - | 2 | 50 |
| 39 | GCBL | 95 | NE | NE | NE | 15 |
| 40 | ABL | 90 | NE | NE | NE | 90 |
| 41 | Type 3 | 90 | F | - | 1 | 85 |
| 42 | GCBL | 90 | - | + | 4 | 15 |
| 43 | GCBL | 95 | N | NE | 2 | 5 |
| 44 | Type 3 | 50 | F | - | 2 | 20 |
| 45 | GCBL | 95 | - | - | 1 | 85 |
| 46 | GCBL | 80 | - | - | 141 | 1 |
NE indicates not evaluable; N, normal; F, failure.
Frequency of t(14;18)(q32;q21), REL amplification, and high-percentage REL-positive nuclei in DLBCL subgroups
DLBCL subgroup . | t(14;18)(q32;q21), no. (%) . | REL amplification with at least 4 copies, no. (%) . | At least 70% of nuclei REL-positive nuclei, no. (%) . |
|---|---|---|---|
| GCBL DLBCL | 7 of 17 (41) | 5 of 18 (28) | 6 of 18 (33) |
| Type 3 DLBCL | 0 of 11 (0) | 2 of 13 (15) | 4 of 10 (40) |
| ABL DLBCL | 2 of 11 (18) | 2 of 12 (17) | 7 of 11 (64) |
DLBCL subgroup . | t(14;18)(q32;q21), no. (%) . | REL amplification with at least 4 copies, no. (%) . | At least 70% of nuclei REL-positive nuclei, no. (%) . |
|---|---|---|---|
| GCBL DLBCL | 7 of 17 (41) | 5 of 18 (28) | 6 of 18 (33) |
| Type 3 DLBCL | 0 of 11 (0) | 2 of 13 (15) | 4 of 10 (40) |
| ABL DLBCL | 2 of 11 (18) | 2 of 12 (17) | 7 of 11 (64) |
Relationship between REL amplification and nuclear accumulation of REL
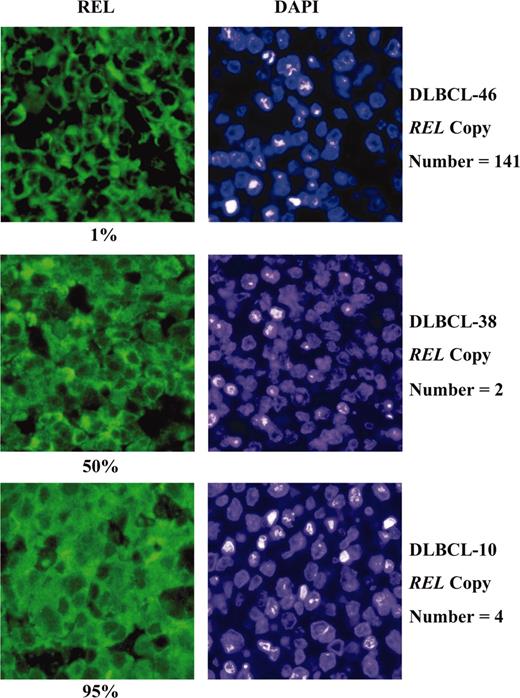
To evaluate the relationship between amplification of the REL locus and REL function, nuclear accumulation of REL was evaluated in DLBCL in vivo by immunofluorescence analysis as a measure of REL activity. Representative immunofluorescence images for 3 of the specimens are shown in Figure 2 along with DAPIcounterstained images clearly revealing nuclei. The percent cells with REL-positive nuclei varied markedly between the 39 analyzable specimens: 17 displayed 20% or less, 5 displayed 25% to 50%, and the remaining 17 displayed 70% to 95% REL-positive nuclei. No clear correlation was evident between REL amplification and REL nuclear accumulation (Table 1). This is exemplified by DLBCL-46, where only 1% of malignant cells were positive for nuclear REL-staining despite more than 70-fold amplification of the REL locus (Figure 2). These data suggest that for the present cohort, REL does not appear to be the functional target gene of the 2p12-16 amplicon.
Expression of REL in DLBCL. Nuclear localization of REL was evaluated by immunofluorescence analysis with DLBCL sections counterstained with DAPI for determination of percent positively stained nuclei. Results obtained for 3 representative DLBCL cases are shown with respective percent positive nuclei and REL copy number. Original magnification, × 400.
Expression of REL in DLBCL. Nuclear localization of REL was evaluated by immunofluorescence analysis with DLBCL sections counterstained with DAPI for determination of percent positively stained nuclei. Results obtained for 3 representative DLBCL cases are shown with respective percent positive nuclei and REL copy number. Original magnification, × 400.
Relationship between REL nuclear accumulation and DLBCL subgroups
ABL DLBCL cell lines have previously been reported to require functional NF-κB for survival; however, it is unknown if this is reflected in vivo.19 To investigate these findings in the DLBCL specimens, nuclear REL accumulation was utilized as a measure of NF-κB activity and levels compared between DLBCL expression signature subgroups. For comparative purposes, specimens with 50% or less REL-positive nuclei (low) and those with 70% or more (high) were grouped separately, because no specimens exhibited expression between 50% and 70%. Upon comparison between DLBCL subgroups, it was evident that ABL DLBCL exhibited a higher incidence of REL-positive nuclei than the other 2 subgroups. These findings are consistent with in vitro data showing elevated NF-κB activity in ABL DLBCL cell lines compared with GCBL DLBCL cell lines.19
Clinicopathologic correlations of REL copy number and nuclear accumulation of REL
In an effort to correlate genetic and biologic features of REL with a clinicopathologic feature of DLBCL, REL copy number and REL nuclear accumulation were statistically compared with multiple clinical parameters. The clinical characteristics of the patients used in the study are listed in Table 3. No significant correlation of high REL copy number or high percentage of REL-positive nuclei was found with age, advanced stage, high LDH, pure extranodal disease, poor performance status, IPI score, response to anthracycline-based chemotherapy, TTF, or OS (P > .05). In the case of REL copy number and extranodal disease, the present result was unlike our previous finding using a cohort of DLBCL comprising 15% relapse biopsies, where high REL copy number correlated with pure extranodal disease, possibly reflecting the 2 overlapping but different cohorts.9
Correlation of REL copy number and REL nuclear accumulation with clinical features
. | . | REL copy no. . | . | % REL-positive nuclei . | . | ||
|---|---|---|---|---|---|---|---|
| Patient characteristics . | All patients . | Less than 4* . | 4 or more† . | 50% or less‡ . | 70% or more§ . | ||
| Male/female, no. | 29/17 | 21/13 | 5/4 | 15/7 | 10/7 | ||
| Median age, y (range) | 62 (25-84) | 63 (29-84) | 63 (41-75) | 63 (29-79) | 67 (25-80) | ||
| Stage | |||||||
| I, no. (%) | 12 (26) | 10 (29) | 2 (22) | 4 (18) | 6 (35) | ||
| II, no. (%) | 14 (30.5) | 8 (24) | 5 (56) | 7 (32) | 5 (29) | ||
| III, no. (%) | 6 (13) | 4 (12) | 0 (0) | 3 (14) | 3 (18) | ||
| IV, no. (%) | 14 (30.5) | 12 (35) | 2 (22) | 8 (36) | 3 (18) | ||
| Pure EN disease, no. (%) | 10 (22) | 7 (21) | 3 (33) | 4 (18) | 4 (24) | ||
| IPI risk group∥ | |||||||
| Low, no. (%) | 13 (29) | 10 (30.5) | 1 (11) | 7 (32) | 6 (35) | ||
| Low-intermediate, no. (%) | 13 (29) | 8 (24) | 5 (56) | 6 (27) | 4 (23.5) | ||
| High-intermediate, no. (%) | 7 (15.5) | 6 (18) | 1 (11) | 3 (14) | 3 (18) | ||
| High, no. (%) | 12 (26.5) | 9 (27.5) | 2 (22) | 6 (27) | 4 (23.5) | ||
. | . | REL copy no. . | . | % REL-positive nuclei . | . | ||
|---|---|---|---|---|---|---|---|
| Patient characteristics . | All patients . | Less than 4* . | 4 or more† . | 50% or less‡ . | 70% or more§ . | ||
| Male/female, no. | 29/17 | 21/13 | 5/4 | 15/7 | 10/7 | ||
| Median age, y (range) | 62 (25-84) | 63 (29-84) | 63 (41-75) | 63 (29-79) | 67 (25-80) | ||
| Stage | |||||||
| I, no. (%) | 12 (26) | 10 (29) | 2 (22) | 4 (18) | 6 (35) | ||
| II, no. (%) | 14 (30.5) | 8 (24) | 5 (56) | 7 (32) | 5 (29) | ||
| III, no. (%) | 6 (13) | 4 (12) | 0 (0) | 3 (14) | 3 (18) | ||
| IV, no. (%) | 14 (30.5) | 12 (35) | 2 (22) | 8 (36) | 3 (18) | ||
| Pure EN disease, no. (%) | 10 (22) | 7 (21) | 3 (33) | 4 (18) | 4 (24) | ||
| IPI risk group∥ | |||||||
| Low, no. (%) | 13 (29) | 10 (30.5) | 1 (11) | 7 (32) | 6 (35) | ||
| Low-intermediate, no. (%) | 13 (29) | 8 (24) | 5 (56) | 6 (27) | 4 (23.5) | ||
| High-intermediate, no. (%) | 7 (15.5) | 6 (18) | 1 (11) | 3 (14) | 3 (18) | ||
| High, no. (%) | 12 (26.5) | 9 (27.5) | 2 (22) | 6 (27) | 4 (23.5) | ||
EN indicates extranodal disease.
n = 34.
n = 9.
n = 22.
n = 17.
One patient had insufficient clinical data to determine IPI score.
Of note, correlation of DLBCL expression signature subgroup assignment with overall survival of the 39 patients who received an anthracycline-containing chemotherapy regimen did not reveal any significant correlation (P = .38). These results are in agreement with the findings of Shipp et al21 but in contrast with those of Rosenwald et al.22
Gene expression signature in DLBCL associated with nuclear localization of REL
The in vivo biologic consequence of nuclear accumulation of REL was examined by determining differentially expressed sequences between DLBCL specimens with high (70% or more positive) and low (50% or less positive) nuclear REL localization using the Wilcoxon rank sum test on a transcript-by-transcript basis. This method identified 71 transcripts with P values less than .005, including 2 reported NF-κB target genes, NQO1 and SELE, with higher expression in specimens with high nuclear REL localization.14 A permutation test found that this was not more than the expected number of transcripts with P values less than .005 (P = .241). Using a lower threshold for low versus high REL-positive nuclei of 25%, a similar analysis revealed fewer differentially expressed transcripts. Thus, a significant expression signature in DLBCL was not found to be associated with nuclear localization of REL.
Discussion
REL maps within 2p12-16, the most frequently amplified region observed in GC-derived B-cell lymphomas.5,6 However, little experimental evidence to date indicates that it is indeed the functional target gene of the amplicon in DLBCL. For the first time, the present study examined the relationship between REL genomic copy number and REL functional localization in the nucleus of DLBCL specimens. It was evident for the present cohort of DLBCL that overt amplification of the REL locus does not appear to lead to functional nuclear accumulation of REL, suggesting that REL is not the functional target of the 2p12-16 amplicon in DLBCL. This is unlike classical Hodgkin lymphoma with constitutive NF-κB activity, where it has recently been reported that Hodgkin and Reed-Sternberg cells with gain of 2p genetic material exhibit nuclear staining for REL.30 In DLBCL, nuclear accumulation of REL varied considerably between specimens and was found to be predominantly cytoplasmic. A recent study reported a rearrangement of the REL locus in a large B-cell lymphoma–derived cell line, resulting in the expression of a hybrid protein where REL lacked a nuclear localization sequence, leading to the suggestion that cytoplasmically located REL may have an oncogenic role.32 However, additional studies did not confirm such a role.33 Another study demonstrated the ability of exogenously expressed REL to directly transform chicken spleen cells utilizing a liquid outgrowth assay to evaluate transformation.34 The subcellular localization of the human REL, however, was not determined in this study. Interestingly, coexpression of BCL2 accelerated the appearance of transformed cultures.34 Further studies are required to distinguish the putative role of nuclear versus cytoplasmically located REL in lymphomagenesis. While in the present study nuclear REL was not found to be associated with a significant expression signature or clinical parameter, the contribution of other individual REL/NF-κB family members to lymphoma biology remains to be examined. Overall, based on the generally accepted notion that nuclear REL is the effector of REL function, our results strongly suggest that the functional consequence of 2p amplification may not be REL activation and that therefore other genes mapped within 2p12-16 may be the target of the amplification event.
Three subgroups of DLBCL have now been described on the basis of the expression signatures of GC and activated peripheral B cells. REL was one of the genes recently identified by Rosenwald et al22 to discriminate between GCBL and ABL DLBCL, being preferentially overexpressed in the former subgroup, where REL amplification was exclusively detected. In the present study, 3 subgroups were similarly identified using a subset of the discriminatory genes but, in contrast, amplification of the REL locus was detected in GCBL as well as ABL subgroups. This finding is not surprising considering that increased copy number of the 2p12-16 region is a frequent genetic alteration observed not only in DLBCL but also in follicular lymphoma and classical Hodgkin disease, being 2 other GC-derived B-cell lymphomas.7,8 The current finding that REL amplification occurs across all DLBCL expression subgroups and not exclusively to GCBL DLBCL is therefore consistent with the molecular and cytogenetic data indicating that amplification of this region is a frequent genetic alteration in GC-derived B-cell lymphomas.
In the present study, no significant association was found between expression subgroup classification and clinical outcome as has been reported by Alizadeh et al20 and Rosenwald et al.22 In those studies, ABL and Type 3 DLBCL exhibited a poor outcome compared with GCBL DLBCL. A lack of such association has also been reported by Shipp et al.21 The reasons for the apparent discrepancies between the studies remain unclear at the present time, though it is feasible that differences in treatment regimens between the studies may influence outcome.
ABL DLBCL–derived cell lines have been reported to exhibit increased NF-κB activity, comprising p50/RELA and p50/REL heterodimers, compared with GCBL DLBCL–derived cell lines with p50/REL heterodimers, as identified by electrophoretic mobility shift assays.19 Furthermore, active NF-κB was found to be necessary for survival of ABL DLBCL cell lines. In the present study, using nuclear-located REL as a measure of NF-κB activity, DLBCLs with higher numbers of REL-positive nuclei were more frequent in the ABL subgroup than in the other 2 subgroups. While this is consistent with the notion that ABL DLBCL exhibit increased NF-κB activity over GCBL DLBCL, determination of the nuclear localization of other REL/NF-κB family members in vivo may further substantiate the in vitro findings. Overall, DLBCLs are heterogeneous with respect to REL and thus NF-κB function, with the precise role of NF-κB in lymphoma biology yet to be fully understood.
Prepublished online as Blood First Edition Paper, November 13, 2003; DOI 10.1182/blood-2003-04-1359.
Supported in part by research grants from National Institutes of Health and the Byrne Fund.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Vladan Miljkovic for dedicated handling of the hybridization, washing, and scanning of the arrays and Ashlyn Celestine for technical assistance with immunofluorescence staining. We also thank Lou Staudt for confirming Lymphochip clone identity.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal